Molecular Transmission Network and Pretreatment Drug Resistance of Newly Diagnosed HIV-1 Infections in Taizhou, a Coastal City in Eastern China, from 2021–2023
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Subjects
2.2. Laboratory Methods
2.3. Subtyping and HIV Drug Resistance Analysis
2.4. Transmission Cluster Construction
2.5. Statistical Analysis
3. Results
3.1. Patient Demographic Characteristics
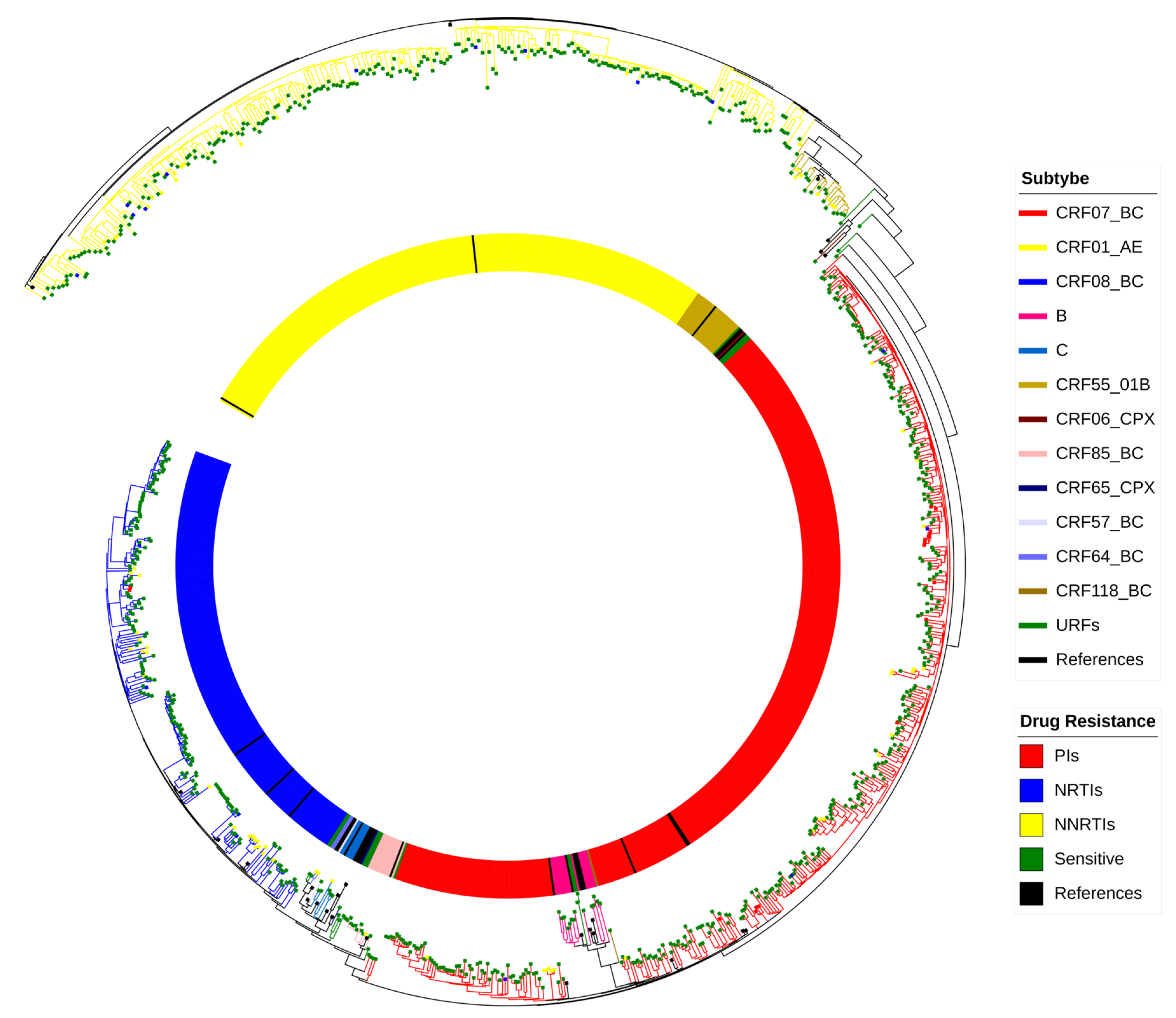
3.2. HIV Subtypes
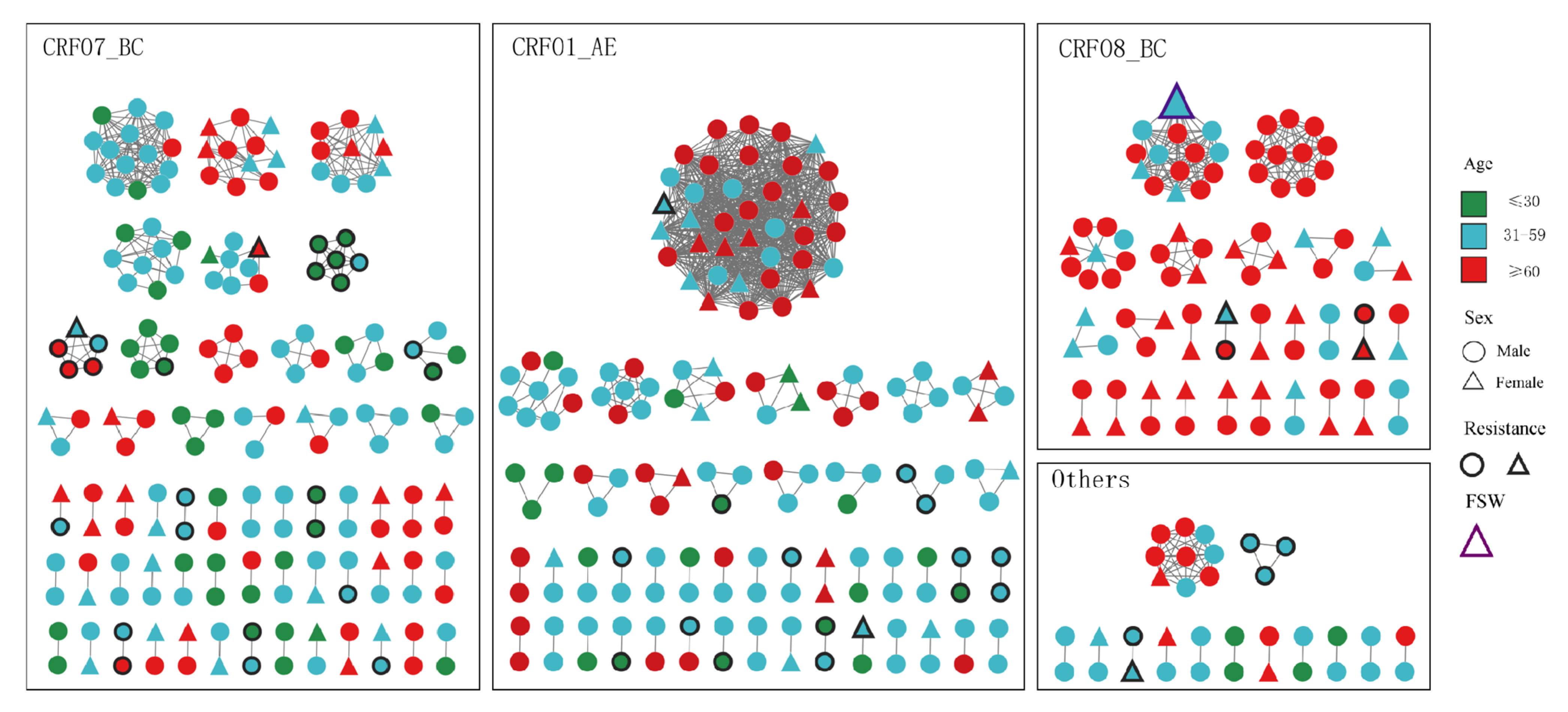
3.3. Characteristics of the HIV Molecular Transmission Network
3.4. Characterization of Large Clusters
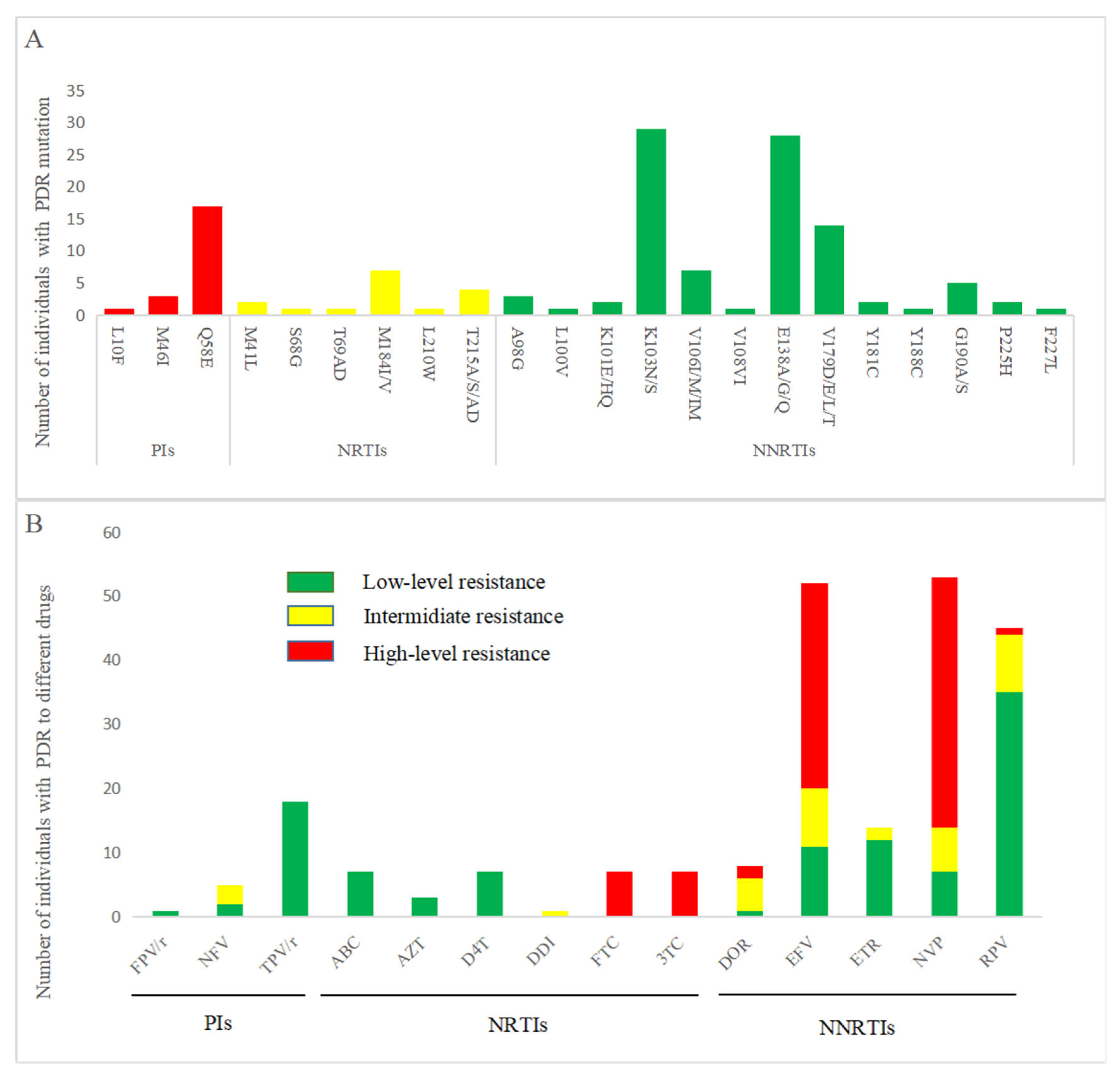
3.5. PDR Distribution and Drug Resistance Mutation Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HIV | Human immunodeficiency virus |
| AIDS | Acquired immunodeficiency syndrome |
| ART | Antiretroviral therapy |
| WHO | World Health Organization |
| PDR | Pretreatment drug resistance |
| PCR | Polymerase chain reaction |
| PI | Protease inhibitor |
| NRTI | Nucleoside reverse transcriptase inhibitor |
| NNRTI | Nonnucleoside reverse transcriptase inhibitor |
| FPV/r | Fosamprenavir/ritonavir |
| NFV | Nelfinavir |
| TPV/r | Ritonavir/ritonavir |
| ABC | Abacavir |
| AZT | Zidovudine |
| D4T | Stavudine |
| DDI | Didanosine |
| FTC | Emtricitabine |
| 3TC | Lamivudine |
| DOR | Doravirine |
| EFV | Efavirenz |
| ETR | Etravirine |
| NVP | Nevirapine |
| RPV | Rilpivirine |
References
- The Joint United Nations Programme on HIV/AIDS. Global HIV & AIDS Statistics—Fact Sheet 2024; The Joint United Nations Programme on HIV/AIDS: Geneva, Switzerland, 2024; Volume 55, pp. 574–578. [Google Scholar]
- Lessells, R.J.; Mutevedzi, P.C.; Iwuji, C.C.; Newell, M.L. Reduction in early mortality on antiretroviral therapy for adults in rural South Africa since change in CD4+ cell count eligibility criteria. J. Acquir. Immune Defic. Syndr. (1999) 2014, 65, e17–e24. [Google Scholar] [CrossRef]
- Cohen, M.S.; Chen, Y.Q.; McCauley, M.; Gamble, T.; Hosseinipour, M.C.; Kumarasamy, N.; Hakim, J.G.; Kumwenda, J.; Grinsztejn, B.; Pilotto, J.H.; et al. Prevention of HIV-1 infection with early antiretroviral therapy. N. Engl. J. Med. 2011, 365, 493–505. [Google Scholar] [CrossRef]
