The Non-Histone Protein FgNhp6 Is Involved in the Regulation of the Development, DON Biosynthesis, and Virulence of Fusarium graminearum
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Protein Identification and Sequence Analysis
2.3. Construction of FgNhp6 Deletion Mutants and Complementary Strains
2.4. Subcellular Localization of FgNhp6
2.5. Evaluation of Conidial Production and Sexual Development
2.6. Pathogenicity Assays
2.7. Assessment of DON Production
2.8. RNA Isolation and RNA-Sequencing Analysis
2.9. qRT-PCR Assays
2.10. Statistical Analyses
3. Results
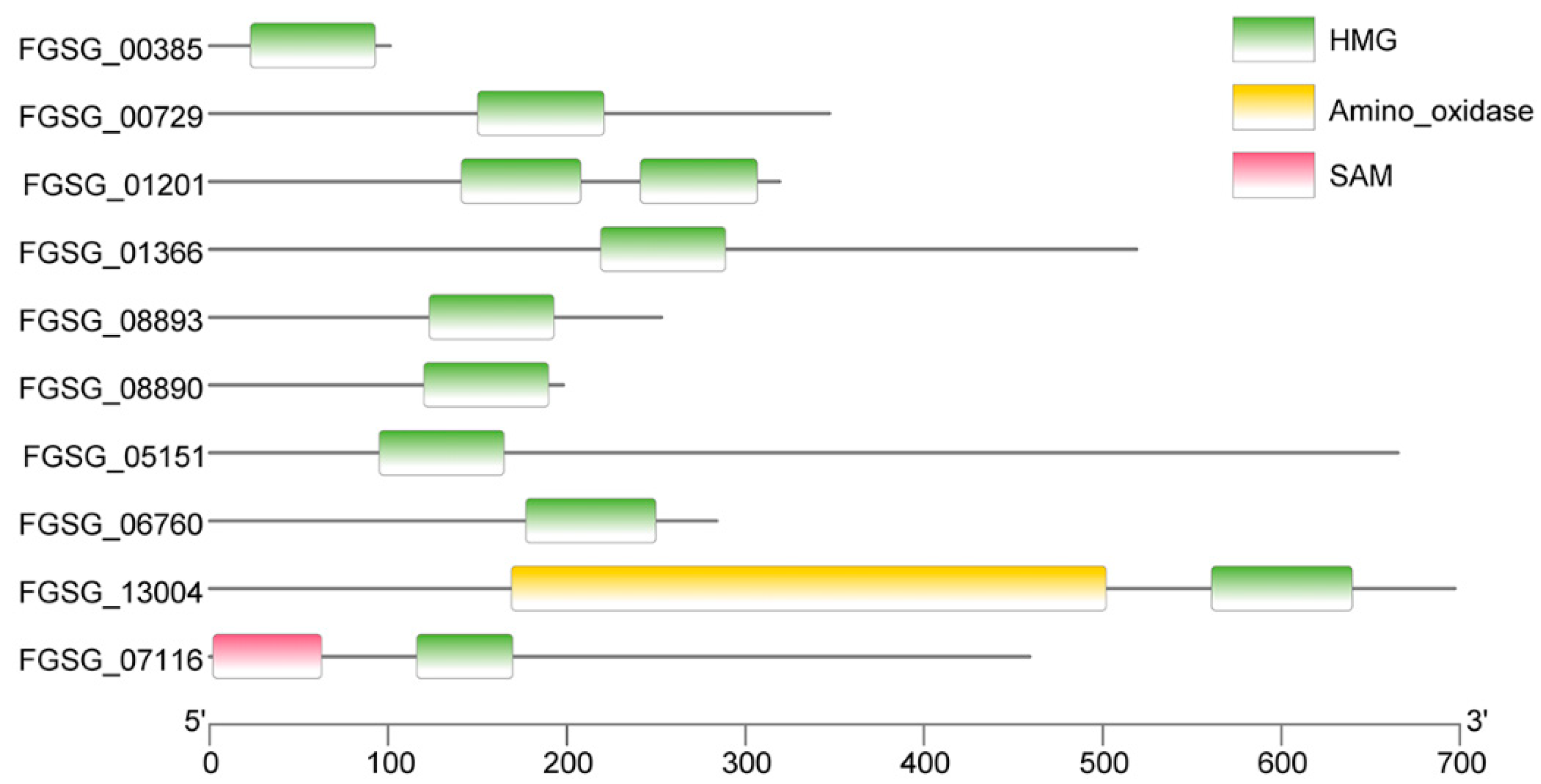
3.1. Identification of the HMG-box Containing Proteins in F. graminearum
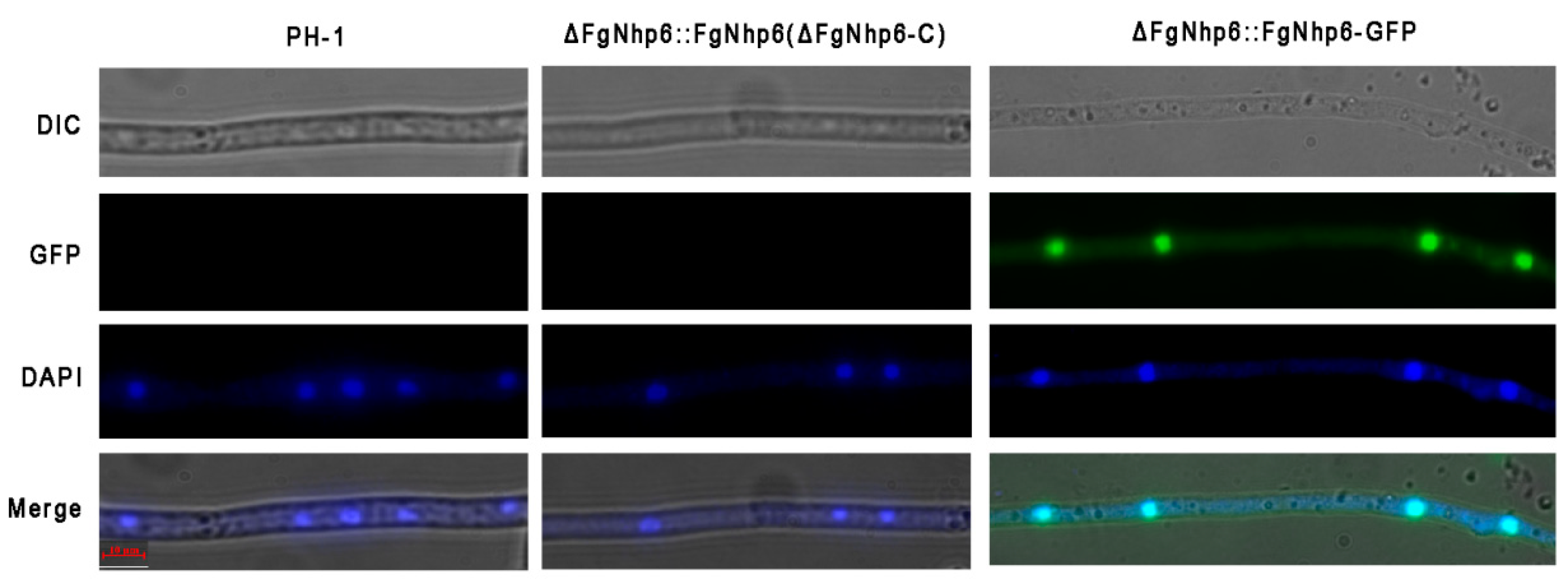
3.2. FgNhp6 Localizes to the Nucleus
3.3. FgNhp6 Is Involved in Vegetative Growth and Pigmentation in F. graminearum
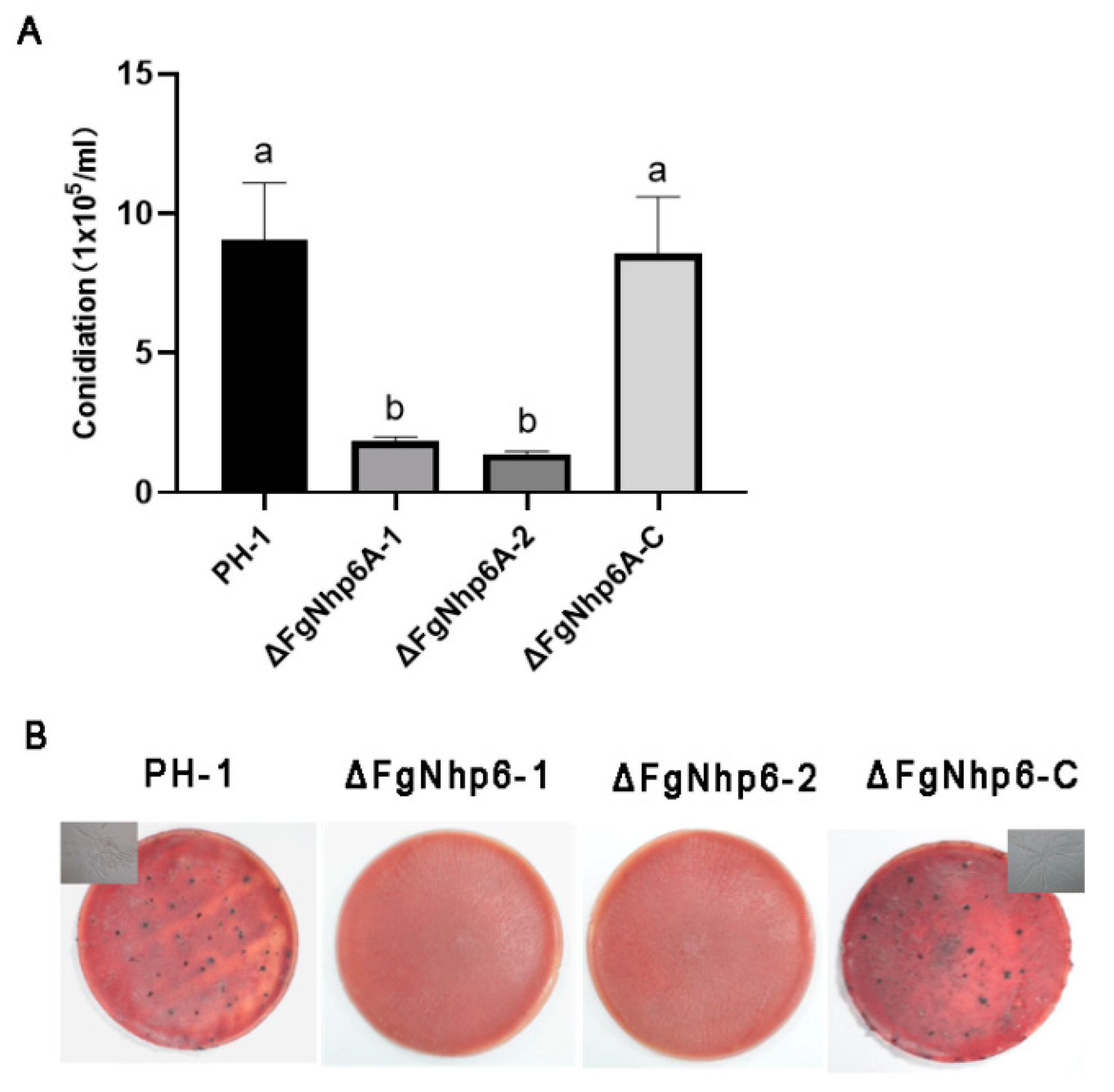
3.4. FgNhp6 Played a Critical Role in Both Asexual and Sexual Developmental Phases
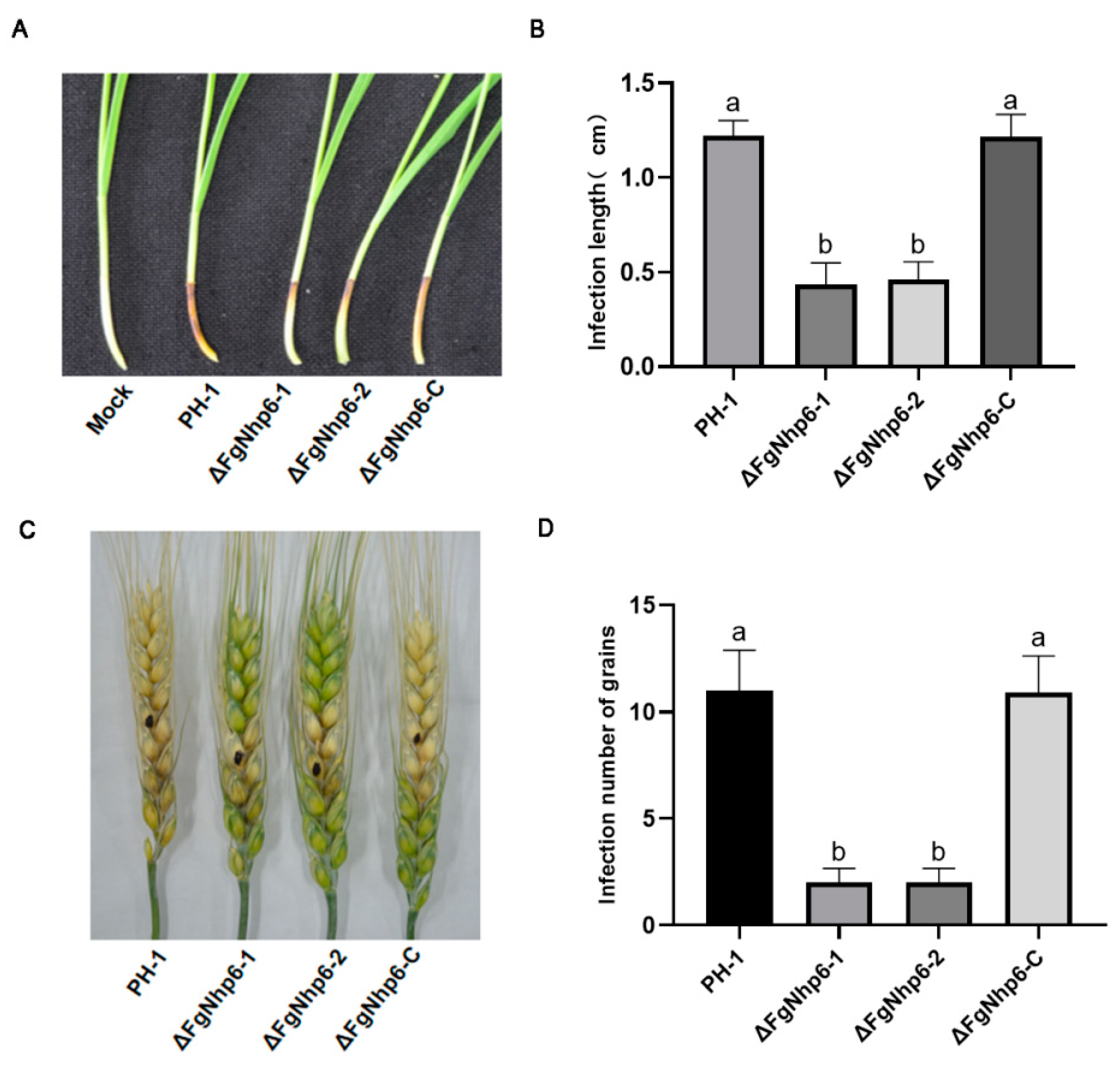
3.5. ΔFgNhp6 Is Indispensable for Full Virulence in F. graminearum
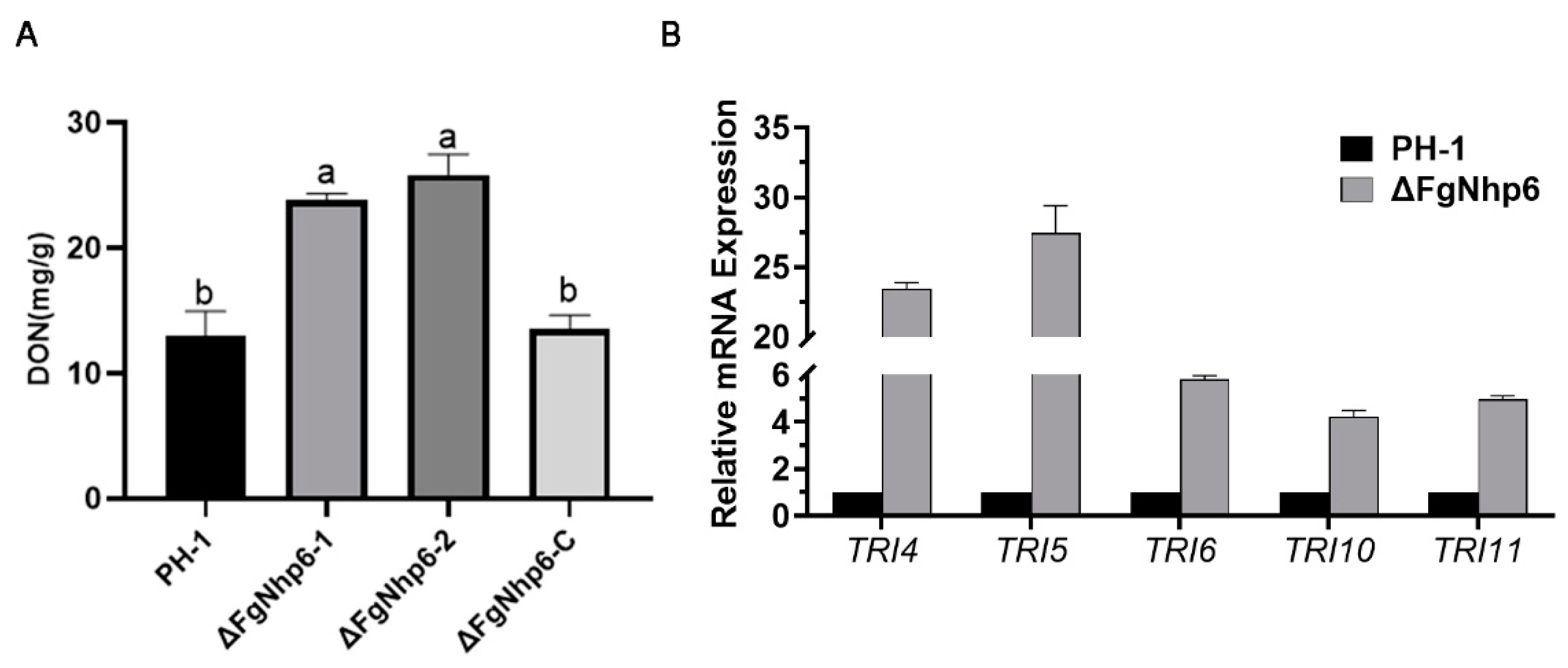
3.6. FgNhp6 Negatively Regulated the DON Biosynthesis
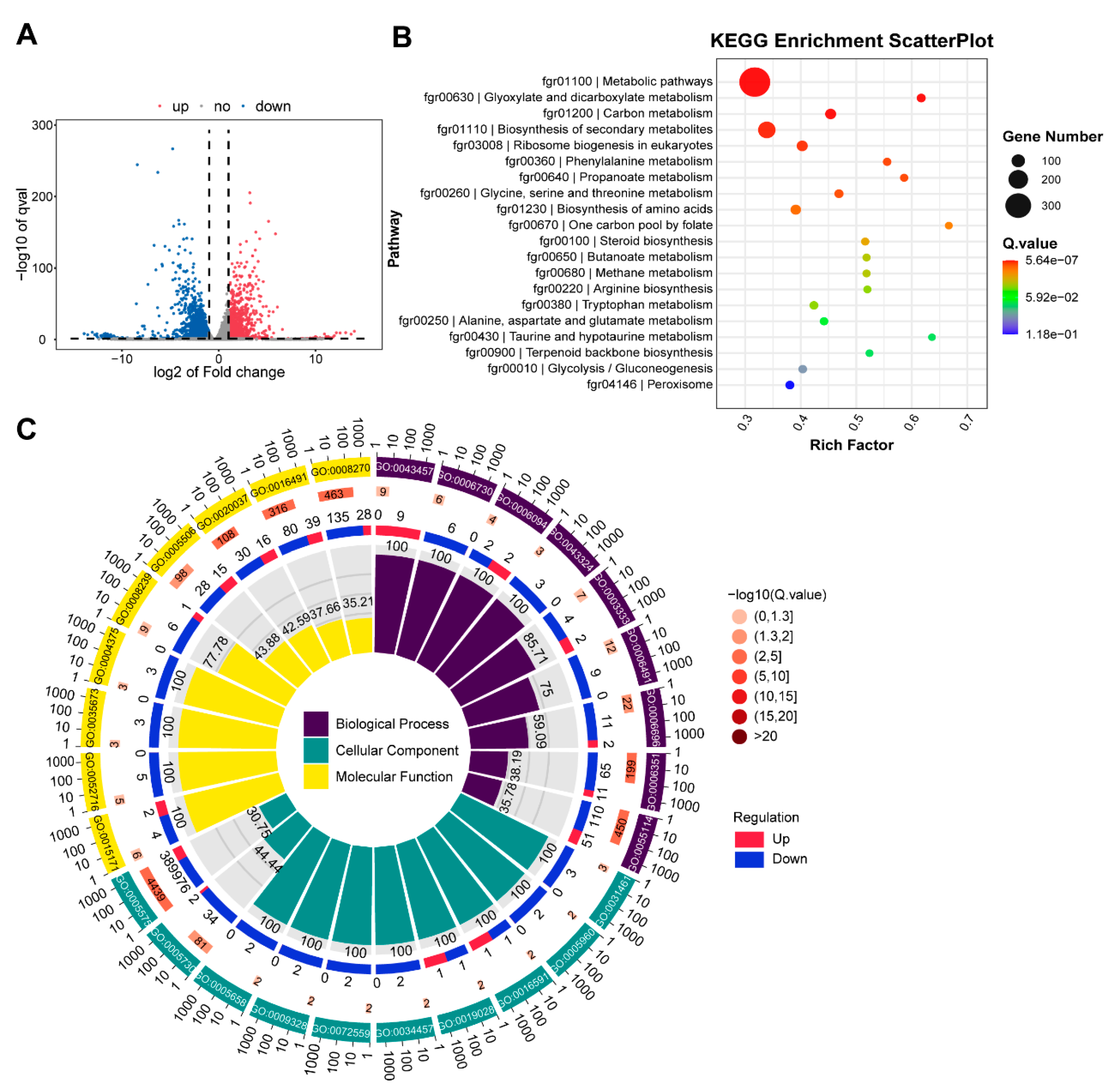
3.7. RNA Seq Analysis with ∆FgNhp6 Mutant
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, A.; Islam, T.; Ma, Z. An Integrated Pest Management Program for Managing Fusarium Head Blight Disease in Cereals. J. Integr. Agric. 2022, 21, 3434–3444. [Google Scholar] [CrossRef]
- Leplat, J.; Friberg, H.; Abid, M.; Steinberg, C. Survival of Fusarium graminearum, the Causal Agent of Fusarium Head Blight. A Review. Agron. Sustain. Dev. 2013, 33, 97–111. [Google Scholar] [CrossRef]
- Chen, Y.; Kistler, H.C.; Ma, Z. Fusarium graminearum Trichothecene Mycotoxins: Biosynthesis, Regulation, and Management. Annu. Rev. Phytopathol. 2019, 57, 15–39. [Google Scholar] [CrossRef] [PubMed]
- Dweba, C.C.; Figlan, S.; Shimelis, H.A.; Motaung, T.E.; Sydenham, S.; Mwadzingeni, L.; Tsilo, T.J. Fusarium Head Blight of Wheat: Pathogenesis and Control Strategies. Crop Prot. 2017, 91, 114–122. [Google Scholar] [CrossRef]
- Bustin, M. Revised Nomenclature for High Mobility Group (HMG) Chromosomal Proteins. Trends Biochem. Sci. 2001, 26, 152–153. [Google Scholar] [CrossRef] [PubMed]
