Establishment of a Luciferase-Based Reporter System to Study Aspects of Human Cytomegalovirus Infection, Replication Characteristics, and Antiviral Drug Efficacy
Abstract
1. Introduction
2. Materials and Methods
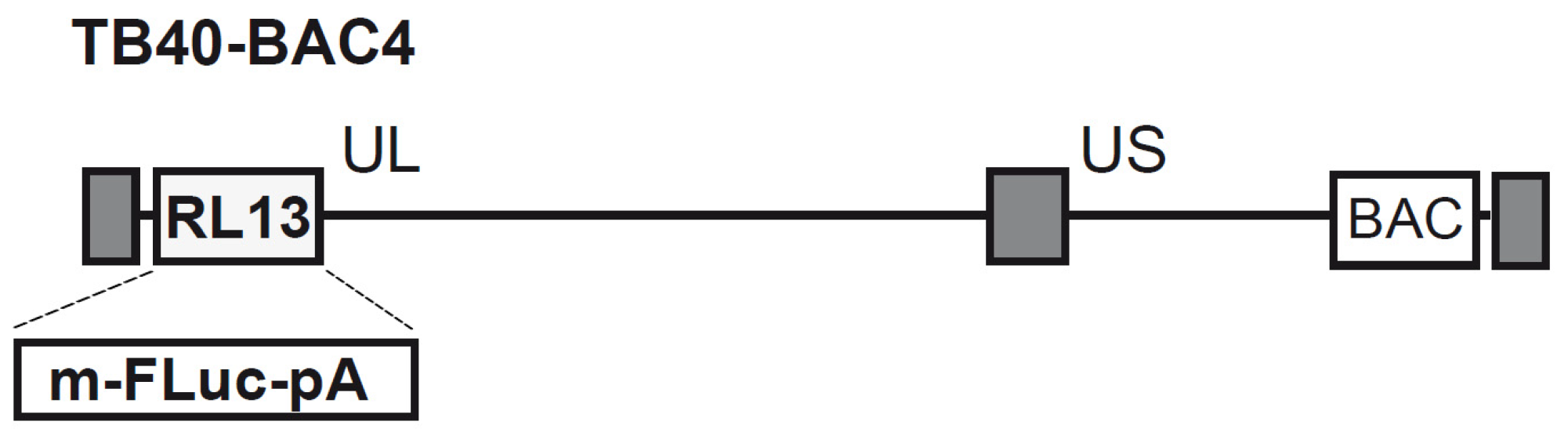
2.1. Genetic Recombination and Reconstitution of Infectious HCMV TB40-FLuc
2.2. Cell Culture and Virus Infection
2.3. Luminescence-Based and Fluorescence-Based HCMV Replication Assays
2.4. Neutral Red Assay
2.5. Quantitative Real-Time PCR (qPCR)
2.6. Indirect Immunofluorescence (IF) Analysis and Confocal Laser-Scanning Microscopy
2.7. SDS Polyacrylamide Gel Electrophoresis (SDS-PAGE) and Western Blot Analysis
2.8. Antiviral Compounds
3. Results and Discussion
3.1. Generation of Recombinant HCMV BAC and Reconstitution of the TB40-FLuc Reporter Virus
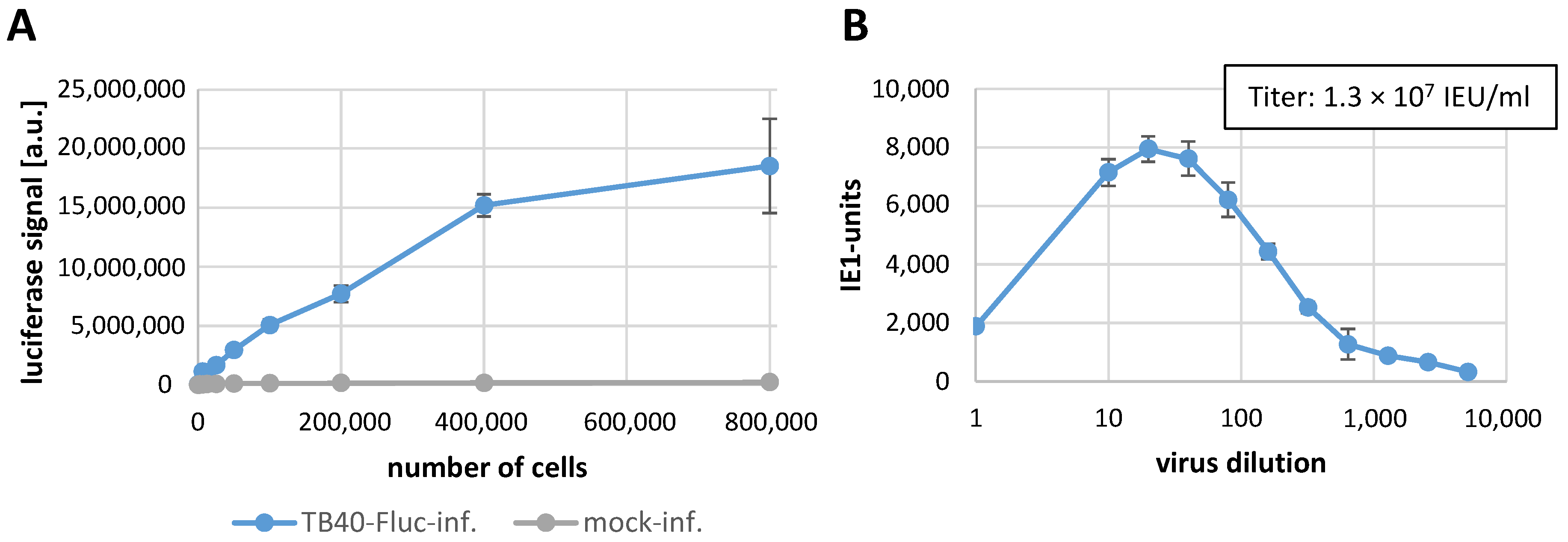
3.2. Establishment of the Infection System with HCMV TB40-FLuc in Primary Human Fibroblasts
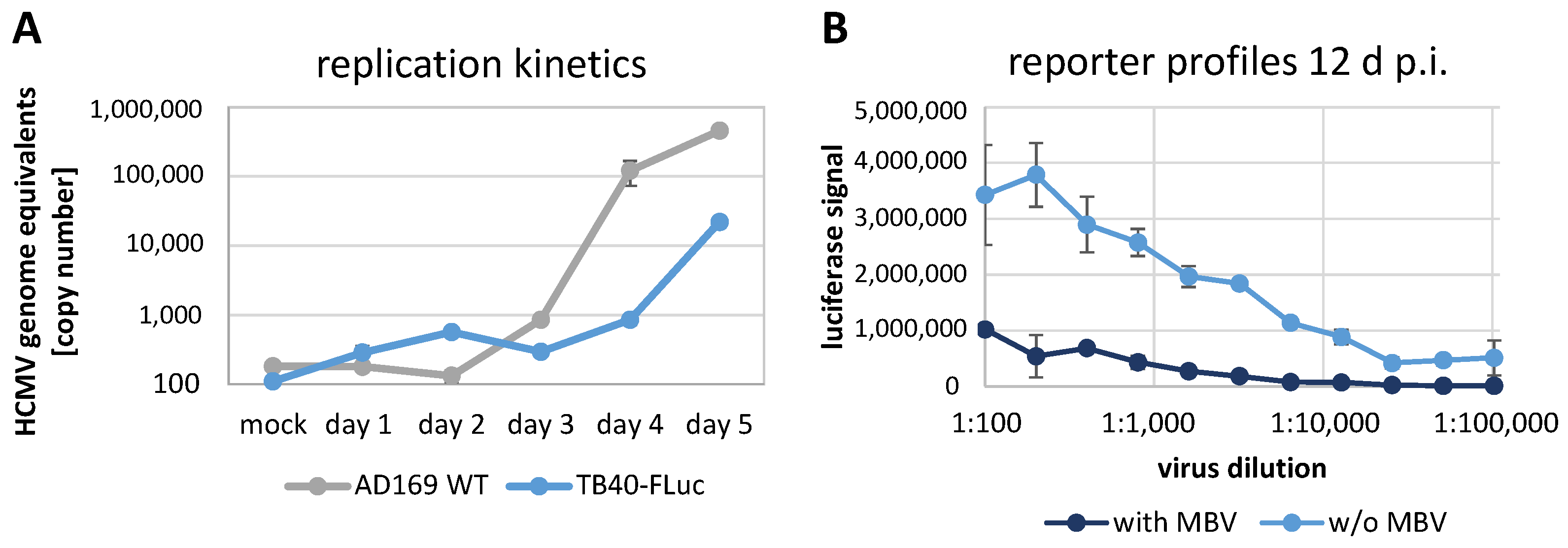
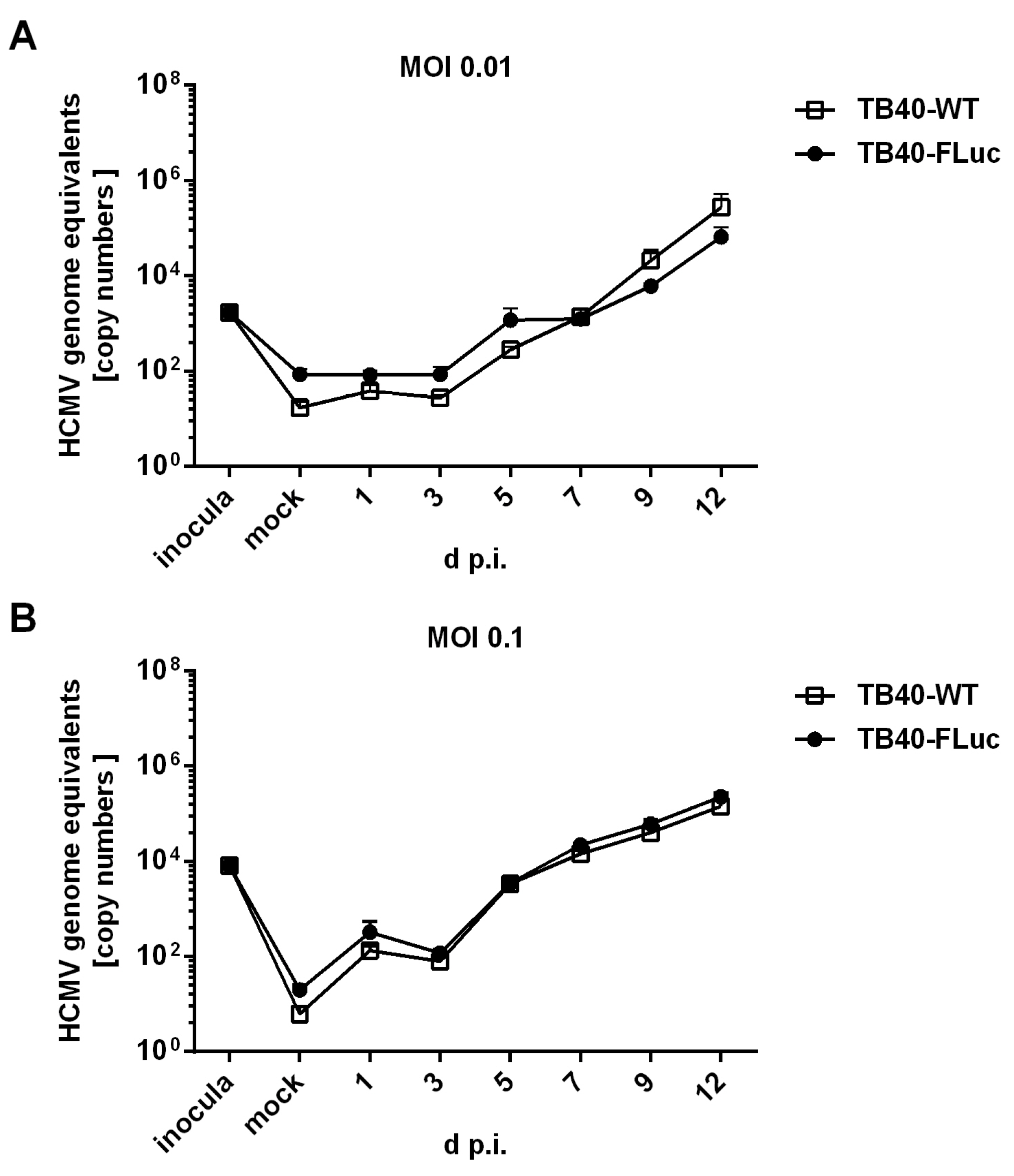
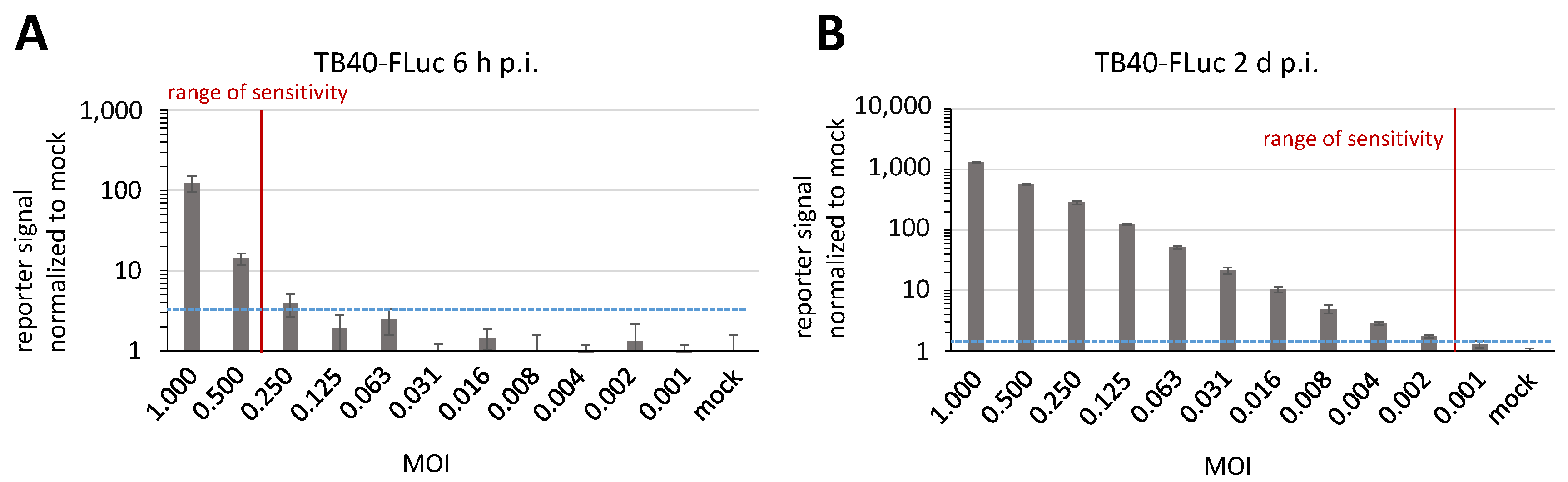
3.3. Replication Kinetics of HCMV TB40-FLuc in Single-Round or Multi-Round Settings
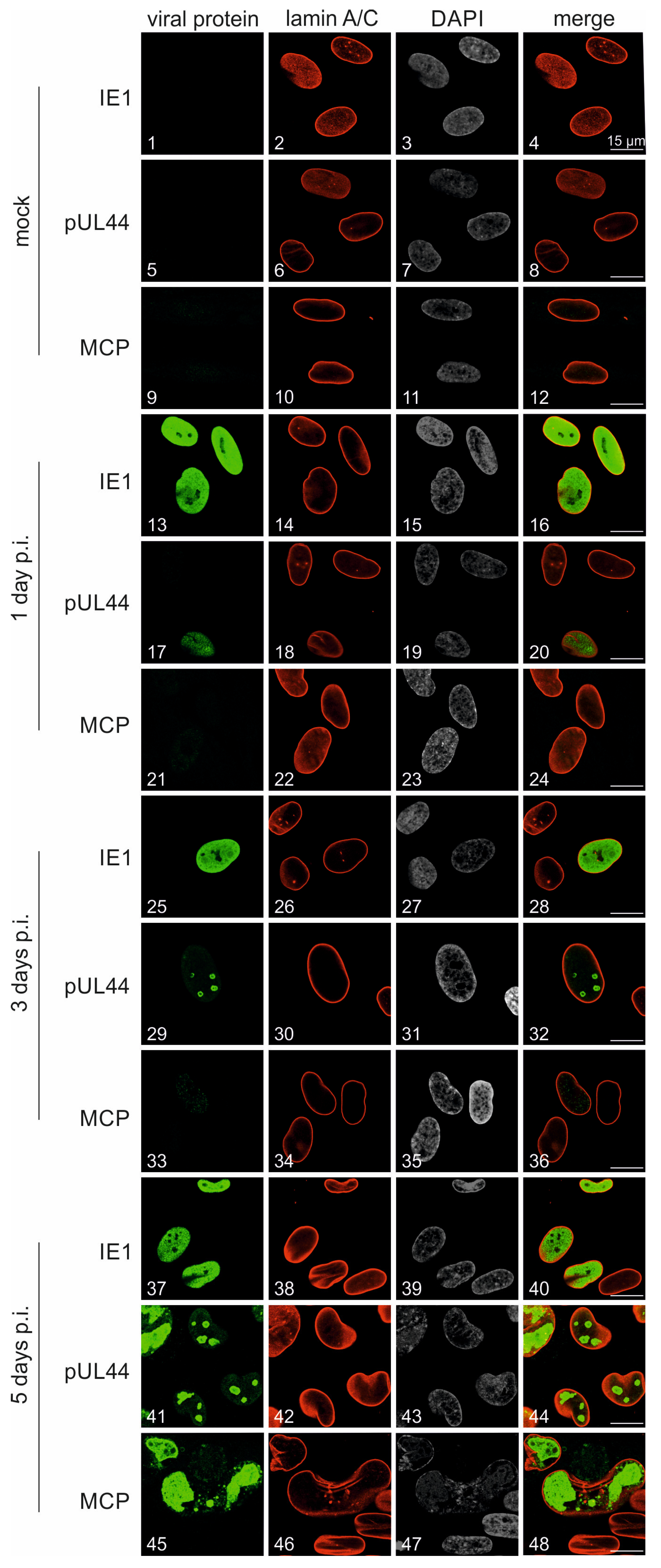
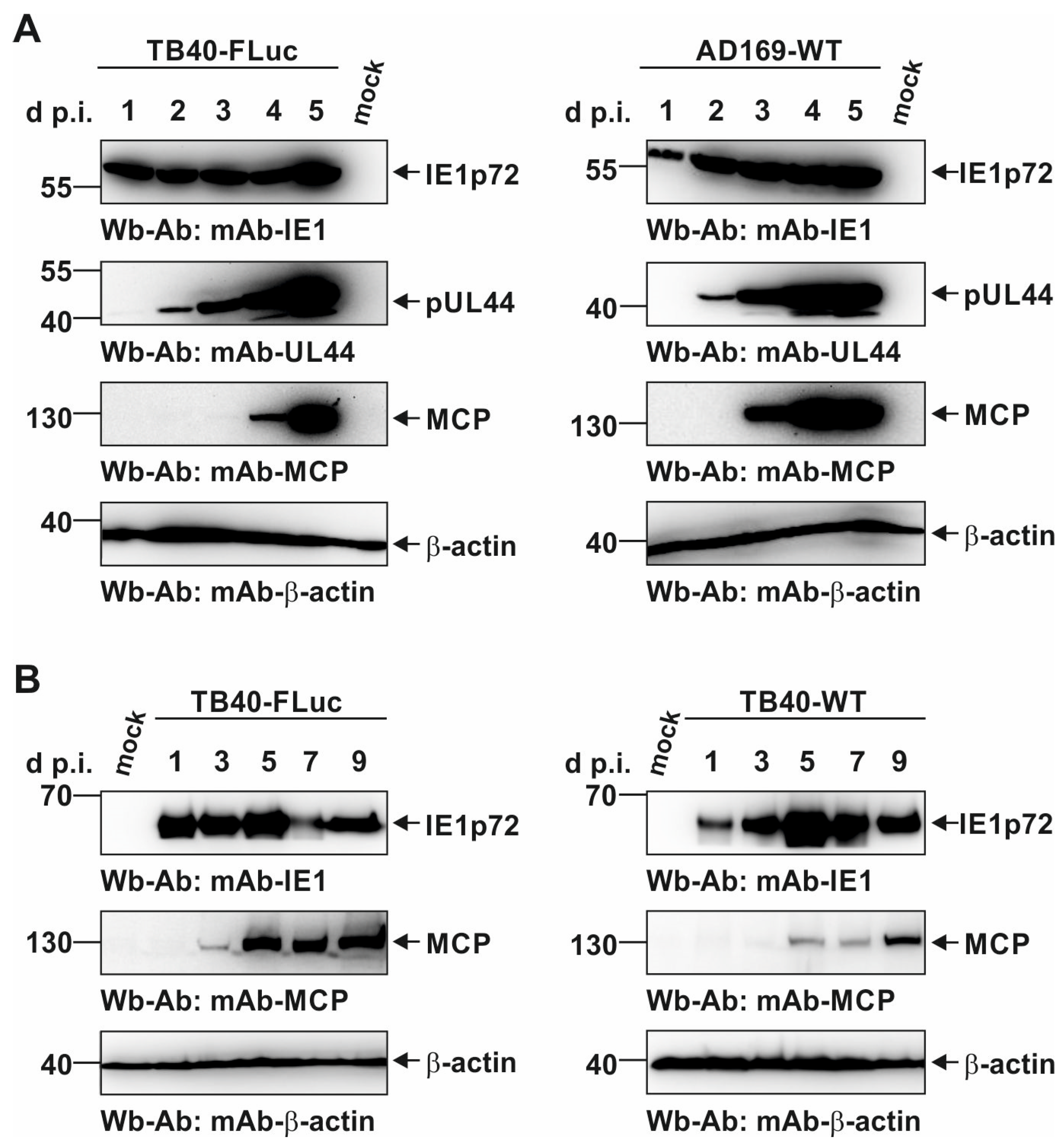
3.4. Characterization of Viral Protein Expression Patterns in HCMV TB40-FLuc-Infected Cells
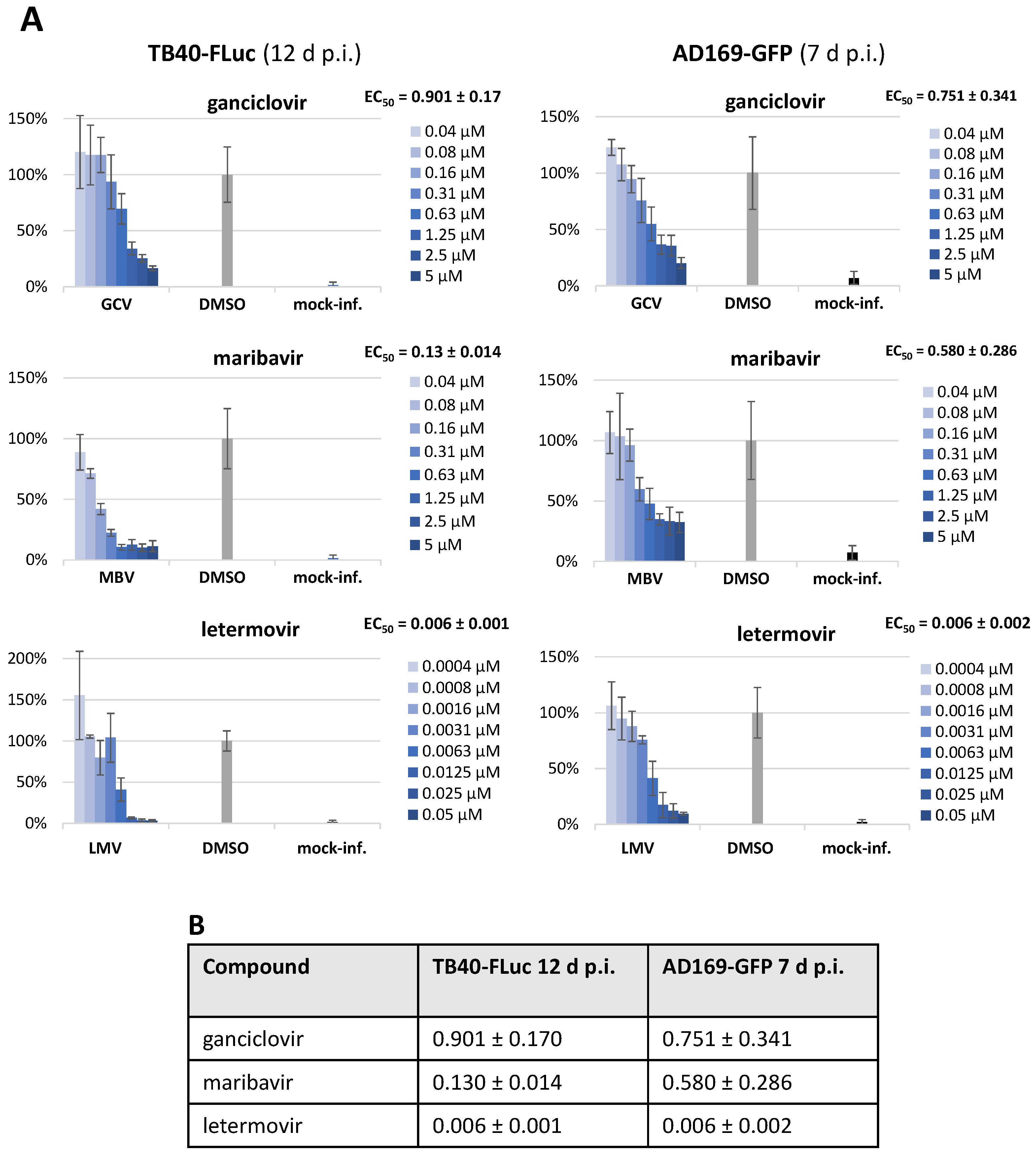
3.5. Assessment of Antiviral Drug Activity in Two HCMV Reporter Systems (TB40-FLuc and AD169-GFP)
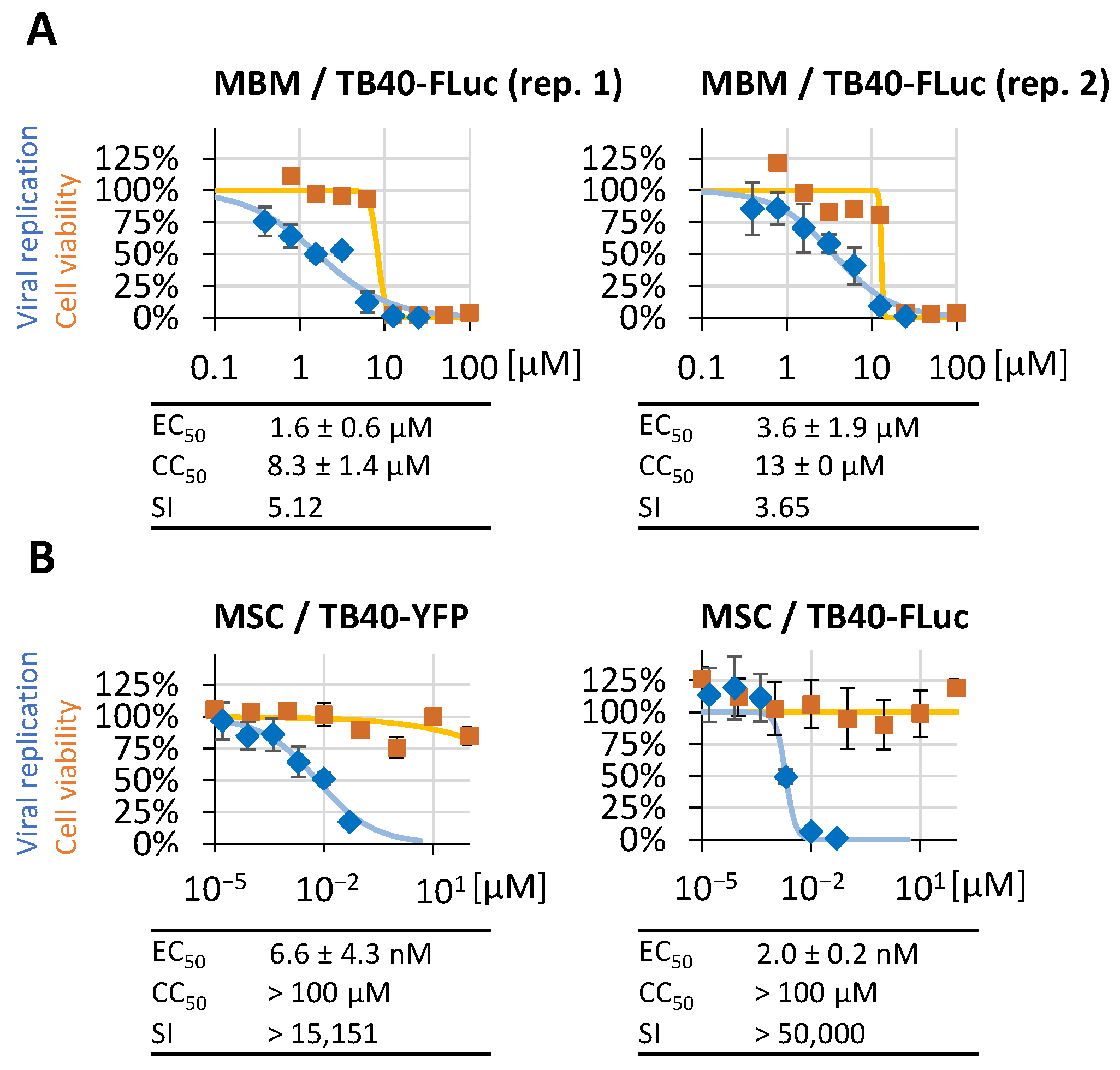
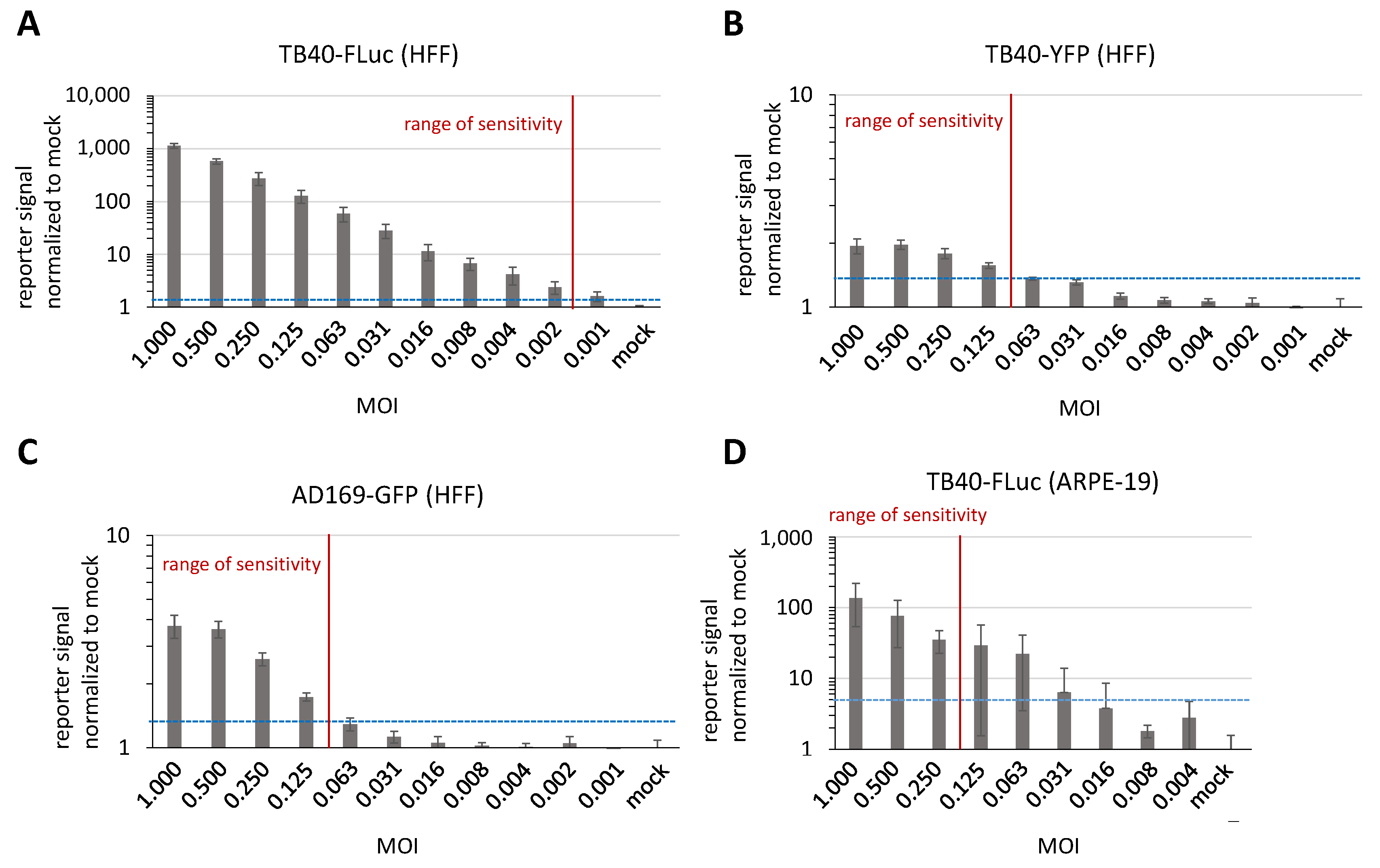
3.6. Determination of Reporter Sensitivity Comparing Different HCMV Infection Systems
3.7. Further Options for the Application of HCMV TB40-FLuc
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carney, W.P.; Hirsch, M.S. Mechanisms of immunosuppression in cytomegalovirus mononucleosis. II. Virus-monocyte interactions. J. Infect. Dis. 1981, 144, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, P.; Baraniak, I.; Reeves, M. The pathogenesis of human cytomegalovirus. J. Pathol. 2015, 235, 288–297. [Google Scholar] [CrossRef]