- van de Vijver, D.A.; Nichols, B.E.; Abbas, U.L.; Boucher, C.A.; Cambiano, V.; Eaton, J.W.; Glaubius, R.; Lythgoe, K.; Mellors, J.; Phillips, A.; et al. Preexposure prophylaxis will have a limited impact on HIV-1 drug resistance in sub-Saharan Africa: A comparison of mathematical models. AIDS 2013, 27, 2943–2951. [Google Scholar] [CrossRef] [PubMed]
- Cambiano, V.; Bertagnolio, S.; Jordan, M.R.; Pillay, D.; Perriëns, J.H.; Venter, F.; Lundgren, J.; Phillips, A. Predicted levels of HIV drug resistance: Potential impact of expanding diagnosis, retention, and eligibility criteria for antiretroviral therapy initiation. AIDS 2014, 28 (Suppl. 1), S15–S23. [Google Scholar] [CrossRef] [PubMed]
- National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention Division of Tuberculosis Elimination. Health Policy 2010. Available online: https://stacks.cdc.gov/view/cdc/5498 (accessed on 8 October 2025).
- Campbell, E.M.; Patala, A.; Shankar, A.; Li, J.F.; Johnson, J.A.; Westheimer, E.; Gay, C.L.; Cohen, S.E.; Switzer, W.M.; Peters, P.J. Phylodynamic Analysis Complements Partner Services by Identifying Acute and Unreported HIV Transmission. Viruses 2020, 12, 145. [Google Scholar] [CrossRef] [PubMed]
- Doan, L.P.; Nguyen, L.H.; Auquier, P.; Boyer, L.; Fond, G.; Nguyen, H.T.; Latkin, C.A.; Vu, G.T.; Hall, B.J.; Ho, C.S.H.; et al. Social network and HIV/AIDS: A bibliometric analysis of global literature. Front. Public Health 2022, 10, 1015023. [Google Scholar] [CrossRef]
- Zhang, J.; Guo, Z.; Yang, J.; Pan, X.; Jiang, J.; Ding, X.; Zhang, W.; Xia, Y.; Xu, Y.; Huang, J. Genetic diversity of HIV-1 and transmitted drug resistance among newly diagnosed individuals with HIV infection in Hangzhou, China. J. Med. Virol. 2015, 87, 1668–1676. [Google Scholar] [CrossRef]
- Shi, H.; Li, X.; Wang, S.; Dong, X.; Qiao, M.; Wu, S.; Wu, R.; Yuan, X.; Wang, J.; Xu, Y.; et al. Molecular transmission network analysis of newly diagnosed HIV-1 infections in Nanjing from 2019 to 2021. BMC Infect. Dis. 2024, 24, 583. [Google Scholar] [CrossRef]
- Tan, T.; Bai, C.; Lu, R.; Chen, F.; Li, L.; Zhou, C.; Xiang, X.; Zhang, W.; Ouyang, L.; Xu, J.; et al. HIV-1 molecular transmission network and drug resistance in Chongqing, China, among men who have sex with men (2018–2021). Virol. J. 2023, 20, 147. [Google Scholar] [CrossRef]
