Detection of Bovine Leukemia Virus in Argentine, Bolivian, Paraguayan and Cuban Native Cattle Using a Quantitative Real-Time PCR Assay-BLV-CoCoMo-qPCR-2
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection, DNA Extraction, and Plasma Isolation
2.2. Detection of BLV Proviral Load Using BLV-CoCoMo-qPCR-2
2.3. Statistical Analysis
2.4. Ethical Approval
3. Results
3.1. BLV Prevalence of Among Latin American Cattle Breeds
3.2. BLV PVL Among Latin American Native Cattle Breeds
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aida, Y.; Murakami, H.; Takahashi, M.; Takeshima, S.N. Mechanisms of pathogenesis induced by bovine leukemia virus as a model for human T-cell leukemia virus. Front. Microbiol. 2013, 4, 328. [Google Scholar] [CrossRef]
- Polat, M.; Takeshima, S.; Aida, Y. Epidemiology and genetic diversity of bovine leukemia virus. Virol. J. 2017, 14, 209. [Google Scholar] [CrossRef] [PubMed]
- OIE. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals: Chapter 2.4.11. In Enzootic Bovine Leukosis, 7th ed.; World Organization for Animal Health: Paris, France, 2012. [Google Scholar]
- Ott, S.; Johnson, R.; Wells, S.J. Association between bovine-leukosis virus seroprevalence and herd-level productivity on US dairy farms. Prev. Vet. Med. 2003, 61, 249–262. [Google Scholar] [CrossRef] [PubMed]
- Ruggiero, V.J.; Norby, B.; Benitez, O.J.; Hutchinson, H.; Sporer, K.; Droscha, C.; Swenson, C.L.; Bartlett, P.C. Controlling bovine leukemia virus in dairy herds by identifying and removing cows with the highest proviral load and lymphocyte counts. J. Dairy Sci. 2019, 102, 9165–9175. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, P.C.; Norby, B.; Byrem, T.M.; Parmelee, A.; Ledergerber, J.T.; Erskine, R.J. Bovine leukemia virus and cow longevity in Michigan dairy herds. J. Dairy Sci. 2013, 96, 1591–1597. [Google Scholar] [CrossRef]
- Kuczewski, A.; Hogeveen, H.; Orsel, K.; Wolf, R.; Thompson, J.; Spackman, E.; van der Meer, F. Economic evaluation of 4 bovine leukemia virus control strategies for Alberta dairy farms. J. Dairy Sci. 2019, 102, 2578–2592. [Google Scholar] [CrossRef]
- Nakada, S.; Fujimoto, Y.; Kohara, J.; Adachi, Y.; Makita, K. Estimation of economic loss by carcass weight reduction of Japanese dairy cows due to infection with bovine leukemia virus. Prev. Vet. Med. 2022, 198, 105528. [Google Scholar] [CrossRef]
- Leisering, A. Hypertrophy der Malpigischen Körpchen der Milz. Ber. Über Vet. Königreich Sachs. 1871, 16, 15–16. [Google Scholar]
- Acaite, J.; Tamosiunas, V.; Lukauskas, K.; Milius, J.; Pieskus, J. The eradication experience of enzootic bovine leukosis from Lithuania. Prev. Vet. Med. 2007, 82, 83–89. [Google Scholar] [CrossRef]
- Nuotio, L.; Rusanen, H.; Sihvonen, L.; Neuvonen, E. Eradication of enzootic bovine leukosis from Finland. Prev. Vet. Med. 2003, 59, 43–49. [Google Scholar] [CrossRef]
- Kettmann, R.; Burny, A.; Callebaut, I.; Droogmans, L.; Mammerickx, M.; Willems, L. Bovine leukemia virus. Retroviridae 1994, 3, 39–81. [Google Scholar]