- Griffiths, P.; Reeves, M. Pathogenesis of human cytomegalovirus in the immunocompromised host. Nat. Rev. Microbiol. 2021, 19, 759–773. [Google Scholar] [CrossRef] [PubMed]
- Dropulic, L.K.; Cohen, J.I. Update on new antivirals under development for the treatment of double-stranded DNA virus infections. Clin. Pharmacol. Ther. 2010, 88, 610–619. [Google Scholar] [CrossRef] [PubMed]
- Marschall, M.; Stamminger, T. Molecular targets for antiviral therapy of cytomegalovirus infections. Future Microbiol. 2009, 4, 731–742. [Google Scholar] [CrossRef] [PubMed]
- Steingruber, M.; Marschall, M. The Cytomegalovirus Protein Kinase pUL97:Host Interactions, Regulatory Mechanisms and Antiviral Drug Targeting. Microorganisms 2020, 8, 515. [Google Scholar] [CrossRef] [PubMed]
- Atabani, S.F.; Smith, C.; Atkinson, C.; Aldridge, R.W.; Rodriguez-Peralvarez, M.; Rolando, N.; Harber, M.; Jones, G.; O’Riordan, A.; Burroughs, A.K.; et al. Cytomegalovirus replication kinetics in solid organ transplant recipients managed by preemptive therapy. Am. J. Transplant. 2012, 12, 2457–2464. [Google Scholar] [CrossRef] [PubMed]
- Zangger, N.; Oxenius, A. T cell immunity to cytomegalovirus infection. Curr. Opin. Immunol. 2022, 77, 102185. [Google Scholar] [CrossRef] [PubMed]
- Kotton, C.N. CMV: Prevention, Diagnosis and Therapy. Am. J. Transplant. 2013, 13 (Suppl. S3), 24–40; quiz 40. [Google Scholar] [CrossRef]
- Crawford, L.B.; Diggins, N.L.; Caposio, P.; Hancock, M.H. Advances in Model Systems for Human Cytomegalovirus Latency and Reactivation. mBio 2022, 13, e0172421. [Google Scholar] [CrossRef]