- Kantzanou, M.; Karalexi, M.A.; Papachristou, H.; Vasilakis, A.; Rokka, C.; Katsoulidou, A. Transmitted drug resistance among HIV-1 drug-naïve patients in Greece. Int. J. Infect. Dis. 2021, 105, 42–48. [Google Scholar] [CrossRef]
- Ma, N.; Chen, X.-H.; Zhao, Y.; Kang, X.; Pan, S.; Yao, W.-Q. HIV-1 molecular transmission network among sexually transmitted populations in Liaoning Province, China. Medicine 2021, 10, e26640. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Dai, J.; Li, Z.; Ma, Y.; Chen, H.; Dong, L.; Jin, X.; Yang, M.; Zeng, Z.; Sun, P.; et al. Using molecular network analysis to explore the characteristics of HIV-1 transmission in a China-Myanmar border area. PLoS ONE 2022, 17, e0268143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xu, K.; Jiang, J.; Fan, Q.; Ding, X.; Zhong, P.; Xing, H.; Chai, C.; Pan, X. Combining molecular network analysis and field epidemiology to quantify local HIV transmission and highlight ongoing epidemics. Int. J. Infect. Dis. 2023, 128, 187–193. [Google Scholar] [CrossRef]
- Zhang, T.; Dou, H.; Ye, H.; Tang, H.; Wang, W.; Hu, W.; Lv, B.; Zhou, M.; Dai, H.; Wang, W.; et al. Transmitted drug resistance and molecular transmission network among treatment-naive HIV-1 patients in Wenzhou, China, 2020–2023. Virol. J. 2024, 21, 257. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.; Tang, C.; Liu, Y.; Jiang, H.; Fang, T.; Xu, G. HIV-1 drug resistance and genetic transmission network among newly diagnosed people living with HIV/AIDS in Ningbo, China between 2018 and 2021. Virol. J. 2023, 20, 233. [Google Scholar] [CrossRef]
- Cao, D.; Xing, H.; Feng, Y.; He, T.; Zhang, J.; Ling, J.; Chen, J.; Zhao, J. Molecular transmission network analysis reveals the challenge of HIV-1 in aging patients in China: Elderly people play a crucial role in the transmission of subtypes and high pretreatment drug resistance in developed Eastern China, 2019–2023. Virol. J. 2024, 21, 199. [Google Scholar] [CrossRef]