- Reeves, R. Molecular Biology of HMGA Proteins: Hubs of Nuclear Function. Gene 2001, 277, 63–81. [Google Scholar] [CrossRef] [PubMed]
- Nanduri, R.; Furusawa, T.; Bustin, M. Biological Functions of HMGN Chromosomal Proteins. Int. J. Mol. Sci. 2020, 21, 449. [Google Scholar] [CrossRef] [PubMed]
- Hardman, C.H.; Broadhurst, R.W.; Raine, A.R.C.; Grasser, K.D.; Thomas, J.O.; Laue, E.D. Structure of the A-Domain of HMG1 and Its Interaction with DNA as Studied by Heteronuclear Three- and Four-Dimensional NMR Spectroscopy. Biochemistry 1995, 34, 16596–16607. [Google Scholar] [CrossRef] [PubMed]
- Masse, J.E.; Wong, B.; Yen, Y.-M.; Allain, F.H.-T.; Johnson, R.C.; Feigon, J. The S. cerevisiae Architectural HMGB Protein NHP6A Complexed with DNA: DNA and Protein Conformational Changes upon Binding. J. Mol. Biol. 2002, 323, 263–284. [Google Scholar] [CrossRef]
- Kastaniotis, A.J.; Zitomer, R.S. Rox1 Medited Repression. In Oxygen Sensing: Molecule to Man; Lahiri, S., Prabhakar, N.R., Forster, R.E., Eds.; Springer: Boston, MA, USA, 2002; pp. 185–195. ISBN 978-0-306-46825-4. [Google Scholar]
- Vizoso-Vázquez, A.; Lamas-Maceiras, M.; Fernández-Leiro, R.; Rico-Díaz, A.; Becerra, M.; Cerdán, M.E. Dual Function of Ixr1 in Transcriptional Regulation and Recognition of Cisplatin-DNA Adducts Is Caused by Differential Binding through Its Two HMG-Boxes. Biochim. Biophys. Acta BBA-Gene Regul. Mech. 2017, 1860, 256–269. [Google Scholar] [CrossRef]
- Ángel, V.-V.; Aida, B.-A.; Agustín, R.-D.; Mónica, L.-M.; Esther, R.-B.; Manuel, B.; Isabel, G.-S.M.; Esperanza, C.M.; Ángel, V.-V.; Aida, B.-A.; et al. HMGB Proteins from Yeast to Human. Gene Regulation, DNA Repair and Beyond. In Old Yeasts-New Questions; IntechOpen: London, UK, 2017; ISBN 978-953-51-3678-1. [Google Scholar]
- Kolodrubetz, D.; Burgum, A. Duplicated NHP6 Genes of Saccharomyces cerevisiae Encode Proteins Homologous to Bovine High Mobility Group Protein 1. J. Biol. Chem. 1990, 265, 3234–3239. [Google Scholar] [CrossRef] [PubMed]
- Biswas, D.; Imbalzano, A.N.; Eriksson, P.; Yu, Y.; Stillman, D.J. Role for Nhp6, Gcn5, and the Swi/Snf Complex in Stimulating Formation of the TATA-Binding Protein-TFIIA-DNA Complex. Mol. Cell. Biol. 2004, 24, 8312–8321. [Google Scholar] [CrossRef] [PubMed]
- Moreira, J.M.A.; Holmberg, S. Chromatin-Mediated Transcriptional Regulation by the Yeast Architectural Factors NHP6A and NHP6B. EMBO J. 2000, 19, 6804–6813. [Google Scholar] [CrossRef] [PubMed]