- Schütz, M.; Müller, R.; Socher, E.; Wangen, C.; Full, F.; Wyler, E.; Wong, D.; Scherer, M.; Stamminger, T.; Chou, S.; et al. Highly Conserved Interaction Profiles between Clinically Relevant Mutants of the Cytomegalovirus CDK-like Kinase pUL97 and Human Cyclins: Functional Significance of Cyclin H. Int. J. Mol. Sci. 2022, 23, 11814. [Google Scholar] [CrossRef] [PubMed]
- Suarez, N.M.; Wilkie, G.S.; Hage, E.; Camiolo, S.; Holton, M.; Hughes, J.; Maabar, M.; Vattipally, S.B.; Dhingra, A.; Gompels, U.A.; et al. Human Cytomegalovirus Genomes Sequenced Directly From Clinical Material: Variation, Multiple-Strain Infection, Recombination, and Gene Loss. J. Infect. Dis. 2019, 220, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Chou, S. Recombinant phenotyping of cytomegalovirus UL97 kinase sequence variants for ganciclovir resistance. Antimicrob. Agents Chemother. 2010, 54, 2371–2378. [Google Scholar] [CrossRef] [PubMed]
- Drebber, U.; Haferkamp, K.; Kern, M.A.; Muller, M.; Zur Hausen, A.; Kasper, H.U.; Odenthal, M.; Dienes, H.P. Induction of early murine cytomegalovirus infection by different reporter gene-associated recombinant viruses. J. Viral Hepat. 2006, 13, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Koshizuka, T.; Sato, Y.; Kobiyama, S.; Oshima, M.; Suzutani, T. A two-step culture method utilizing secreted luciferase recombinant virus for detection of anti-cytomegalovirus compounds. Microbiol. Immunol. 2018, 62, 651–658. [Google Scholar] [CrossRef] [PubMed]
- Marschall, M.; Freitag, M.; Weiler, S.; Sorg, G.; Stamminger, T. Recombinant green fluorescent protein-expressing human cytomegalovirus as a tool for screening antiviral agents. Antimicrob. Agents Chemother. 2000, 44, 1588–1597. [Google Scholar] [CrossRef] [PubMed]