- Pluta, A.; Jaworski, J.P.; Droscha, C.; VanderWeele, S.; Taxis, T.M.; Valas, S.; Brnić, D.; Jungić, A.; Ruano, M.J.; Sánchez, A.; et al. Inter laboratory comparison of eleven quantitative or digital PCR assays for detection of proviral bovine leukemia virus in blood samples. BMC Vet. Res. 2024, 20, 381. [Google Scholar] [CrossRef] [PubMed]
- Kuczewski, A.; Orsel, K.; Barkema, H.W.; Kelton, D.F.; Hutchins, W.A.; van der Meer, F.J.U. Short communication: Evaluation of 5 different ELISA for the detection of bovine leukemia virus antibodies. J. Dairy Sci. 2018, 101, 2433–2437. [Google Scholar] [CrossRef] [PubMed]
- Lv, G.; Wang, J.; Lian, S.; Wang, H.; Wu, R. The global epidemiology of bovine leukemia virus: Current trends and future implications. Animals 2024, 14, 297. [Google Scholar] [CrossRef] [PubMed]
- D’Angelino, J.; Garcia, M.; Birgel, E. Epidemiological study of enzootic bovine leukosis in Brazil. Trop. Anim. Health Prod. 1998, 30, 13–15. [Google Scholar] [CrossRef]
- Marín, C.; de López, N.M.; Álvarez, L.; Lozano, O.; España, W.; Castaños, H.; León, A. Epidemiology of bovine leukemia in Venezuela. Ann. Rech. Vet. 1978, 9, 4. [Google Scholar]
- Alfonso, R.; Almansa, J.E.; Barrera, J.D. Serological prevalence and evaluation of risk factors of enzootic bovine leukosis in the Sabana de Bogota region and the Ubate and Chiquinquira valleys of Colombia. Rev. Sci. Tech. Off. Int. Epizoot. 1998, 17, 723–732. [Google Scholar] [CrossRef]
- Felmer, R.; Munoz, G.; Zuniga, J.; Recabal, M. Molecular analysis of a 444 bp fragment of the bovine leukaemia virus gp51 env gene reveals a high frequency of non-silent point mutations and suggests the presence of two subgroups of BLV in Chile. Vet. Microbiol. 2005, 108, 39–47. [Google Scholar] [CrossRef]
- Moratorio, G.; Obal, G.; Dubra, A.; Correa, A.; Bianchi, S.; Buschiazzo, A.; Cristina, J.; Pritsch, O. Phylogenetic analysis of bovine leukemia viruses isolated in South America reveals diversification in seven distinct genotypes. Arch. Virol. 2010, 155, 481–489. [Google Scholar] [CrossRef]
- Hernández-Herrera, D.Y.; Posso-Terranova, A.; Benavides, J.; Muñoz-Florez, J.; Giovambattista, G.; Álvarez-Franco, L. Bovine leukosis virus detection in Creole Colombian breeds using nested-PCR. Acta Agronómica 2011, 60, 311–317. [Google Scholar]
- Benavides, B.; Quevedo, D.; Cruz, M. Epidemiological study of bovine leukemia virus in dairy cows in six herds in the municipality of Pasto Nariño. Rev. Lasallista Investig. 2013, 10, 18–26. [Google Scholar]
- Polat, M.; Takeshima, S.N.; Hosomichi, K.; Kim, J.; Miyasaka, T.; Yamada, K.; Arainga, M.; Murakami, T.; Matsumoto, Y.; de la Barra Diaz, V.; et al. A new genotype of bovine leukemia virus in South America identified by NGS-based whole genome sequencing and molecular evolutionary genetic analysis. Retrovirology 2016, 13, 4. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Kelly, P.J.; Bai, J.; Zhang, R.; Wang, C. First molecular characterization of bovine leukemia virus infections in the Caribbean. PLoS ONE 2016, 11, e0168379. [Google Scholar] [CrossRef] [PubMed]
- Corredor-Figueroa, A.P.; Salas, S.; Olaya-Galán, N.N.; Quintero, J.S.; Fajardo, Á.; Soñora, M.; Moreno, P.; Cristina, J.; Sánchez, A.; Tobón, J.; et al. Prevalence and molecular epidemiology of bovine leukemia virus in Colombian cattle. Infect. Genet. Evol. 2020, 80, 104171. [Google Scholar] [CrossRef] [PubMed]