- Paull, T.T.; Carey, M.; Johnson, R.C. Yeast HMG Proteins NHP6A/B Potentiate Promoter-Specific Transcriptional Activation in Vivo and Assembly of Preinitiation Complexes in Vitro. Genes Dev. 1996, 10, 2769–2781. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Eriksson, P.; Bhoite, L.T.; Stillman, D.J. Regulation of TATA-Binding Protein Binding by the SAGA Complex and the Nhp6 High-Mobility Group Protein. Mol. Cell. Biol. 2003, 23, 1910–1921. [Google Scholar] [CrossRef] [PubMed]
- Rhoades, A.R.; Ruone, S.; Formosa, T. Structural Features of Nucleosomes Reorganized by Yeast FACT and Its HMG Box Component, Nhp6. Mol. Cell. Biol. 2004, 24, 3907–3917. [Google Scholar] [CrossRef] [PubMed]
- Kruppa, M.; Moir, R.D.; Kolodrubetz, D.; Willis, I.M. Nhp6, an HMG1 Protein, Functions in SNR6 Transcription by RNA Polymerase III in S. cerevisiae. Mol. Cell 2001, 7, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Lopez, S.; Livingstone-Zatchej, M.; Jourdain, S.; Thoma, F.; Sentenac, A.; Marsolier, M.-C. High-Mobility-Group Proteins NHP6A and NHP6B Participate in Activation of the RNA Polymerase III SNR6 Gene. Mol. Cell. Biol. 2001, 21, 3096–3104. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Costigan, C.; Kolodrubetz, D.; Snyder, M. NHP6A and NHP6B, Which Encode HMG1-like Proteins, Are Candidates for Downstream Components of the Yeast SLT2 Mitogen-Activated Protein Kinase Pathway. Mol. Cell. Biol. 1994, 14, 2391–2403. [Google Scholar] [CrossRef] [PubMed]
- Stillman, D.J. Nhp6: A Small but Powerful Effector of Chromatin Structure in Saccharomyces cerevisiae. Biochim. Biophys. Acta BBA-Gene Regul. Mech. 2010, 1799, 175–180. [Google Scholar] [CrossRef]
- Lu, J.-P.; Feng, X.-X.; Liu, X.-H.; Lu, Q.; Wang, H.-K.; Lin, F.-C. Mnh6, a Nonhistone Protein, Is Required for Fungal Development and Pathogenicity of Magnaporthe grisea. Fungal Genet. Biol. 2007, 44, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER Web Server: 2018 Update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent Updates, New Developments and Status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-Data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Catlett, N.L.; Lee, B.-N.; Yoder, O.C.; Turgeon, B.G. Split-Marker Recombination for Efficient Targeted Deletion of Fungal Genes. Fungal Genet. Rep. 2003, 50, 9–11. [Google Scholar] [CrossRef]