- Sinzger, C.; Hahn, G.; Digel, M.; Katona, R.; Sampaio, K.L.; Messerle, M.; Hengel, H.; Koszinowski, U.; Brune, W.; Adler, B. Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J. Gen. Virol. 2008, 89, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Marquardt, A.; Halle, S.; Seckert, C.K.; Lemmermann, N.A.W.; Veres, T.Z.; Braun, A.; Maus, U.A.; Forster, R.; Reddehase, M.J.; Messerle, M.; et al. Single cell detection of latent cytomegalovirus reactivation in host tissue. J. Gen. Virol. 2011, 92, 1279–1291. [Google Scholar] [CrossRef]
- Tischer, B.K.; Smith, G.A.; Osterrieder, N. En passant mutagenesis: A two step markerless red recombination system. Methods Mol. Biol. 2010, 634, 421–430. [Google Scholar] [CrossRef]
- Hammer, Q.; Ruckert, T.; Borst, E.M.; Dunst, J.; Haubner, A.; Durek, P.; Heinrich, F.; Gasparoni, G.; Babic, M.; Tomic, A.; et al. Peptide-specific recognition of human cytomegalovirus strains controls adaptive natural killer cells. Nat. Immunol. 2018, 19, 453–463. [Google Scholar] [CrossRef]
- Elbasani, E.; Gabaev, I.; Steinbruck, L.; Messerle, M.; Borst, E.M. Analysis of essential viral gene functions after highly efficient adenofection of cells with cloned human cytomegalovirus genomes. Viruses 2014, 6, 354–370. [Google Scholar] [CrossRef] [PubMed]
- Rowe, W.P.; Hartley, J.W.; Waterman, S.; Turner, H.C.; Huebner, R.J. Cytopathogenic agent resembling human salivary gland virus recovered from tissue cultures of human adenoids. Proc. Soc. Exp. Biol. Med. 1956, 92, 418–424. [Google Scholar]
- Wagenknecht, N.; Reuter, N.; Scherer, M.; Reichel, A.; Muller, R.; Stamminger, T. Contribution of the Major ND10 Proteins PML, hDaxx and Sp100 to the Regulation of Human Cytomegalovirus Latency and Lytic Replication in the Monocytic Cell Line THP-1. Viruses 2015, 7, 2884–2907. [Google Scholar] [CrossRef]
- Repetto, G.; del Peso, A.; Zurita, J.L. Neutral red uptake assay for the estimation of cell viability/cytotoxicity. Nat. Protoc. 2008, 3, 1125–1131. [Google Scholar] [CrossRef] [PubMed]
