A Rapid and Inexpensive PCR Test for Mastitis Diagnosis Based on NGS Data
Abstract
1. Introduction
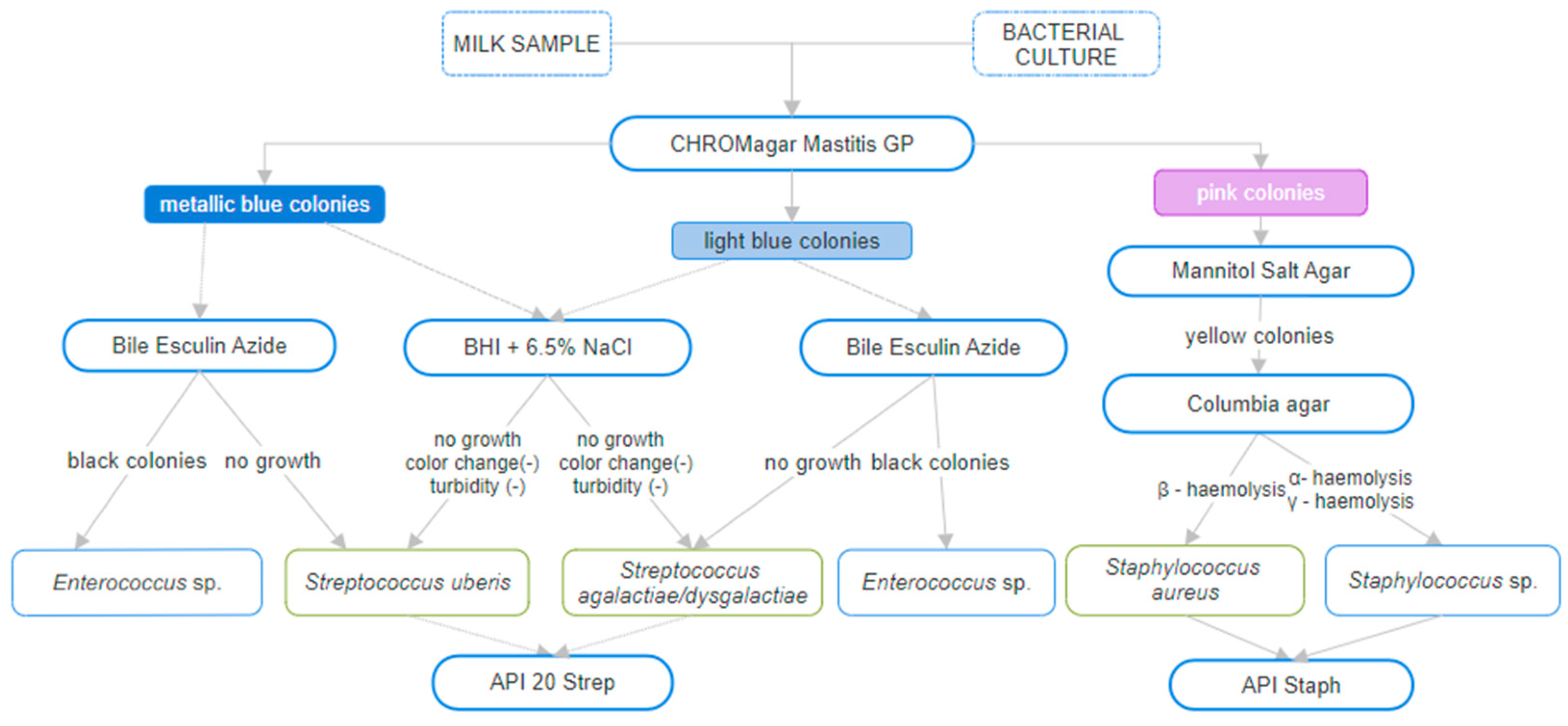
2. Materials and Methods
2.1. Bacterial Strains
2.2. Extraction of Bacterial Genomic DNA
2.3. Next Generation Sequencing
2.4. Bioinformatic Analysis of NGS Data
2.5. Multiplex PCR Primer Design
2.6. Single PCR Conditions
2.7. Multiplex PCR Conditions
3. Results
3.1. Taxonomy Analysis of NGS Data Analysed Bacterial Strains
3.2. Primer Design
3.3. Establishment of Single PCR Conditions
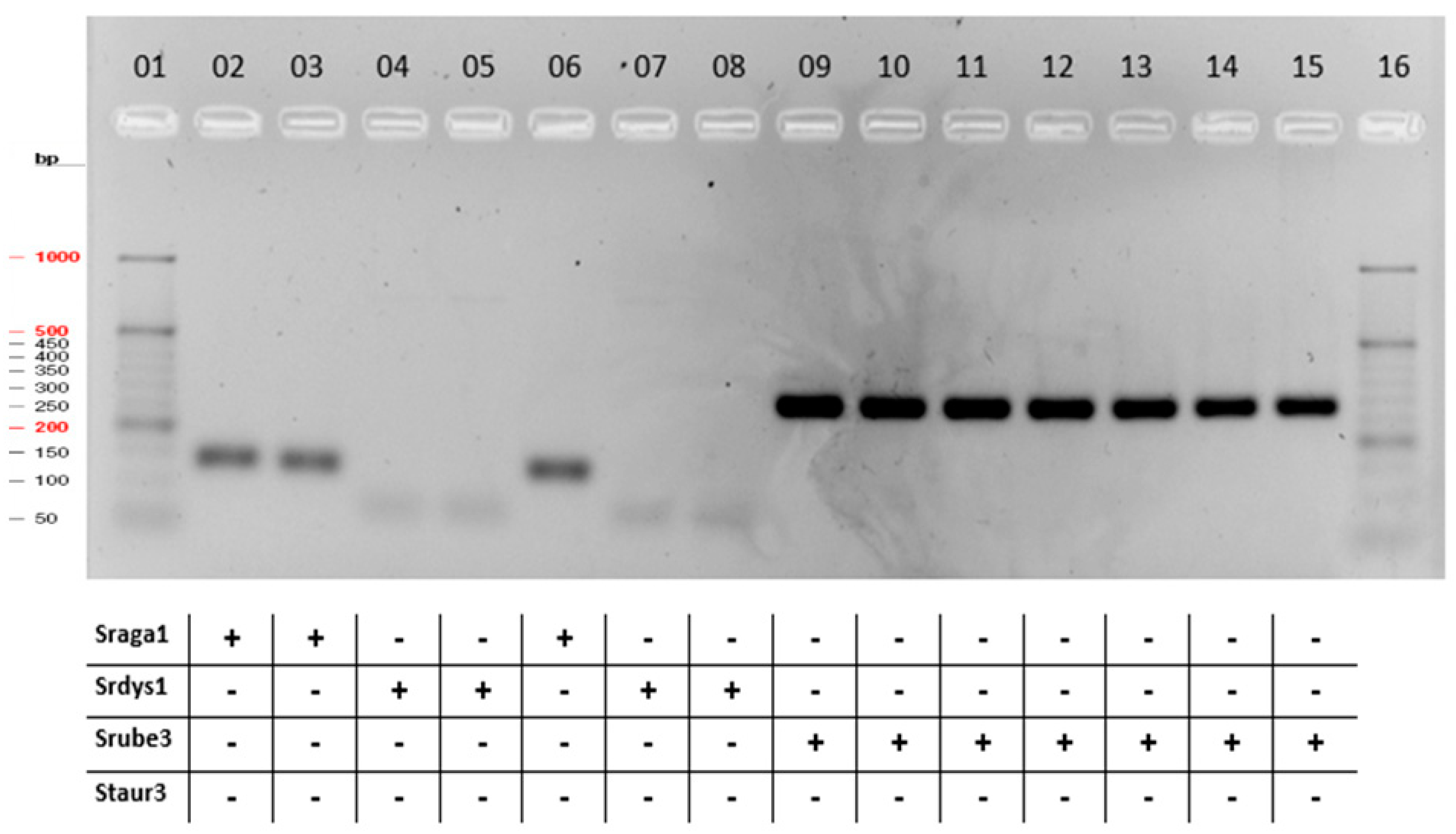
3.4. Multiplex PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abebe, R.; Markos, A.; Abera, M.; Mekbib, B. Incidence Rate, Risk Factors, and Bacterial Causes of Clinical Mastitis on Dairy Farms in Hawassa City, Southern Ethiopia. Sci. Rep. 2023, 13, 10945. [Google Scholar] [CrossRef]
- Fukushima, Y.; Kino, E.; Furutani, A.; Minamino, T.; Mikurino, Y.; Horii, Y.; Honkawa, K.; Sasaki, Y. Epidemiological Study to Investigate the Incidence and Prevalence of Clinical Mastitis, Peracute Mastitis, Metabolic Disorders and Peripartum Disorders, on a Dairy Farm in a Temperate Zone in Japan. BMC Vet. Res. 2020, 16, 389. [Google Scholar] [CrossRef]
- Kaczorek-Łukowska, E.; Małaczewska, J.; Wójcik, R.; Naumowicz, K.; Blank, A.; Siwicki, A.K. Streptococci as the New Dominant Aetiological Factors of Mastitis in Dairy Cows in North-Eastern Poland: Analysis of the Results Obtained in 2013–2019. Ir. Vet. J. 2021, 74, 2. [Google Scholar] [CrossRef]
- Gonçalves, J.L.; de Campos, J.L.; Steinberger, A.J.; Safdar, N.; Kates, A.; Sethi, A.; Shutske, J.; Suen, G.; Goldberg, T.; Cue, R.I.; et al. Incidence and Treatments of Bovine Mastitis and Other Diseases on 37 Dairy Farms in Wisconsin. Pathogens 2022, 11, 1282. [Google Scholar] [CrossRef]
- Leite de Campos, J.; Gonçalves, J.L.; Kates, A.; Steinberger, A.; Sethi, A.; Suen, G.; Shutske, J.; Safdar, N.; Goldberg, T.; Ruegg, P.L. Variation in Partial Direct Costs of Treating Clinical Mastitis among 37 Wisconsin Dairy Farms. J. Dairy Sci. 2023, 106, 9276–9286. [Google Scholar] [CrossRef]
- Zadoks, R.N.; Middleton, J.R.; McDougall, S.; Katholm, J.; Schukken, Y.H. Molecular Epidemiology of Mastitis Pathogens of Dairy Cattle and Comparative Relevance to Humans. J. Mammary Gland. Biol. Neoplas. 2011, 16, 357–372. [Google Scholar] [CrossRef]