- Zhao, Y.; Sun, H.; Li, J.; Ju, C.; Huang, J. The Transcription Factor FgAtrR Regulates Asexual and Sexual Development, Virulence, and DON Production and Contributes to Intrinsic Resistance to Azole Fungicides in Fusarium graminearum. Biology 2022, 11, 326. [Google Scholar] [CrossRef] [PubMed]
- Menke, J.; Dong, Y.; Kistler, H.C. Fusarium graminearum Tri12p Influences Virulence to Wheat and Trichothecene Accumulation. Mol. Plant. Microbe Interact. 2012, 25, 1408–1418. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- The Gene Ontology Consortium. The Gene Ontology Resource: Enriching a GOld Mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating Viruses and Cellular Organisms. Nucleic Acids Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, C.; Wang, L.; Sun, S.; Liu, A.; Liang, Y.; Yu, J.; Dong, H. FgPEX1 and FgPEX10 Are Required for the Maintenance of Woronin Bodies and Full Virulence of Fusarium graminearum. Curr. Genet. 2019, 65, 1383–1396. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.-B.; Li, H.-P.; Zhang, J.-B.; Song, B.; Chen, F.-F.; Duan, X.-J.; Xu, H.-Q.; Liao, Y.-C. Disruption of the Chitin Synthase Gene CHS1 from Fusarium asiaticum Results in an Altered Structure of Cell Walls and Reduced Virulence. Fungal Genet. Biol. 2010, 47, 205–215. [Google Scholar] [CrossRef]
- Fan, J.; Urban, M.; Parker, J.E.; Brewer, H.C.; Kelly, S.L.; Hammond-Kosack, K.E.; Fraaije, B.A.; Liu, X.; Cools, H.J. Characterization of the Sterol 14α-Demethylases of Fusarium graminearum Identifies a Novel Genus-Specific CYP51 Function. New Phytol. 2013, 198, 821–835. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Jiang, J.; Yin, Y.; Ma, Z. Involvement of FgERG4 in Ergosterol Biosynthesis, Vegetative Differentiation and Virulence in Fusarium graminearum. Mol. Plant Pathol. 2013, 14, 71–83. [Google Scholar] [CrossRef]
- Frandsen, R.J.N.; Nielsen, N.J.; Maolanon, N.; Sørensen, J.C.; Olsson, S.; Nielsen, J.; Giese, H. The Biosynthetic Pathway for Aurofusarin in Fusarium graminearum Reveals a Close Link between the Naphthoquinones and Naphthopyrones. Mol. Microbiol. 2006, 61, 1069–1080. [Google Scholar] [CrossRef]
- Becher, R.; Weihmann, F.; Deising, H.B.; Wirsel, S.G. Development of a Novel Multiplex DNA Microarray for Fusarium graminearum and Analysis of Azole Fungicide Responses. BMC Genom. 2011, 12, 52. [Google Scholar] [CrossRef]