- Lorz, K.; Hofmann, H.; Berndt, A.; Tavalai, N.; Mueller, R.; Schlotzer-Schrehardt, U.; Stamminger, T. Deletion of open reading frame UL26 from the human cytomegalovirus genome results in reduced viral growth, which involves impaired stability of viral particles. J. Virol. 2006, 80, 5423–5434. [Google Scholar] [CrossRef] [PubMed]
- Häge, S.; Buscher, N.; Pakulska, V.; Hahn, F.; Adrait, A.; Krauter, S.; Borst, E.M.; Schlotzer-Schrehardt, U.; Coute, Y.; Plachter, B.; et al. The Complex Regulatory Role of Cytomegalovirus Nuclear Egress Protein pUL50 in the Production of Infectious Virus. Cells 2021, 10, 3119. [Google Scholar] [CrossRef] [PubMed]
- Milbradt, J.; Sonntag, E.; Wagner, S.; Strojan, H.; Wangen, C.; Lenac Rovis, T.; Lisnic, B.; Jonjic, S.; Sticht, H.; Britt, W.J.; et al. Human Cytomegalovirus Nuclear Capsids Associate with the Core Nuclear Egress Complex and the Viral Protein Kinase pUL97. Viruses 2018, 10, 35. [Google Scholar] [CrossRef] [PubMed]
- Webel, R.; Milbradt, J.; Auerochs, S.; Schregel, V.; Held, C.; Nobauer, K.; Razzazi-Fazeli, E.; Jardin, C.; Wittenberg, T.; Sticht, H.; et al. Two isoforms of the protein kinase pUL97 of human cytomegalovirus are differentially regulated in their nuclear translocation. J. Gen. Virol. 2011, 92, 638–649. [Google Scholar] [CrossRef]
- Dargan, D.J.; Douglas, E.; Cunningham, C.; Jamieson, F.; Stanton, R.J.; Baluchova, K.; McSharry, B.P.; Tomasec, P.; Emery, V.C.; Percivalle, E.; et al. Sequential mutations associated with adaptation of human cytomegalovirus to growth in cell culture. J. Gen. Virol. 2010, 91, 1535–1546. [Google Scholar] [CrossRef]
- Stanton, R.J.; Baluchova, K.; Dargan, D.J.; Cunningham, C.; Sheehy, O.; Seirafian, S.; McSharry, B.P.; Neale, M.L.; Davies, J.A.; Tomasec, P.; et al. Reconstruction of the complete human cytomegalovirus genome in a BAC reveals RL13 to be a potent inhibitor of replication. J. Clin. Investig. 2010, 120, 3191–3208. [Google Scholar] [CrossRef]