- Dufour, S.; Labrie, J.; Jacques, M. The Mastitis Pathogens Culture Collection. Microbiol. Resour. Announc. 2019, 8, e00133-19. [Google Scholar] [CrossRef]
- Haxhiaj, K.; Wishart, D.S.; Ametaj, B.N. Mastitis: What It Is, Current Diagnostics, and the Potential of Metabolomics to Identify New Predictive Biomarkers. Dairy 2022, 3, 722–746. [Google Scholar] [CrossRef]
- Cheng, W.N.; Han, S.G. Bovine Mastitis: Risk Factors, Therapeutic Strategies, and Alternative Treatments—A Review. Asian Australas. J. Anim. Sci. 2020, 33, 1699–1713. [Google Scholar] [CrossRef]
- McKernan, C.; Benson, T.; Farrell, S.; Dean, M. Antimicrobial Use in Agriculture: Critical Review of the Factors Influencing Behaviour. JAC Antimicrob. Resist. 2021, 3, dlab178. [Google Scholar] [CrossRef]
- Tommasoni, C.; Fiore, E.; Lisuzzo, A.; Gianesella, M. Mastitis in Dairy Cattle: On-Farm Diagnostics and Future Perspectives. Animals 2023, 13, 2538. [Google Scholar] [CrossRef]
- Adkins, P.R.F.; Middleton, J.R. Methods for Diagnosing Mastitis. Vet. Clin. N. Am. Food Anim. Pract. 2018, 34, 479–491. [Google Scholar] [CrossRef]
- Ashraf, A.; Imran, M. Diagnosis of Bovine Mastitis: From Laboratory to Farm. Trop. Anim. Health Prod. 2018, 50, 1193–1202. [Google Scholar] [CrossRef]
- Borelli, E.; Ellis, K.; Pamphilis, N.M.; Tomlinson, M.; Hotchkiss, E. Factors Influencing Scottish Dairy Farmers’ Antimicrobial Usage, Knowledge and Attitude towards Antimicrobial Resistance. Prev. Vet. Med. 2023, 221, 106073. [Google Scholar] [CrossRef]
- Bexiga, R.; Koskinen, M.T.; Holopainen, J.; Carneiro, C.; Pereira, H.; Ellis, K.A.; Vilela, C.L. Diagnosis of Intramammary Infection in Samples Yielding Negative Results or Minor Pathogens in Conventional Bacterial Culturing. J. Dairy Res. 2011, 78, 49–55. [Google Scholar] [CrossRef]
- Anis, E.; Hawkins, I.K.; Ilha, M.R.S.; Woldemeskel, M.W.; Saliki, J.T.; Wilkes, R.P. Evaluation of Targeted Next-Generation Sequencing for Detection of Bovine Pathogens in Clinical Samples. J. Clin. Microbiol. 2018, 56, e00399-18. [Google Scholar] [CrossRef]