- Ma, T.; Li, Y.; Lou, Y.; Shi, J.; Sun, K.; Ma, Z.; Yan, L.; Yin, Y. The Drug H+ Antiporter FgQdr2 Is Essential for Multiple Drug Resistance, Ion Homeostasis, and Pathogenicity in Fusarium graminearum. J. Fungi 2022, 8, 1009. [Google Scholar] [CrossRef] [PubMed]
- Riedelberger, M.; Penninger, P.; Tscherner, M.; Seifert, M.; Jenull, S.; Brunnhofer, C.; Scheidl, B.; Tsymala, I.; Bourgeois, C.; Petryshyn, A. Type I Interferon Response Dysregulates Host Iron Homeostasis and Enhances Candida glabrata Infection. Cell Host Microbe 2020, 27, 454–466. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Tong, Q.; Zhang, C.; Ding, K. The Transcription Factor FgCrz1A Is Essential for Fungal Development, Virulence, Deoxynivalenol Biosynthesis and Stress Responses in Fusarium graminearum. Curr. Genet. 2019, 65, 153–166. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, X.; Liu, X.; Fu, C.; Han, X.; Yin, Y.; Ma, Z. The Chitin Synthase FgChs2 and Other FgChss Co-Regulate Vegetative Development and Virulence in F. graminearum. Sci. Rep. 2016, 6, 34975. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Hu, J.; Liu, Z.; Liang, W.; Song, L. Sir2-mediated Cytoplasmic Deacetylation Facilitates Pathogenic Fungi Infection in Host Plants. New Phytol. 2024, 241, 1732–1746. [Google Scholar] [CrossRef] [PubMed]
- Lange, S.S.; Mitchell, D.L.; Vasquez, K.M. High Mobility Group Protein B1 Enhances DNA Repair and Chromatin Modification after DNA Damage. Proc. Natl. Acad. Sci. USA 2008, 105, 10320–10325. [Google Scholar] [CrossRef] [PubMed]
- Reeves, R. High Mobility Group (HMG) Proteins: Modulators of Chromatin Structure and DNA Repair in Mammalian Cells. DNA Repair 2015, 36, 122–136. [Google Scholar] [CrossRef] [PubMed]
- Ueda, T.; Yoshida, M. HMGB Proteins and Transcriptional Regulation. Biochim. Biophys. Acta BBA-Gene Regul. Mech. 2010, 1799, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.-A.; Wang, G.-H.; Chen, A.-L.; Li, Y.-F.; Liu, J.-P.; Li, Y.-Y.; Bolotin-Fukuhara, M.; Bao, W.-G. Gene Responses to Oxygen Availability in Kluyveromyces lactis: An Insight on the Evolution of the Oxygen-Responding System in Yeast. PLoS ONE 2009, 4, e7561. [Google Scholar] [CrossRef] [PubMed]