- Jaimes-Dueñez, J.; Goyeneche-Ortiz, E.; Tique-Oviedo, M.; Ortiz-Pineda, M.C.; Cardenas-Pinto, L.; Jimenez-Leaño, A.P.; Ruiz-Saenz, J. Molecular frequency of bovine leukemia virus in Creole cattle of Eastern Colombia. Vet. Anim. Sci. 2024, 25, 100372. [Google Scholar] [CrossRef]
- Giovambattista, G.; Ripoli, M.V.; Peral-García, P.; Bouzat, J.L. Indigenous domestic breeds as reservoirs of genetic diversity: The Argentinean Creole cattle. Anim. Genet. 2001, 32, 240–247. [Google Scholar] [CrossRef]
- Ortega, M.F.; Giovambattista, G.; Cutullé, C.; Santos, D.D.; Nava, S.; Bonamy, M.; Holgado, F. Phenotypic evaluation of genetic resistance to the tick Rhipicephalus (Boophilus) microplus in Argentine Creole cattle. Ticks Tick Borne Dis. 2023, 14, 102223. [Google Scholar] [CrossRef]
- Jacobsen, K.L.; Kaneene, J.B.; Miller, J.M.; Bull, R.W. Comparison of the commercial agar-gel immunodiffusion test and radioimmunoprecipitation assay for detection of antibodies to bovine leukemia virus. Vet. Microbiol. 1985, 46, 1430–1433. [Google Scholar] [CrossRef]
- Klintevall, K.; Ballagi-Pordány, A.; Näslund, K.; Belák, S. Bovine leukaemia virus: Rapid detection of proviral DNA by nested PCR in blood and organs of experimentally infected calves. Vet. Microbiol. 1994, 42, 191–204. [Google Scholar] [CrossRef]
- Martin, D.; Arjona, A.; Soto, I.; Barquero, N.; Viana, M.; Gómez-Lucía, E. Comparative study of PCR as a direct assay and ELISA and AGID as indirect assays for the detection of bovine leukaemia virus. J. Vet. Med. B Infect. Dis. Vet. Public Health 2001, 48, 611–619. [Google Scholar] [CrossRef]
- Rola, M.; Kuzmak, J. The detection of bovine leukemia virus proviral DNA by PCR–ELISA. J. Virol. Methods 2002, 101, 33–40. [Google Scholar] [CrossRef]
- Jaworski, J.P.; Pluta, A.; Rola-Łuszczak, M.; McGowan, S.L.; Finnegan, C.; Heenemann, K.; Carignano, H.A.; Alvarez, I.; Murakami, K.; Willems, L.; et al. Interlaboratory Comparison of Six Real-Time PCR Assays for Detection of Bovine Leukemia Virus Proviral DNA. J. Clin. Microbiol. 2018, 56, e00304-18. [Google Scholar] [CrossRef] [PubMed]
- Ruggiero, V.J.; Bartlett, P.C. Control of Bovine Leukemia Virus in Three US Dairy Herds by Culling ELISA-Positive Cows. Vet. Med. Int. 2019, 2019, 3202184. [Google Scholar] [CrossRef] [PubMed]
- Jimba, M.; Takeshima, S.N.; Matoba, K.; Endoh, D.; Aida, Y. BLV-CoCoMo-qPCR: Quantitation of bovine leukemia virus proviral load using the CoCoMo algorithm. Retrovirology 2010, 7, 91. [Google Scholar] [CrossRef]
- Kobayashi, T.; Inagaki, Y.; Ohnuki, N.; Sato, R.; Murakami, S.; Imakawa, K. Increasing bovine leukemia virus (BLV) proviral load is a risk factor for progression of enzootic bovine leucosis: A prospective study in Japan. Prev. Vet. Med. 2019, 178, 104680. [Google Scholar] [CrossRef] [PubMed]
- Somura, Y.; Sugiyama, E.; Fujikawa, H.; Murakami, K. Comparison of the copy numbers of bovine leukemia virus in the lymph nodes of cattle with enzootic bovine leukosis and cattle with latent infection. Arch. Virol. 2014, 159, 2693–2697. [Google Scholar] [CrossRef]