- El-Sayed, A.; Awad, W.; Abdou, N.E.; Castañeda Vázquez, H. Molecular Biological Tools Applied for Identification of Mastitis Causing Pathogens. Int. J. Vet. Sci. Med. 2017, 5, 89–97. [Google Scholar] [CrossRef]
- Sonowal, D.; Ghatak, S.; Barua, A.G.; Kandhan, S.; Hazarika, R.A.; Sen, A.; Das, S.; Sonowal, S.; Sharma, R.K.; Tamuly, S.; et al. Livestock, Pets and Humans as Carriers of Methicillin-Resistant Staphylococcus aureus and Comparative Evaluation of Two PCR Protocols for Detection. Vet. Res. Forum 2023, 14, 351–358. [Google Scholar] [CrossRef]
- Quijada, N.M.; Rodríguez-Lázaro, D.; Eiros, J.M.; Hernández, M.; Valencia, A. TORMES: An Automated Pipeline for Whole Bacterial Genome Analysis. Bioinformatics 2019, 35, 4207–4212. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved Metagenomic Analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of RRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R. Shifting the Genomic Gold Standard for the Prokaryotic Species Definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.G.; Toth, I.K. Genomics and Taxonomy in Diagnostics for Food Security: Soft-Rotting Enterobacterial Plant Pathogens. Analy. Methods 2015, 8, 12–24. [Google Scholar] [CrossRef]
- Ciufo, S.; Kannan, S.; Sharma, S.; Badretdin, A.; Clark, K.; Turner, S.; Brover, S.; Schoch, C.L.; Kimchi, A.; DiCuccio, M. Using Average Nucleotide Identity to Improve Taxonomic Assignments in Prokaryotic Genomes at the NCBI. Int. J. Syst. Evol. Microbiol. 2018, 68, 2386–2392. [Google Scholar] [CrossRef]
- Thomsen, M.C.F.; Hasman, H.; Westh, H.; Kaya, H.; Lund, O. RUCS: Rapid Identification of PCR Primers for Unique Core Sequences. Bioinformatics 2017, 33, 3917–3921. [Google Scholar] [CrossRef]
- Kibbe, W.A. OligoCalc: An Online Oligonucleotide Properties Calculator. Nucleic Acids Res. 2007, 35, W43–W46. [Google Scholar] [CrossRef]
- Lu, J.; Johnston, A.; Berichon, P.; Ru, K.L.; Korbie, D.; Trau, M. PrimerSuite: A High-Throughput Web-Based Primer Design Program for Multiplex Bisulfite PCR. Sci. Rep. 2017, 7, srep41328. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A Unified Bioinformatics Toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Ruiz-Ripa, L.; Simón, C.; Ceballos, S.; Ortega, C.; Zarazaga, M.; Torres, C.; Gómez-Sanz, E.S. Pseudintermedius and S. Aureus Lineages with Transmission Ability Circulate as Causative Agents of Infections in Pets for Years. BMC Vet. Res. 2021, 17, 42. [Google Scholar] [CrossRef] [PubMed]