- Li, K.; Liu, M.; Chen, H.; Li, J.; Liang, Y.; Feng, Y.; Xing, H.; Shao, Y. Using molecular transmission networks to understand the epidemic characteristics of HIV-1 CRF08_BC across China. Emerg. Microbes Infect. 2021, 10, 497–506. [Google Scholar] [CrossRef]
- Feng, Y.; Takebe, Y.; Wei, H.; He, X.; Hsi, J.H.; Li, Z.; Xing, H.; Ruan, Y.; Yang, Y.; Li, F.; et al. Geographic origin and evolutionary history of China’s two predominant HIV-1 circulating recombinant forms, CRF07_BC and CRF08_BC. Sci. Rep. 2016, 6, 19279. [Google Scholar] [CrossRef]
- Li, X.-J.; Kusagawa, S.; Xia, X.; Yang, C.; Wang, Q.; Yokota, Y.; Hoshina, Y.; Onogi, T.; Nohtomi, K.; Imamura, Y.; et al. Molecular epidemiology of the heterosexual HIV-1 transmission in Kunming, Yunnan Province of China suggests origin from the local IDU epidemic. AIDS Res. Hum. Retroviruses 2005, 21, 977–980. [Google Scholar] [CrossRef]
- Chen, M.; Chen, H.; Dai, J.; Dong, L.; Ma, Y.; Jia, M.; Ding, W. Identification of a Novel Second-Generation HIV-1 Circulating Recombinant Form (CRF174_0708) Arising from CRF07_BC and CRF08_BC in Yunnan, China. AIDS Res. Hum. Retroviruses 2025, 41, 322–326. [Google Scholar] [CrossRef]
- HIV Drug Resistance Report 2019. Available online: https://iris.who.int/handle/10665/325891 (accessed on 6 September 2025).
- Zhang, M.; Ma, Y.; Wang, Z.; Wang, G.; Wang, Q.; Li, X.; Lin, F.; Zhang, C. Prevalence and transmission of pretreatment drug resistance in people living with HIV-1 in Shanghai China, 2017–2021. Virulence 2024, 15, 2373105. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.; Du, Z.; Zhou, J.; Ye, L.; Su, L.; Yang, H.; Yuan, F.; Li, Y.; Liu, H.; Zhai, W.; et al. HIV-1 subtype diversity, drug resistance, and genetic transmission networks in men who have sex with men with virologic failure in antiretroviral therapy in Sichuan, China, 2011 to 2017. Medicine 2019, 98, e17585. [Google Scholar] [CrossRef] [PubMed]
- Parczewski, M.; Sulkowska, E.; Urbańska, A.; Scheibe, K.; Serwin, K.; Grabarczyk, P. Transmitted HIV drug resistance and subtype patterns among blood donors in Poland. Sci. Rep. 2021, 11, 12734. [Google Scholar] [CrossRef] [PubMed]
- Macdonald, V.; Mbuagbaw, L.; Jordan, M.R.; Mathers, B.; Jay, S.; Baggaley, R.; Verster, A.; Bertagnolio, S. Prevalence of pretreatment HIV drug resistance in key populations: A systematic review and meta-analysis. J. Int. AIDS Soc. 2020, 23, e25656. [Google Scholar] [CrossRef]