- Juliarena, M.A.; Barrios, C.N.; Ceriani, M.C.; Esteban, E.N. Hot topic: Bovine leukemia virus (BLV)-infected cows with low proviral load are not a source of infection for BLV-free cattle. J. Dairy Sci. 2016, 99, 4586–4589. [Google Scholar] [CrossRef]
- Sato, H.; Watanuki, S.; Murakami, H.; Sato, R.; Ishizaki, H.; Aida, Y. Development of a luminescence syncytium induction assay (LuSIA) for easily detecting and quantitatively measuring bovine leukemia virus infection. Arch. Virol. 2018, 163, 1519–1530. [Google Scholar] [CrossRef]
- Bai, L.; Borjigin, L.; Sato, H.; Takeshima, S.N.; Asaji, S.; Ishizaki, H.; Kawashima, K.; Obuchi, Y.; Sunaga, S.; Ando, A.; et al. Kinetic Study of BLV Infectivity in BLV Susceptible and Resistant Cattle in Japan from 2017 to 2019. Pathogens 2021, 10, 1281. [Google Scholar] [CrossRef]
- Borjigin, L.; Watanuki, S.; Hamada, R.; Bai, L.; Hirose, T.; Sato, H.; Yoneyama, S.; Yasui, A.; Yasuda, S.; Yamanaka, R.; et al. Effectiveness of integrated bovine leukemia virus eradication strategies utilizing cattle carrying resistant and susceptible histocompatibility complex class II DRB3 alleles. J. Dairy Sci. 2023, 106, 9393–9409. [Google Scholar] [CrossRef]
- Fechner, H.; Blankenstein, P.; Looman, A.C.; Elwert, J.; Geue, L.; Albrecht, C.; Kurg, A.; Beier, D.; Marquardt, O.; Ebner, D. Provirus variants of the bovine leukemia virus and their relation to the serological status of naturally infected cattle. Virology 1997, 237, 261–269. [Google Scholar] [CrossRef]
- Rodríguez, S.M.; Golemba, M.D.; Campos, R.H.; Trono, K.; Jones, L.R. Bovine leukemia virus can be classified into seven genotypes: Evidence for the existence of two novel clades. J. Gen. Virol. 2009, 90, 2788–2797. [Google Scholar] [CrossRef]
- Hernández, D.; Muñoz, J.; Álvarez, L. Dinámica de la Leucosis Bovina en el ganado criollo Hartón del Valle en infección natural. Arch. Zootec. 2016, 65, 365. [Google Scholar] [CrossRef]
- Hernández, D.; Montes, D.; De La Ossa-V, J. The Proviral Load of the Bovine Leukosis Virus is Associated with the Polymorphisms of the BoLA-DRB3 Gene in the Harton del Valle Breed. Indian J. Sci. Technol. 2018, 11, 1–7. [Google Scholar] [CrossRef]
- Takeshima, S.N.; Kitamura-Muramatsu, Y.; Yuan, Y.; Polat, M.; Saito, S.; Aida, Y. BLV-CoCoMo-qPCR-2: Improvements to the BLV-CoCoMo-qPCR assay for bovine leukemia virus by reducing primer degeneracy and constructing an optimal standard curve. Arch. Virol. 2015, 160, 1325–1332. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.; Hidano, A.; Tsutsui, T.; Yamamoto, T.; Hayama, Y.; Nishida, T.; Muroga, N.; Konishi, M.; Kameyama, K.; Murakami, K. Analysis of risk factors associated with bovine leukemia virus seropositivity within dairy and beef breeding farms in Japan: A nationwide survey. Res. Vet. Sci. 2014, 96, 47–53. [Google Scholar] [CrossRef]
- Ohno, A.; Takeshima, S.N.; Matsumoto, Y.; Aida, Y. Risk factors associated with increased bovine leukemia virus proviral load in infected cattle in Japan from 2012 to 2014. Virus Res. 2015, 210, 283–290. [Google Scholar] [CrossRef]