- Miyakawa, I.; Okamuro, A.; Kinsky, S.; Visacka, K.; Tomaska, L.; Nosek, J. Mitochondrial Nucleoids from the Yeast Candida parapsilosis: Expansion of the Repertoire of Proteins Associated with Mitochondrial DNA. Microbiology 2009, 155, 1558–1568. [Google Scholar] [CrossRef]
- Visacka, K.; Gerhold, J.M.; Petrovicova, J.; Kinsky, S.; Jõers, P.; Nosek, J.; Sedman, J.; Tomaska, L. Novel Subfamily of Mitochondrial HMG Box-Containing Proteins: Functional Analysis of Gcf1p from Candida albicans. Microbiology 2009, 155, 1226–1240. [Google Scholar] [CrossRef] [PubMed]
- Benkhali, J.A.; Coppin, E.; Brun, S.; Peraza-Reyes, L.; Martin, T.; Dixelius, C.; Lazar, N.; van Tilbeurgh, H.; Debuchy, R. A Network of HMG-Box Transcription Factors Regulates Sexual Cycle in the Fungus Podospora anserina. PLoS Genet. 2013, 9, e1003642. [Google Scholar] [CrossRef] [PubMed]
- Ámon, J.; Varga, G.; Pfeiffer, I.; Farkas, Z.; Karácsony, Z.; Hegedűs, Z.; Vágvölgyi, C.; Hamari, Z. The Role of the Aspergillus nidulans High Mobility Group B Protein HmbA, the Orthologue of Saccharomyces cerevisiae Nhp6p. Sci. Rep. 2022, 12, 17334. [Google Scholar] [CrossRef] [PubMed]
- Bokor, E.; Ámon, J.; Keisham, K.; Karácsony, Z.; Vágvölgyi, C.; Hamari, Z. HMGB Proteins Are Required for Sexual Development in Aspergillus nidulans. PLoS ONE 2019, 14, e0216094. [Google Scholar] [CrossRef] [PubMed]
- Karácsony, Z.; Gácser, A.; Vágvölgyi, C.; Scazzocchio, C.; Hamari, Z. A Dually Located Multi-HMG-Box Protein of Aspergillus nidulans Has a Crucial Role in Conidial and Ascospore Germination. Mol. Microbiol. 2014, 94, 383–402. [Google Scholar] [CrossRef] [PubMed]
- Franco-Losilla, M.; Nordzieke, S.; Feldmann, I.; Limón, M.C.; Avalos, J. HmbC, a Protein of the HMG Family, Participates in the Regulation of Carotenoid Biosynthesis in Fusarium fujikuroi. Genes 2023, 14, 1661. [Google Scholar] [CrossRef] [PubMed]
- Perincherry, L.; Lalak-Kańczugowska, J.; Stępień, Ł. Fusarium-Produced Mycotoxins in Plant-Pathogen Interactions. Toxins 2019, 11, 664. [Google Scholar] [CrossRef]
- Hou, S.; Ma, J.; Cheng, Y.; Wang, H.; Sun, J.; Yan, Y. The Toxicity Mechanisms of DON to Humans and Animals and Potential Biological Treatment Strategies. Crit. Rev. Food Sci. Nutr. 2023, 63, 790–812. [Google Scholar] [CrossRef] [PubMed]