- Ourahmane, A.; Hertel, L.; McVoy, M.A. The RL13 Temperance Factor Represses Replication of the Highly Cell Culture-Adapted Towne Strain of Human Cytomegalovirus. Viruses 2023, 15, 1023. [Google Scholar] [CrossRef] [PubMed]
- Schultz, E.P.; Lanchy, J.M.; Day, L.Z.; Yu, Q.; Peterson, C.; Preece, J.; Ryckman, B.J. Specialization for Cell-Free or Cell-to-Cell Spread of BAC-Cloned Human Cytomegalovirus Strains Is Determined by Factors beyond the UL128-131 and RL13 Loci. J. Virol. 2020, 94, 10–128. [Google Scholar] [CrossRef] [PubMed]
- Weiler, N.; Laib Sampaio, K.; Stanton, R.J.; Sinzger, C. Combined knockdown of RL13 and UL128 for release of cell-free infectivity from recent HCMV isolates. J. Virol. Methods 2022, 305, 114537. [Google Scholar] [CrossRef] [PubMed]
- Krosky, P.M.; Baek, M.C.; Coen, D.M. The human cytomegalovirus UL97 protein kinase, an antiviral drug target, is required at the stage of nuclear egress. J. Virol. 2003, 77, 905–914. [Google Scholar] [CrossRef] [PubMed]
- Wild, M.; Karner, D.; Eickhoff, J.; Wagner, S.; Kicuntod, J.; Chang, W.; Barry, P.; Jonjic, S.; Lenac Rovis, T.; Marschall, M. Combined Treatment with Host-Directed and Anticytomegaloviral Kinase Inhibitors: Mechanisms, Synergisms and Drug Resistance Barriers. Pharmaceutics 2023, 15, 2680. [Google Scholar] [CrossRef]
- Alkhashrom, S.; Kicuntod, J.; Hage, S.; Schweininger, J.; Muller, Y.A.; Lischka, P.; Marschall, M.; Eichler, J. Exploring the Human Cytomegalovirus Core Nuclear Egress Complex as a Novel Antiviral Target: A New Type of Small Molecule Inhibitors. Viruses 2021, 13, 471. [Google Scholar] [CrossRef] [PubMed]
- Kicuntod, J.; Alkhashrom, S.; Hage, S.; Diewald, B.; Muller, R.; Hahn, F.; Lischka, P.; Sticht, H.; Eichler, J.; Marschall, M. Properties of Oligomeric Interaction of the Cytomegalovirus Core Nuclear Egress Complex (NEC) and Its Sensitivity to an NEC Inhibitory Small Molecule. Viruses 2021, 13, 462. [Google Scholar] [CrossRef] [PubMed]