- Russo, T.P.; Borrelli, L.; Minichino, A.; Fioretti, A.; Dipineto, L. Occurrence and Antimicrobial Resistance of Staphylococcus Aureus Isolated from Healthy Pet Rabbits. Vector Borne Zoonotic Dis. 2023, 24, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Saeed, S.I.; Kamaruzzaman, N.F.; Gahamanyi, N.; Nguyen, T.T.H.; Hossain, D.; Kahwa, I. Confronting the Complexities of Antimicrobial Management for Staphyloccous Aureus Causing Bovine Mastitis: An Innovative Paradigm. Ir. Vet. J. 2024, 77, 4. [Google Scholar] [CrossRef]
- Belay, N.; Mohammed, N.; Seyoum, W. Bovine Mastitis: Prevalence, Risk Factors, and Bacterial Pathogens Isolated in Lactating Cows in Gamo Zone, Southern Ethiopia. Vet. Med. Res. Rep. 2022, 13, 9–19. [Google Scholar] [CrossRef]
- Pokorska, J.; Kułaj, D.; Dusza, M.; Żychlińska-Buczek, J.; Makulska, J. New Rapid Method of DNA Isolation from Milk Somatic Cells. Anim. Biotechnol. 2016, 27, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Schmenger, A.; Krömker, V. Characterization, Cure Rates and Associated Risks of Clinical Mastitis in Northern Germany. Vet. Sci. 2020, 7, 170. [Google Scholar] [CrossRef]
- Dobrut, A.; Wójcik-Grzybek, D.; Młodzińska, A.; Pietras-Ożga, D.; Michalak, K.; Tabacki, A.; Mroczkowska, U.; Brzychczy-Włoch, M. Detection of Immunoreactive Proteins of Escherichia Coli, Streptococcus Uberis, and Streptococcus Agalactiae Isolated from Cows with Diagnosed Mastitis. Front. Cell. Infect. Microbiol. 2023, 13, 987842. [Google Scholar] [CrossRef]
- Viveiros, S.; Rodrigues, M.; Albuquerque, D.; Martins, S.A.M.; Cardoso, S.; Martins, V.C. Multiple Bacteria Identification in the Point-of-Care: An Old Method Serving a New Approach. Sensors 2020, 20, 3351. [Google Scholar] [CrossRef]
- Michira, L.; Kagira, J.; Maina, N.; Waititu, K.; Kiboi, D.; Ongera, E.; Ngotho, M. Prevalence of Subclinical Mastitis, Associated Risk Factors and Antimicrobial Susceptibility Pattern of Bacteria Isolated from Milk of Dairy Cattle in Kajiado Central Sub-County, Kenya. Vet. Med. Sci. 2023, 9, 2885–2892. [Google Scholar] [CrossRef]