- Zhu, M.; Sun, Z.; Zhang, X.; Luo, W.; Wu, S.; Ye, L.; Xu, K.; Chen, J. Epidemiological dynamics and molecular characterization of HIV drug resistance in eastern China from 2020 to 2023. Front. Microbiol. 2024, 15, 1475548. [Google Scholar] [CrossRef]
- Zhao, B.; Song, W.; An, M.; Dong, X.; Li, X.; Wang, L.; Liu, J.; Tian, W.; Wang, Z.; Ding, H.; et al. Priority Intervention Targets Identified Using an In-Depth Sampling HIV Molecular Network in a Non-Subtype B Epidemics Area. Front. Cell. Infect. Microbiol. 2021, 11, 642903. [Google Scholar] [CrossRef]
- AIDS and Hepatitis C Professional Group SoID, Chinese Medical Association: Chinese Center for Disease Control and Prevention. Chinese guidelines for diagnosis and treatment of HIV/AIDS (2018). Zhonghua Nei Ke Za Zhi 2018, 52, 867–884. [Google Scholar]
- Lake Yimer, B. HIV/AIDS risk-reduction options as predictor of female sex workers’ sexual behaviour. Women’s Health 2022, 18, 17455057221118167. [Google Scholar] [CrossRef]
- Shushtari, Z.J.; Hosseini, S.A.; Sajjadi, H.; Salimi, Y.; Latkin, C.; Snijders, T.A.B. Social network and HIV risk behaviors in female sex workers: A systematic review. BMC Public Health 2018, 18, 1020. [Google Scholar] [CrossRef]
- Wensing, A.M.; Calvez, V.; Ceccherini-Silberstein, F.; Charpentier, C.; Günthard, H.F.; Paredes, R.; Shafer, R.W.; Richman, D.D. 2019 update of the drug resistance mutations in HIV-1. Top Antivir Med. 2019, 27, 111–121. [Google Scholar]
- Frieden, T.R.; Foti, K.E.; Mermin, J. Applying Public Health Principles to the HIV Epidemic--How Are We Doing? N. Engl. J. Med. 2015, 373, 2281–2287. [Google Scholar] [CrossRef]
| Cases (%) (n = 937) | HIV Subtypes (%) | χ2 | p | ||||
|---|---|---|---|---|---|---|---|
| CRF07_BC (n = 395) | CRF01_AE (n = 259) | CRF08_BC (n = 209) | Others (n = 74) | ||||
| Collection time | 10.337 | 0.111 | |||||
| 2021 | 344 (36.7) | 150 (38.0) | 92 (35.5) | 67 (32.1) | 35 (47.3) | ||
| 2022 | 289 (30.8) | 114 (28.9) | 85 (32.8) | 65 (31.1) | 25 (33.8) | ||
| 2023 | 304 (32.4) | 131 (33.2) | 82 (31.7) | 77 (36.8) | 14 (18.9) | ||
| Sex | 23.381 | p < 0.001 | |||||