- Murakami, K.; Kobayashi, S.; Konishi, M.; Kameyama, K.; Tsutsui, T. Nationwide survey of bovine leukemia virus infection among dairy and beef breeding cattle in Japan from 2009–2011. J. Vet. Med. Sci. 2013, 75, 1123–1126. [Google Scholar] [CrossRef]
- Chung, A.H. Bovine leukaemia virus infection in Peru. Trop. Anim. Health Prod. 1983, 15, 61. [Google Scholar] [CrossRef]
- Ducreux, F.; Arrieta, E.; Jiminez, C.; Moreno, E.; Rodríguez, L. Estudios sobre leucosis viral bovina en ganado Bos indicus en Costa Rica. Cienc. Vet. 1987, 9, 95–99. [Google Scholar] [CrossRef]
- Castro, R.S.; Leite, R.C.; Abreu, J.J.; Lage, A.P.; Ferraz, I.B.; Lobato, Z.I.; Balsamão, S.L. Prevalence of antibodies to selected viruses in bovine embryo donors and recipients from Brazil, and its implications in international embryo trade. Trop. Anim. Health Prod. 1992, 24, 173–176. [Google Scholar] [CrossRef]
- Molnár, É.; Molnár, L.; Dias, H.; Silva, A.; Vale, W. Occurrence of enzootic bovine leukosis in the State of Pará, Brazil. Pesqui. Vet. Bras. 1999, 19, 7–11. [Google Scholar] [CrossRef]
- Trono, K.G.; Pérez-Filgueira, D.M.; Duffy, S.; Borca, M.V.; Carrillo, C. Seroprevalence of bovine leukemia virus in dairy cattle in Argentina: Comparison of sensitivity and specificity of different detection methods. Vet. Microbiol. 2001, 83, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Monti, G.; Schrijver, R.; Beier, D. Genetic diversity and spread of Bovine leukaemia virus isolates in Argentine dairy cattle. Arch. Virol. 2005, 150, 443–458. [Google Scholar] [CrossRef] [PubMed]
- Monti, G.E.; Frankena, K.; Engel, B.; Buist, W.; Tarabla, H.D.; de Jong, M.C.M. Evaluation of a new antibody-based enzyme-linked immunosorbent assay for the detection of bovine leukemia virus infection in dairy cattle. J. Vet. Diagn. Investig. 2005, 17, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Delgado, I.; Alfonso, A.; Martínez, N.; Abeledo, M.; Rodrigues, M.; Barrera, M. Presencia de anticuerpos al virus de la leucosis bovina en rebaños pertenecientes a las provincias occidentales y centrales de Cuba. Rev. Salud Anim. 2009, 31, 24–28. [Google Scholar]
- Heinecke, N.; Tórtora, J.; Martínez, H.A.; González-Fernández, V.D.; Ramírez, H. Detection and genotyping of bovine leukemia virus in Mexican cattle. Arch. Virol. 2017, 162, 3191–3196. [Google Scholar] [CrossRef]
- Freitas, T.M.S.; Dias, J.M.; Veríssimo, A.C.F.; Lobo, J.R.; Costa, G.L.; Moura, M.I.; Landi, V.; Martínez, A.M.; Carmo, A.S.; Soares, F.M.C. Population Structure of Curraleiro Pé-Duro Cattle and its Relationship with the Serological Profile Against Pathogens of Economic and Zoonotic Interest. Front. Genet. 2022, 13, 872660. [Google Scholar] [CrossRef]
- Fonteque, G.V.; Casa, M.S.; Ribeiro, G.S.N.; Silva, Z.; Vogel, C.I.G.; Fonteque, J.H.; Costa, U.M.; Saito, M.E.; Giovambattista, G.; Takeshima, S.-N.; et al. Quantifying the enzootic leukosis virus threat: How prevalent is the natural infection in Crioulo Lageano cattle in southern Brazil? Braz. J. Biol. 2025, 85, 291923. [Google Scholar] [CrossRef]
- Villalobos-Cortes, A.; Franco, S.; González, R.; Jaén, M. Nested polymerase chain reaction (nPCR) based diagnosis of bovine leukemia virus in Panama. Afr. J. Biotechnol. 2017, 16, 528–535. [Google Scholar] [CrossRef][Green Version]