- Sayed, R.H.; Soliman, R.T.; Elsaady, S.A. Development of a Lateral Flow Device for Rapid Simultaneous Multiple Detections of Some Common Bacterial Causes of Bovine Mastitis. J. Adv. Vet. Anim. Res. 2023, 10, 292–300. [Google Scholar] [CrossRef]
- Deb, R.; Chaudhary, P.; Pal, P.; Tomar, R.S.; Roshan, M.; Parmanand; Ludri, A.; Gupta, V.K.; De, S. Development of an On-Site Lateral Flow Immune Assay Based on Mango Leaf Derived Colloidal Silver Nanoparticles for Rapid Detection of Staphylococcus Aureus in Milk. J. Food Sci. Technol. 2023, 60, 132–146. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, S.; Virchow, F.; Torgerson, P.R.; Bischoff, M.; Biner, B.; Hartnack, S.; Rüegg, S.R. Test Characteristics of Milk Amyloid A ELISA, Somatic Cell Count, and Bacteriological Culture for Detection of Intramammary Pathogens That Cause Subclinical Mastitis. J. Dairy Sci. 2017, 100, 7419–7426. [Google Scholar] [CrossRef] [PubMed]
- Puggioni, G.M.G.; Tedde, V.; Uzzau, S.; Guccione, J.; Ciaramella, P.; Pollera, C.; Moroni, P.; Bronzo, V.; Addis, M.F. Evaluation of a Bovine Cathelicidin ELISA for Detecting Mastitis in the Dairy Buffalo: Comparison with Milk Somatic Cell Count and Bacteriological Culture. Res. Vet. Sci. 2020, 128, 129–134. [Google Scholar] [CrossRef]
- Alhussien, M.N.; Dang, A.K. Sensitive and Rapid Lateral-Flow Assay for Early Detection of Subclinical Mammary Infection in Dairy Cows. Sci. Rep. 2020, 10, 3351. [Google Scholar] [CrossRef]
- Chen, R.; Wang, H.; Zhao, Y.; Nan, X.; Wei, W.; Du, C.; Zhang, F.; Luo, Q.; Yang, L.; Xiong, B. Quantitative Detection of Mastitis Factor IL-6 in Dairy Cow Using the SERS Improved Immunofiltration Assay. Nanomaterials 2022, 12, 1091. [Google Scholar] [CrossRef]
- Sajid, M.; Kawde, A.N.; Daud, M. Designs, Formats and Applications of Lateral Flow Assay: A Literature Review. J. Saudi Chem. Soc. 2015, 19, 689–705. [Google Scholar] [CrossRef]
- Koskinen, M.T.; Holopainen, J.; Pyörälä, S.; Bredbacka, P.; Pitkälä, A.; Barkema, H.W.; Bexiga, R.; Roberson, J.; Sølverød, L.; Piccinini, R.; et al. Analytical Specificity and Sensitivity of a Real-Time Polymerase Chain Reaction Assay for Identification of Bovine Mastitis Pathogens. J. Dairy Sci. 2009, 92, 952–959. [Google Scholar] [CrossRef]