| Male | 761 (81.2) | 335 (84.8) | 220 (84.9) | 146 (69.9) | 60 (81.1) | ||
| Female | 176 (18.8) | 60 (15.2) | 39 (15.1) | 63 (30.1) | 14 (18.9) | ||
| Age (years) | 99.917 | p < 0.001 | |||||
| ≤30 | 155 (16.5) | 97 (24.6) | 40 (15.4) | 7 (3.3) | 11 (14.9) | ||
| 31–59 | 497 (53.0) | 210 (53.2) | 154 (59.5) | 87 (41.6) | 46 (62.2) | ||
| ≥60 | 285 (30.4) | 88 (22.3) | 65 (25.1) | 115 (55.0) | 17 (23.0) | ||
| Married status | 68.190 | p < 0.001 | |||||
| Singal | 272 (29.0) | 152 (38.5) | 82 (31.7) | 18 (8.6) | 20 (27.0) | ||
| Married | 442 (47.2) | 173 (43.8) | 103 (39.8) | 128 (61.2) | 38 (51.4) | ||
| Divorced/Widowed | 223 (23.8) | 70 (17.7) | 74 (28.6) | 63 (30.1) | 16 (21.6) | ||
| Education | 122.43 | p < 0.001 | |||||
| Primary school/illiterate | 421 (44.9) | 130 (32.9) | 106 (40.9) | 158 (75.6) | 27 (36.5) | ||
| Junior middle school | 275 (29.3) | 129 (32.7) | 79 (30.5) | 37 (17.7) | 30 (40.5) | ||
| High or technical secondary school | 147 (15.7) | 74 (18.7) | 47 (18.1) | 12 (5.7) | 14 (18.9) | ||
| Junior college or above | 94 (10.0) | 62 (15.7) | 27 (10.4) | 2 (1.0) | 3 (4.1) | ||
| Sexual contact | 112.59 | p < 0.001 | |||||
| Heterosexual | 612 (65.3) | 215 (54.4) | 149 (57.5) | 198 (94.7) | 50 (67.6) | ||
| Homosexual | 320 (34.2) | 179 (45.3) | 108 (41.7) | 9 (4.3) | 24 (32.4) | ||
| Others | 5 (0.5) | 1 (0.3) | 2 (0.8) | 2 (1.0) | 0 (0.0) | ||
| CD4+ T lymphocyte count | 10.895 | 0.092 | |||||
| <200 | 378 (40.3) | 140 (35.4) | 121 (46.7) | 81 (38.8) | 36 (48.6) | ||
| 200–499 | 478 (51.0) | 220 (55.7) | 117 (45.2) | 109 (52.2) | 32 (43.2) | ||
| ≥500 | 81 (8.6) | 35 (8.9) | 21 (8.1) | 19 (9.1) | 6 (8.1) | ||
| LC | Subtype | Nodes | Growing Case (%) | Age ≥ 60 (%) | Male (%) | Heterosexual (%) | Taizhou Household Registration (%) | ||
|---|---|---|---|---|---|---|---|---|---|
| 2021 | 2022 | 2023 | |||||||
| LC1 | CRF01_AE | 38 | 16 (42.1) | 8 (21.1) | 14 (36.8) | 25 (65.7) | 26 (68.4) | 35 (92.1) | 29 (76.3) |
| LC2 | CRF07_BC | 14 | 6 (42.8) | 4 (28.6) | 4 (28.6) | 14 (40.0) | 26 (74.2) | 2 (14.2) | 7 (50.0) |
| LC3 | CRF08_BC | 14 | 1 (7.2) | 5 (35.7) | 8 (57.1) | 7 (50) | 11 (78.5) | 14 (100.0) | 10 (71.4) |