- Rocha, O.; Ansari, K.; Doohan, F.M. Effects of Trichothecene Mycotoxins on Eukaryotic Cells: A Review. Food Addit. Contam. 2005, 22, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Desjardins, A.E.; Proctor, R.H. Molecular Biology of Fusarium Mycotoxins. Int. J. Food Microbiol. 2007, 119, 47–50. [Google Scholar] [CrossRef]
- Proctor, R.H.; Hohn, T.M.; McCormick, S.P. Reduced Virulence of Gibberella zeae Caused by Disruption of a Trichothecene Toxin Biosynthetic Gene. Mol. Plant-Microbe Interact. MPMI 1995, 8, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, D.M.; Kazan, K.; Manners, J.M. Nutrient Profiling Reveals Potent Inducers of Trichothecene Biosynthesis in Fusarium graminearum. Fungal Genet. Biol. 2009, 46, 604–613. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, D.M.; Kazan, K.; Praud, S.; Torney, F.J.; Rusu, A.; Manners, J.M. Early Activation of Wheat Polyamine Biosynthesis during Fusarium Head Blight Implicates Putrescine as an Inducer of Trichothecene Mycotoxin Production. BMC Plant Biol. 2010, 10, 289. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Guo, Z.; Zhang, M.; Chen, H.; Peng, M.; Abubakar, Y.S.; Zheng, H.; Yun, Y.; Zheng, W.; Wang, Z.; et al. Golgi-localized Calcium/Manganese Transporters FgGdt1 and FgPmr1 Regulate Fungal Development and Virulence by Maintaining Ca2+ and Mn2+ Homeostasis in Fusarium graminearum. Environ. Microbiol. 2022, 24, 4623–4640. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Yu, Z.; Yuan, Y.; Sun, D.; Abubakar, Y.S.; Zhou, J.; Wang, Z.; Zheng, H. The GTPase-Activating Protein FgGyp1 Is Important for Vegetative Growth, Conidiation, and Virulence and Negatively Regulates DON Biosynthesis in Fusarium graminearium. Front. Microbiol. 2021, 12, 621519. [Google Scholar] [CrossRef]
- Shi, D.; Zhang, Y.; Wang, J.; Ren, W.; Zhang, J.; Mbadianya, J.I.; Zhu, Y.; Chen, C.; Ma, H. S-Adenosyl-L-Homocysteine Hydrolase FgSah1 Is Required for Fungal Development and Virulence in Fusarium graminearum. Virulence 2021, 12, 2171–2185. [Google Scholar] [CrossRef] [PubMed]
- Bui, D.-C.; Son, H.; Shin, J.Y.; Kim, J.-C.; Kim, H.; Choi, G.J.; Lee, Y.-W. The FgNot3 Subunit of the Ccr4-Not Complex Regulates Vegetative Growth, Sporulation, and Virulence in Fusarium graminearum. PLoS ONE 2016, 11, e0147481. [Google Scholar] [CrossRef]
- Zhang, N.; Wang, S.; Tian, H.; Li, S.; Liu, L.; Li, J.; Chen, D.; Zhao, S.; Yan, X.; Niaz, M.; et al. Functions of Lysine 2-hydroxyisobutyrylation and Future Perspectives on Plants. Proteomics 2023, 23, 2300045. [Google Scholar] [CrossRef]