- Johnson, J.S.; Spakowicz, D.J.; Hong, B.-Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M.; et al. Evaluation of 16S RRNA Gene Sequencing for Species and Strain-Level Microbiome Analysis. Nat. Commun. 2019, 10, 5029. [Google Scholar] [CrossRef]
| Species | Primer | Sequence | Product [nt] | Detected Conservative Region | GenBank ID |
|---|---|---|---|---|---|
| S. agalactiae | F | CTGTGCTTAGTCCACTTGAA | 130 | zinc ABC transporter substrate-binding protein AdcA | 66885516 |
| R | GGAGCTACTTCTTTACCTGC | ||||
| S. dysgalactiae | F | CTTGCTCCTTTGAAGTAATCA | 66 | CDP-diacylglycerol—glycerol-3-phosphate 3-phosphatidyltransferase | 83689676 |
| R | TGTCTTTCTCTATGTTGCTCT | ||||
| S. uberis | F | CGTGAAGATGAAGATGTCCTA | 272 | Two genes encoding: YitT family protein and ABC-F family ATPase | 58021974, 58021975 |
| R | GTGGGTTCAATGTCTCCAG | ||||
| S. aureus | F | CCTTGACTCGCAATGTTAAG | 362 | phage infection protein | 3921461 |
| R | ATTGAAGAAAATGTGCCGAC |
| Species | Primer | Length [nt] | Tm (basic) [°C] | GC [%] | Self | Self-3′ | Hairpin |
|---|---|---|---|---|---|---|---|
| S. agalactiae | F | 20 | 49.7 | 45 | 4 | 3 | No |
| R | 20 | 51.8 | 50 | 4 | 2 | No | |
| S. dysgalactiae | F | 21 | 48.5 | 38.1 | 4 | 2 | No |
| R | 21 | 48.5 | 38.1 | 2 | 0 | No | |
| S. uberis | F | 21 | 50.5 | 42.86 | 2 | 2 | No |
| R | 19 | 51.1 | 52.63 | 3 | 2 | No | |
| S. aureus | F | 20 | 49.7 | 45 | 6 | 6 | No |
| R | 20 | 47.7 | 40 | 3 | 3 | No |
| Primers | ||||
|---|---|---|---|---|
| Sample | Sraga1_F/R | Srdys1_F/R | Srube3_F/R | Staur3_F/R |
| Staphylococcus warneri ATCC 27836 | ||||
| Staphylococcus epidermidis ATCC 14990 | ||||
| Streptococcus dysgalactiae ATCC 12394 | + | |||
| Aerococcus viridans ATCC 11563 | ||||
| Enterococcus faecalis ATCC 29212 | ||||
| Staphylococcus aureus ATCC 6538P | + | |||
| Staphylococcus aureus PCM 2267 | + | |||
| Staphylococcus aureus PCM 458/2195 | + | |||
| Staphylococcus aureus PCM 2101 | + | |||
| Staphylococcus aureus PCM 2054 | + | |||
| Staphylococcus aureus PCM 1650 | + | |||
| Staphylococcus aureus PCM 1116 | + | |||
| Staphylococcus aureus PCM 1115 | + | |||
| Staphylococcus aureus PCM 1102 | + | |||
| Staphylococcus aureus PCM 565 | + | |||
| Staphylococcus aureus PCM 502 | + | |||
| Staphylococcus aureus PCM 1937 | + | |||
| Streptococcus agalactiae 023PP2021 | + | |||