- Wild, M.; Hahn, F.; Bruckner, N.; Schutz, M.; Wangen, C.; Wagner, S.; Sommerer, M.; Strobl, S.; Marschall, M. Cyclin-Dependent Kinases (CDKs) and the Human Cytomegalovirus-Encoded CDK Ortholog pUL97 Represent Highly Attractive Targets for Synergistic Drug Combinations. Int. J. Mol. Sci. 2022, 23, 2493. [Google Scholar] [CrossRef]
- Klenovsek, K.; Weisel, F.; Schneider, A.; Appelt, U.; Jonjic, S.; Messerle, M.; Bradel-Tretheway, B.; Winkler, T.H.; Mach, M. Protection from CMV infection in immunodeficient hosts by adoptive transfer of memory B cells. Blood 2007, 110, 3472–3479. [Google Scholar] [CrossRef]
- Sonntag, E.; Hahn, F.; Bertzbach, L.D.; Seyler, L.; Wangen, C.; Muller, R.; Tannig, P.; Grau, B.; Baumann, M.; Zent, E.; et al. In vivo proof-of-concept for two experimental antiviral drugs, both directed to cellular targets, using a murine cytomegalovirus model. Antivir. Res. 2019, 161, 63–69. [Google Scholar] [CrossRef]
- Wild, M.; Bertzbach, L.D.; Tannig, P.; Wangen, C.; Muller, R.; Herrmann, L.; Frohlich, T.; Tsogoeva, S.B.; Kaufer, B.B.; Marschall, M.; et al. The trimeric artesunate derivative TF27 exerts strong anti-cytomegaloviral efficacy: Focus on prophylactic efficacy and oral treatment of immunocompetent mice. Antivir. Res. 2020, 178, 104788. [Google Scholar] [CrossRef] [PubMed]
- Maier, A.K.; Jung, R.; Villinger, C.; Schubert, A.; Walther, P.; Sinzger, C.; Lieber, D. A Luciferase Gene Driven by an Alphaherpesviral Promoter Also Responds to Immediate Early Antigens of the Betaherpesvirus HCMV, Allowing Comparative Analyses of Different Human Herpesviruses in One Reporter Cell Line. PLoS ONE 2017, 12, e0169580. [Google Scholar] [CrossRef] [PubMed]