| LC4 | CRF07_BC | 11 | 4 (36.4) | 2 (18.2) | 5 (45.4) | 8 (72.7) | 5 (54.5) | 11 (100.0) | 10 (90.9) |
| LC5 | CRF08_BC | 11 | 2 (18.2) | 5 (45.4) | 4 (36.4) | 11 (100.0) | 11 (100.0) | 10 (90.9) | 11 (100.0) |
| LC6 | CRF07_BC | 10 | 5 (50.0) | 3 (30.0) | 2 (20.0) | 5 (50.0) | 6 (60.0) | 10 (100.0) | 10 (100.0) |
| Total N (%) a | Not Included in a Large Cluster (<10) N (%) b | Included in a Large Cluster (≥10) N (%) b | Univariate Analysis | Multivariate Analysis | |||
|---|---|---|---|---|---|---|---|
| OR (95CI) | p | AOR (95CI) | p | ||||
| HIV subtypes | |||||||
| CRF07_BC | 395 (42.2) | 360 (42.9) | 35 (35.7) | ||||
| CRF01_AE | 259 (27.6) | 221 (26.3) | 38 (38.8) | 1.77(1.08–2.88) | 0.022 | 1.50 (0.89–2.53) | p = 0.130 |
| CRF08_BC | 209 (22.3) | 184 (21.9) | 25 (25.5) | 1.40(0.81–2.41) | 0.227 | 0.67 (0.37–1.20) | p = 0.177 |
| Others | 74 (7.9) | 74 (8.8) | 0 (0.0) | 0 | 0.974 | 0 | p = 0.988 |
| Collection time | |||||||
| 2021 | 344 (36.7) | 310 (36.9) | 34 (34.7) | ||||
| 2022 | 289 (30.8) | 262 (31.2) | 27 (27.6) | 0.94 (0.55–1.60) | p = 0.818 | ||
| 2023 | 304 (32.4) | 267 (31.8) | 37 (37.8) | 1.26 (0.77–2.07) | p = 0.353 | ||
| Gender | |||||||
| Male | 761 (81.2) | 687 (81.9) | 74 (75.5) | ||||
| Female | 176 (18.8) | 152 (18.1) | 24 (24.5) | 1.47 (0.90–2.40) | p = 0.128 | ||
| Age (years) | |||||||
| ≤30 | 155 (16.5) | 153 (18.2) | 2 (2.0) | ||||
| 31–59 | 497 (53.0) | 458 (54.6) | 39 (39.8) | 6.51 (1.55–27.30) | p = 0.010 | 3.21 (0.67–15.43) | p = 0.145 |
| ≥60 | 285 (30.4) | 228 (27.2) | 57 (58.2) | 19.12 (4.60–79.50) | p < 0.001 | 6.91 (1.34–35.53) | p = 0.021 |
| Married status | |||||||
| Singal | 272 (29.0) | 261 (31.1) | 11 (11.2) | ||||
| Married | 442 (47.2) | 392 (46.7) | 50 (51.0) | 3.03 (1.55–5.92) | p = 0.001 | 0.82 (0.37–1.82) | p = 0.632 |
| Divorced/Widowed | 223 (23.8) | 186 (22.2) | 37 (37.8) | 4.72 (2.35–9.49) | p < 0.001 | 1.25 (0.55–2.81) | p = 0.597 |
| Education | |||||||
| Primary school/illiterate | 421 (44.9) | 350 (41.7) | 71 (72.4) | ||||
| Junior middle school | 275 (29.3) | 256 (30.5) | 19 (19.4) | 0.37 (0.22–0.62) | p < 0.001 | 0.60 (0.33–1.08) | p = 0.087 |
| High or technical secondary school | 147 (15.7) | 139 (16.6) | 8 (8.2) | 0.28 (0.13–0.60) | p = 0.001 | 0.62 (0.27–1.46) | p = 0.277 |