| Streptococcus agalactiae 024PP2021 | + | |||
| Streptococcus dysgalactiae 004PP2021 | + | |||
| Streptococcus dysgalactiae 005PP2021 | + | |||
| Streptococcus agalactiae 027PP2021 | + | |||
| Streptococcus dysgalactiae 001PP2016 | ||||
| Streptococcus dysgalactiae 00PP2016 | + | |||
| Streptococcus uberis 120PP2022 | + | |||
| Streptococcus uberis 041PP2021 | + | |||
| Streptococcus uberis 045PP2021 | + | |||
| Streptococcus uberis 059PP2021 | + | |||
| Streptococcus uberis 067PP2021 | + | |||
| Streptococcus uberis 071PP2021 | + | |||
| Streptococcus uberis 103PP2021 | + | |||
| Streptococcus uberis 105PP2021 | + | |||
| Streptococcus uberis 112PP2021 | + | |||
| Aerococcus urinaeequi 001PP2021 | ||||
| Aerococcus urinaeequi 002PP2021 | ||||
| Mammaliicoccus sciuri 004PP2021 | ||||
| Enterococcus faecium 003PP2021 | ||||
| Enterococcus faecium 004PP2021 | ||||
| Enterococcus faecalis 022PP2021 | ||||
| Staphylococcus haemolyticus 004PP2022 | ||||
| Staphylococcus sciuri 001PP2020 | ||||
| Streptococcus agalactiae 028PP2022 | + | |||
| Staphylococcus equorum 003PP2022 | ||||
| Staphylococcus equorum 001PP2022 | ||||
| Staphylococcus equorum 002PP2022 | ||||
| Mammaliicoccus vitulinus 001PP2022 | ||||
| Enterococcus sp.009PP2022 | ||||
| Enterococcus sp. 010PP2022 | ||||
| Enterococcus sp. 011PP2022 | ||||
| Enterococcus sp. 012PP2022 | ||||
| Enterococcus sp. 013PP2022 | ||||
| Staphylococcus sp. 008PP2022 | + | |||
| Staphylococcus sp. 009PP2022 | + | |||
| Staphylococcus sp. 010PP2022 | ||||
| Staphylococcus sp. 011PP2022 | + | |||
| Staphylococcus sp. 012PP2022 | ||||
| Staphylococcus sp. 013PP2022 | + | |||
| Staphylococcus sp. 014PP2022 | + | |||
| Staphylococcus sp. 015PP2022 | + | |||
| Staphylococcus sp. 016PP2022 | + | |||
| Staphylococcus sp. 017PP2022 | ||||
| Staphylococcus sp. 018PP2022 | + | |||
| Staphylococcus sp. 019PP2022 | + | |||
| Staphylococcus sp. 023PP2022 | ||||
| Staphylococcus sp. 025PP2022 | + | |||
| Staphylococcus sp. 026PP2022 | ||||
| Staphylococcus sp. 027PP2022 | ||||
| Staphylococcus sp. 030PP2022 | ||||
| Staphylococcus sp. 031PP2022 | ||||
| Staphylococcus sp. 032PP2022 | ||||
| Staphylococcus sp. 034PP2022 | ||||
| Staphylococcus sp. 038PP2022 | ||||
| Staphylococcus sp. 039PP2022 | ||||