| Protein Name | Gene Locus | Log2FC (RNA-seq) | Log2FC (qPCR) | Reference |
|---|---|---|---|---|
| Sterol-biosynthesis-related genes | ||||
| FgCYP51A | FGSG_04092 | −6.30 | −4.54 | [39] |
| FgCYP51B | FGSG_01000 | −1.69 | −1.43 | [39] |
| FgERG9 | FGSG_09381 | −2.76 | −2.87 | [40] |
| FgCYP51C | FGSG_11024 | −0.48 | −0.48 | [39] |
| Aurofusarin biosynthesis gene cluster | ||||
| GIP1 | FGSG_02328 | −3.36 | −3.06 | [41] |
| AurJ | FGSG_02326 | −3.34 | −2.90 | [41] |
| AurF | FGSG_02327 | −3.36 | −3.49 | [41] |
| PKS12 | FGSG_02324 | −4.18 | −4.45 | [41] |
| Transporters | ||||
| FGSG_08308 | −2.11 | −2.15 | [42] | |
| FGSG_08309 | −2.79 | −2.61 | [42] | |
| FGSG_08749 | −2.42 | −3.10 | [43] | |
| FGSG_03882 | 5.85 | 5.00 | [42] | |
| FGSG_05096 | 2.21 | 1.72 | [44] | |
| Other genes related to virulence | ||||
| FgPex1 | FGSG_07104 | −1.75 | −2.11 | [37] |
| FgCrz1A | FGSG_13711 | −1.65 | −1.30 | [45] |
| FgChs1 | FGSG_10327 | −1.77 | −1.71 | [38] |
| FgChs2 | FGSG_02483 | −1.53 | −1.74 | [46] |
| FgSIRT2 | FGSG_09218 | −1.40 | −1.42 | [47] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, J.; Lv, J.; Zhang, L.; Li, H.; Ma, H.; Zhao, Y.; Huang, J. The Non-Histone Protein FgNhp6 Is Involved in the Regulation of the Development, DON Biosynthesis, and Virulence of Fusarium graminearum. Pathogens 2024, 13, 592. https://doi.org/10.3390/pathogens13070592
Cao J, Lv J, Zhang L, Li H, Ma H, Zhao Y, Huang J. The Non-Histone Protein FgNhp6 Is Involved in the Regulation of the Development, DON Biosynthesis, and Virulence of Fusarium graminearum. Pathogens. 2024; 13(7):592. https://doi.org/10.3390/pathogens13070592
Chicago/Turabian StyleCao, Jiakuo, Junbo Lv, Limin Zhang, Heng Li, Hao Ma, Yanxiang Zhao, and Jinguang Huang. 2024. "The Non-Histone Protein FgNhp6 Is Involved in the Regulation of the Development, DON Biosynthesis, and Virulence of Fusarium graminearum" Pathogens 13, no. 7: 592. https://doi.org/10.3390/pathogens13070592
APA StyleCao, J., Lv, J., Zhang, L., Li, H., Ma, H., Zhao, Y., & Huang, J. (2024). The Non-Histone Protein FgNhp6 Is Involved in the Regulation of the Development, DON Biosynthesis, and Virulence of Fusarium graminearum. Pathogens, 13(7), 592. https://doi.org/10.3390/pathogens13070592