| Junior college or above | 94 (10.0) | 94 (11.2) | 0 (0.0) | 0 | p = 0.980 | 0 | p = 0.987 |
| Sexual contact | |||||||
| Heterosexual | 612 (65.3) | 530 (63.2) | 82 (83.7) | ||||
| Homosexual | 320 (34.2) | 304 (36.2) | 16 (16.3) | 0.34 (0.20–0.59) | p < 0.001 | 0.61 (0.32–1.14) | p = 0.122 |
| Others | 5 (0.5) | 5 (0.6) | 0 (0.0) | 0 | p = 0.983 | 0 | p = 0.997 |
| CD4+ T lymphocyte count | |||||||
| <200 | 378 (40.3) | 333 (39.7) | 45 (45.9) | ||||
| 200–499 | 478 (51.0) | 433 (51.6) | 45 (45.9) | 0.77 (0.50–1.19) | p = 0.239 | ||
| ≥500 | 81 (8.6) | 73 (8.7) | 8 (8.2) | 0.81 (0.37–1.79) | p = 0.605 | ||
| PDR mutation | |||||||
| Yes | 106 (12.6) | 1 (1.0) | |||||
| No | 733 (87.4) | 97 (99.0) | 14.03 (1.94–101.65) | p = 0.009 | 10.16 (1.38–74.75) | p = 0.023 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, J.; Lin, H.; Li, G.; Wang, S.; Wang, T.; Meng, Q.; Hua, T.; Xie, Y.; Zhang, J.; Shen, W. Molecular Transmission Network and Pretreatment Drug Resistance of Newly Diagnosed HIV-1 Infections in Taizhou, a Coastal City in Eastern China, from 2021–2023. Pathogens 2025, 14, 1030. https://doi.org/10.3390/pathogens14101030
Lin J, Lin H, Li G, Wang S, Wang T, Meng Q, Hua T, Xie Y, Zhang J, Shen W. Molecular Transmission Network and Pretreatment Drug Resistance of Newly Diagnosed HIV-1 Infections in Taizhou, a Coastal City in Eastern China, from 2021–2023. Pathogens. 2025; 14(10):1030. https://doi.org/10.3390/pathogens14101030
Chicago/Turabian StyleLin, Junxiao, Haijiang Lin, Guixia Li, Shanling Wang, Tingting Wang, Qiguo Meng, Tingting Hua, Yali Xie, Jiafeng Zhang, and Weiwei Shen. 2025. "Molecular Transmission Network and Pretreatment Drug Resistance of Newly Diagnosed HIV-1 Infections in Taizhou, a Coastal City in Eastern China, from 2021–2023" Pathogens 14, no. 10: 1030. https://doi.org/10.3390/pathogens14101030
APA StyleLin, J., Lin, H., Li, G., Wang, S., Wang, T., Meng, Q., Hua, T., Xie, Y., Zhang, J., & Shen, W. (2025). Molecular Transmission Network and Pretreatment Drug Resistance of Newly Diagnosed HIV-1 Infections in Taizhou, a Coastal City in Eastern China, from 2021–2023. Pathogens, 14(10), 1030. https://doi.org/10.3390/pathogens14101030