| Staphylococcus sp. 040PP2022 | ||||
| Staphylococcus sp. 041PP2022 | ||||
| Staphylococcus sp. 042PP2022 | ||||
| Staphylococcus sp. 043PP2022 | + | + | ||
| Staphylococcus sp. 044PP2022 | + | |||
| Staphylococcus sp. 045PP2022 | ||||
| Staphylococcus sp. 046PP2022 | ||||
| Staphylococcus sp. 057PP2022 | + | |||
| Mammalicoccus sciuri 005PP2022 | ||||
| Streptococcus agalactiae 021PP2018 | + | |||
| Streptococcus agalactiae 022PP2021 | + | |||
| Streptococcus sp. 003PP2017 | + | |||
| Streptococcus sp. 004PP2017 | + | |||
| Streptococcus sp. 006PP2017 | ||||
| Streptococcus sp. 008PP2018 | + | |||
| Streptococcus sp. 009PP2019 | + | |||
| Streptococcus sp. 014PP2022 | + | |||
| Streptococcus sp. 017PP2022 | ||||
| Streptococcus sp. 018PP2022 | ||||
| Staphylococcus pseudintermedius 001PP2023 | ||||
| Staphylococcus pseudintermedius 002PP2023 | ||||
| Staphylococcus simulans 002PP2022 | ||||
| Staphylococcus pseudintermedius 003PP2023 | ||||
| Staphylococcus pseudintermedius 004PP2023 | ||||
| Staphylococcus pseudintermedius 005PP2023 | ||||
| Staphylococcus pseudintermedius 006PP2023 | ||||
| Staphylococcus pseudintermedius 007PP2023 | ||||
| Staphylococcus pseudintermedius 011PP2023 | ||||
| Staphylococcus pseudintermedius 012PP2023 | ||||
| NTC (No Template Control) | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kajdanek, A.; Kluska, M.; Matusiak, R.; Kazimierczak, J.; Dastych, J. A Rapid and Inexpensive PCR Test for Mastitis Diagnosis Based on NGS Data. Pathogens 2024, 13, 423. https://doi.org/10.3390/pathogens13050423
Kajdanek A, Kluska M, Matusiak R, Kazimierczak J, Dastych J. A Rapid and Inexpensive PCR Test for Mastitis Diagnosis Based on NGS Data. Pathogens. 2024; 13(5):423. https://doi.org/10.3390/pathogens13050423
Chicago/Turabian StyleKajdanek, Agnieszka, Magdalena Kluska, Rafał Matusiak, Joanna Kazimierczak, and Jarosław Dastych. 2024. "A Rapid and Inexpensive PCR Test for Mastitis Diagnosis Based on NGS Data" Pathogens 13, no. 5: 423. https://doi.org/10.3390/pathogens13050423
APA StyleKajdanek, A., Kluska, M., Matusiak, R., Kazimierczak, J., & Dastych, J. (2024). A Rapid and Inexpensive PCR Test for Mastitis Diagnosis Based on NGS Data. Pathogens, 13(5), 423. https://doi.org/10.3390/pathogens13050423