- Reinhard, H.; Le, V.T.; Ohlin, M.; Hengel, H.; Trilling, M. Exploitation of herpesviral transactivation allows quantitative reporter gene-based assessment of virus entry and neutralization. PLoS ONE 2011, 6, e14532. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Wagner, S.; Schutz, M.; Jeon, Y.; Seo, M.; Kim, J.; Bruckner, N.; Kicuntod, J.; Tillmanns, J.; Wangen, C.; et al. An Antiherpesviral Host-Directed Strategy Based on CDK7 Covalently Binding Drugs: Target-Selective, Picomolar-Dose, Cross-Virus Reactivity. Pharmaceutics 2024, 16, 158. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tillmanns, J.; Kicuntod, J.; Ehring, A.; Elbasani, E.; Borst, E.M.; Obergfäll, D.; Müller, R.; Hahn, F.; Marschall, M. Establishment of a Luciferase-Based Reporter System to Study Aspects of Human Cytomegalovirus Infection, Replication Characteristics, and Antiviral Drug Efficacy. Pathogens 2024, 13, 645. https://doi.org/10.3390/pathogens13080645
Tillmanns J, Kicuntod J, Ehring A, Elbasani E, Borst EM, Obergfäll D, Müller R, Hahn F, Marschall M. Establishment of a Luciferase-Based Reporter System to Study Aspects of Human Cytomegalovirus Infection, Replication Characteristics, and Antiviral Drug Efficacy. Pathogens. 2024; 13(8):645. https://doi.org/10.3390/pathogens13080645
Chicago/Turabian StyleTillmanns, Julia, Jintawee Kicuntod, Antonia Ehring, Endrit Elbasani, Eva Maria Borst, Debora Obergfäll, Regina Müller, Friedrich Hahn, and Manfred Marschall. 2024. "Establishment of a Luciferase-Based Reporter System to Study Aspects of Human Cytomegalovirus Infection, Replication Characteristics, and Antiviral Drug Efficacy" Pathogens 13, no. 8: 645. https://doi.org/10.3390/pathogens13080645
APA StyleTillmanns, J., Kicuntod, J., Ehring, A., Elbasani, E., Borst, E. M., Obergfäll, D., Müller, R., Hahn, F., & Marschall, M. (2024). Establishment of a Luciferase-Based Reporter System to Study Aspects of Human Cytomegalovirus Infection, Replication Characteristics, and Antiviral Drug Efficacy. Pathogens, 13(8), 645. https://doi.org/10.3390/pathogens13080645