- Saa, L.R.; Guzmán, L.T.; Fierro, N.C.; Castro, L.M.; Reyes-Bueno, F.; Carbonero, A. Seroprevalence and risk factors associated with bovine leukemia virus (BLV) seropositivity in cattle herds from Ecuador. Rev. Colomb. Cienc. Pecu. 2021, 34, 177–188. [Google Scholar] [CrossRef]
- Lirón, J.P.; Peral-García, P.; Giovambattista, G. Genetic characterization of Argentine and Bolivian Creole cattle breeds assessed through microsatellites. J. Hered. 2006, 97, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Ginja, C.; Gama, L.T.; Cortés, O.; Burriel, I.M.; Vega-Pla, J.L.; Penedo, C.; Sponenberg, P.; Cañón, J.; Sanz, A.; do Egito, A.A.; et al. The genetic ancestry of American Creole cattle inferred from uniparental and autosomal genetic markers. Sci. Rep. 2019, 9, 11486, Erratum in Sci. Rep. 2020, 10, 16930. https://doi.org/10.1038/s41598-020-73066-4. [Google Scholar] [CrossRef] [PubMed]
- Marcuzzi, O.; Calcaterra, F.; Loza Vega, A.; Ortega Masaguém, M.F.; Armstrong, E.; Pereira Rico, J.A.; Jara, E.; Olivera, L.H.; Peral García, P.; Giovambattista, G. Genomic analysis of inbreeding level, kinship and breed relationships in Creole cattle from South America. Anim. Genet. 2024, 55, 527–539. [Google Scholar] [CrossRef]
- Rahman, M.M.; Badr, Y.; Kamatari, Y.O.; Kitamura, Y.; Shimizu, K.; Okada, A.; Inoshima, Y. Data on proteomic analysis of milk extracellular vesicles from bovine leukemia virus-infected cattle. Data Brief 2020, 33, 106510. [Google Scholar] [CrossRef]
- Rahman, M.M.; Ishikawa, H.; Yamauchi, M.; Takashima, S.; Kamatari, Y.O.; Shimizu, K.; Okada, A.; Inoshima, Y. Characterization of mRNA Signature in Milk Small Extracellular Vesicles from Cattle Infected with Bovine Leukemia Virus. Pathogens 2023, 12, 1239. [Google Scholar] [CrossRef]
- Lew, A.E.; Bock, R.E.; Miles, J.; Cuttell, L.B.; Steer, P.; Nadin-Davis, S.A. Sensitive and specific detection of bovine immunodeficiency virus and bovine syncytial virus by 5′ Taq nuclease assays with fluorescent 3′ minor groove binder-DNA probes. J. Virol. Methods 2004, 116, 1–9. [Google Scholar] [CrossRef]
- Yoneyama, S.; Kobayashi, S.; Matsunaga, T.; Tonosaki, K.; Leng, D.; Sakai, Y.; Yamada, S.; Kimura, A.; Ichijo, T.; Hikono, H.; et al. Comparative Evaluation of Three Commercial Quantitative Real-Time PCRs Used in Japan for Bovine Leukemia Virus. Viruses 2022, 14, 1182. [Google Scholar] [CrossRef]
- Jimba, M.; Takeshima, S.N.; Murakami, H.; Kohara, J.; Kobayashi, N.; Matsuhashi, T.; Ohmori, T.; Nunoya, T.; Aida, Y. BLV-CoCoMo-qPCR: A useful tool for evaluating bovine leukemia virus infection status. BMC Vet. Res. 2012, 8, 167. [Google Scholar] [CrossRef]
- Watanuki, S.; Shoji, K.; Izawa, M.; Okami, M.; Ye, Y.; Bao, A.; Liu, Y.; Saitou, E.; Sugiyama, K.; Endo, M.; et al. Development of a dry- and liquid-duplex-reagent mix-based polymerase chain reaction assay as a novel tool for the rapid and easy quantification of bovine leukemia virus (BLV) proviral loads. Viruses 2024, 16, 1016. [Google Scholar] [CrossRef]
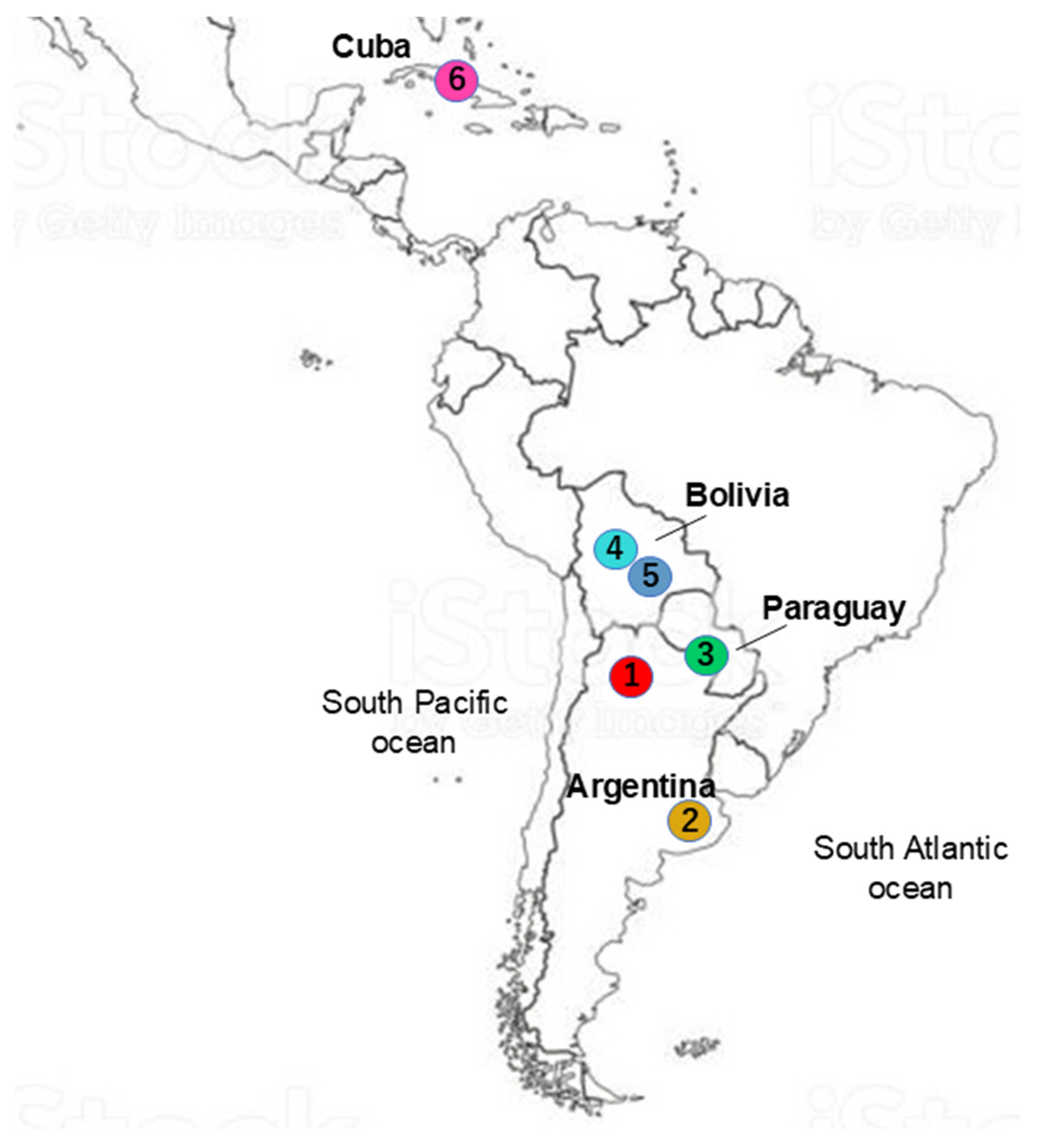

| Breed | Sample Size | Breed Origin | Sample Site | Production Purpose | Environment |
|---|---|---|---|---|---|
| Argentine Creole (CrAr) | 112 | Argentina | Tucumán province | Beef | Subtropical to temperate |
| Argentine Patagonian Creole | 23 | Argentina | Buenos Aires province | Beef | Temperate to cold |
| (CrArPat) | |||||
| Bolivian Highland Creole (CrCoch) | 13 | Bolivia | Cochabamba department | Beef | Highland plain |
| Saavedreño Creole (CrSaa) | 10 | Bolivia | Santa Cruz department | Beef and dairy | Subtropical plain |
| Pampa Chaqueño Creole (CrPaCh) | 37 | Paraguay | Paraguayan Chaco | Beef | Dry forest |
| Siboney de Cuba (Sib) | 49 | Cuba | La Havana province, Cuba | Dairy | Subtropical plain |
| Breed | Positive No. a/Tested Samples No. (Positive %) |
|---|---|
| Argentine Creole (CrAr) | 11/112 (9.8) |
| Argentine Patagonian Creole (CrArPat) | 10/23 (43.5) |
| Saavedreño Creole (CrSaa) | 7/10 (70.0) |
| Bolivian Highland Creole (CrCoch) | 3/13 (23.8) |
| Pampa Chaqueño (CrPaCh) | 31/37 (83.8) |
| Siboney (Sib) | 14/49 (28.6) |
| Total breeds | 76/244 (31.1) |
| Country | Positive No.a/Tested Samples No. (Positive %) |
|---|---|
| Argentina | 21/135 (15.6) |
| Bolivia | 10/23 (43.5) |
| Paraguay | 31/37 (83.8) |
| Cuba | 14/49 (28.6) |
| Total breeds | 76/244 (31.1) |
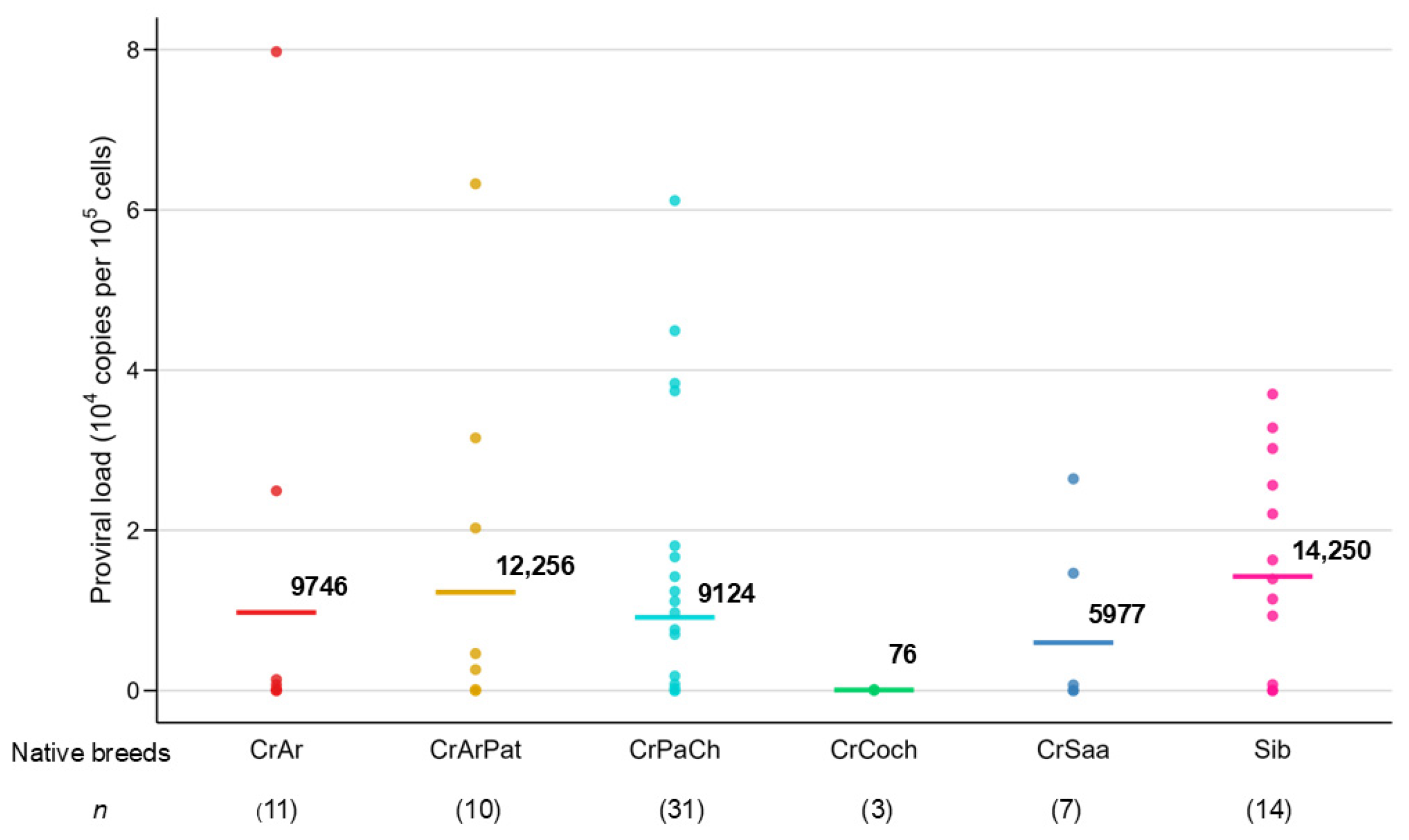
| Breed | BLV– Positive n a | PVL b (Copy/105 Cells) | Mean Value (Range) | ||
|---|---|---|---|---|---|
| 1–1000 n (%) | 1001–9999 n (%) | ≥10,000 n (%) | |||
| CrAr | 11 | 8 (72.7) | 1 (9.1) | 2 (18.2) | 9746 (3–79,740) |
| CrArPat | 10 | 5 (50.0) | 2 (20.0) | 3 (30.0) | 12,256 (13–63,263) |
| CrSaa | 7 | 5 (71.4) | 2 (28.6) | 0 (0.0) | 5977 (5–26,435) |
| CrCoch | 3 | 3 (100.0) | 0 (0.0) | 0 (0.0) | 76 (21–119) |
| CrPaCh | 31 | 18 (58.1) | 4 (12.9) | 9 (29.0) | 9124 (1–61,161) |
| Sib | 14 | 5 (35.7) | 1 (7.2) | 8 (57.1) | 14,250 (1–37,009) |
| Total | 76 | 44 (57.9) | 10 (13.2) | 22 (28.9) | 9923 (1–79,740) |
| Country | BLV– Positive n a | PVL b (Copy/105 Cells) | Mean Value (Range) | ||
|---|---|---|---|---|---|
| 1–1000 n (%) | 1001–9999 n (%) | ≥10,000 n (%) | |||
| Argentina | 21 | 13 (61.9) | 3 (14.29) | 5 (23.8) | 9746 (3–79,740) |
| Bolivia | 10 | 8 (80.0) | 2 (20.0) | 0 (0.0) | 4207 (5–26,435) |
| Paraguay | 31 | 18 (58.1) | 4 (12.9) | 9 (29.0) | 9124 (1–61,161) |
| Cuba | 14 | 5 (35.7) | 1 (7.2) | 8 (57.1) | 14,250 (1–37,009) |
| Total | 76 | 44 (57.9) | 10 (13.2) | 22 (29.0) | 9923 (1–79,740) |
| Breed | Typed | Country | Used Method | n a | BLV b Prevalence (%) | Reference |
|---|---|---|---|---|---|---|
| Argentine Creole | Creole | Argentina | CoCoMo qPCR | 112 | 9.8 (11/112) | This work |
| Argentine Patagonian Creole | Creole | Argentina | CoCoMo qPCR | 23 | 43.5 (10/23) | This work |
| Pampa Chaqueño | Creole | Paraguay | CoCoMo qPCR | 37 | 83.2 (31/37) | This work |
| Saavedreño | Creole | Bolivia | CoCoMo qPCR | 10 | 70 (7/10) | This work |
| Cochabamba Creole | Creole | Bolivia | CoCoMo qPCR | 13 | 23.1 (3/13) | This work |
| Yacumeño | Creole | Bolivia | CoCoMo qPCR | 130 | 20.37 (22/108) | [23] |
| Lageano | Creole | Brazil | standard PCR | 308 | 36.69 (113/308) | [60] |
| Curraleiro Pé-Duro | Creole | Brazil | ELISA | 596 | 18.29 (109/596) | [59] |
| Blanco Orijinegro | Creole | Colombia | nested PCR | 30 | 0 (0/30) | [21] |
| Casareño | Creole | Colombia | nested PCR | 30 | 26.7 (8/30) | [21] |
| Casareño | Creole | Colombia | nested PCR | 73 | 16.4 (12/73) | [26] |
| Costeño con Cuernos | Creole | Colombia | nested PCR | 30 | 23.3 (7/30) | [21] |
| Chino Santandereano | Creole | Colombia | nested PCR | 30 | 60 (18/30) | [21] |
| Chino Santandereano | Creole | Colombia | nested PCR | 108 | 35.1 (38/108) | [26] |
| Caqueño | Creole | Colombia | nested PCR | 30 | 16.7 (5/30) | [21] |
| Hartón del Valle | Creole | Colombia | nested PCR | 30 | 83.3 (25/30) | [21] |
| Hartón del Valle | Creole | Colombia | nested PCR, qPCR | 93 | 18.9 (18/93) | [44] |
| Romosinuano | Creole | Colombia | nested PCR | 30 | 0 (0/30) | [21] |
| Sanmartineano | Creole | Colombia | nested PCR | 30 | 0 (0/30) | [21] |
| Sanmartineano | Creole | Colombia | nested PCR | 72 | 23.6 (17/72) | [26] |
| Peruvian Creole | Creole | Perú | AGID | nd | nd | [24] |
| Siboney | Composite c | Cuba | CoCoMo qPCR | 49 | 28.6 (14/49) | This work |
| Lucerna | Composite d | Colombia | nested PCR | 30 | 50 (15/30) | [21] |
| Lucerna | Composite d | Colombia | nested PCR, qPCR | 24 | 48.5 (17/72) | [44] |
| Velásquez | Composite e | Colombia | nested PCR | 30 | 50 (15/30) | [21] |
| Creole | Creole | Ecuador | ELISA | 1604 | 10.6 (170/1604) | [62] |
| Guaymí | Creole | Panamá | nested PCR | 40 | 80 (32/40) | [61] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giovambattista, G.; Bao, A.; Marcuzzi, O.; Vega, A.L.; Rico, J.A.P.; Ortega Masague, M.F.; Rojas, L.A.C.; Martinez, R.D.; Reinosa, O.U.; Aida, Y. Detection of Bovine Leukemia Virus in Argentine, Bolivian, Paraguayan and Cuban Native Cattle Using a Quantitative Real-Time PCR Assay-BLV-CoCoMo-qPCR-2. Pathogens 2025, 14, 1005. https://doi.org/10.3390/pathogens14101005
Giovambattista G, Bao A, Marcuzzi O, Vega AL, Rico JAP, Ortega Masague MF, Rojas LAC, Martinez RD, Reinosa OU, Aida Y. Detection of Bovine Leukemia Virus in Argentine, Bolivian, Paraguayan and Cuban Native Cattle Using a Quantitative Real-Time PCR Assay-BLV-CoCoMo-qPCR-2. Pathogens. 2025; 14(10):1005. https://doi.org/10.3390/pathogens14101005
Chicago/Turabian StyleGiovambattista, Guillermo, Aronggaowa Bao, Olivia Marcuzzi, Ariel Loza Vega, Juan Antonio Pereira Rico, Maria Florencia Ortega Masague, Liz Aurora Castro Rojas, Ruben Dario Martinez, Odalys Uffo Reinosa, and Yoko Aida. 2025. "Detection of Bovine Leukemia Virus in Argentine, Bolivian, Paraguayan and Cuban Native Cattle Using a Quantitative Real-Time PCR Assay-BLV-CoCoMo-qPCR-2" Pathogens 14, no. 10: 1005. https://doi.org/10.3390/pathogens14101005
APA StyleGiovambattista, G., Bao, A., Marcuzzi, O., Vega, A. L., Rico, J. A. P., Ortega Masague, M. F., Rojas, L. A. C., Martinez, R. D., Reinosa, O. U., & Aida, Y. (2025). Detection of Bovine Leukemia Virus in Argentine, Bolivian, Paraguayan and Cuban Native Cattle Using a Quantitative Real-Time PCR Assay-BLV-CoCoMo-qPCR-2. Pathogens, 14(10), 1005. https://doi.org/10.3390/pathogens14101005







