Loop-Mediated Isothermal Amplification for the Fast Detection of Bonamia ostreae and Bonamia exitiosa in Flat Oysters
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Controls
2.2. LAMP Primers Assay Design
2.3. LAMP Quantification DNA Standards
2.4. LAMP Assay Protocol
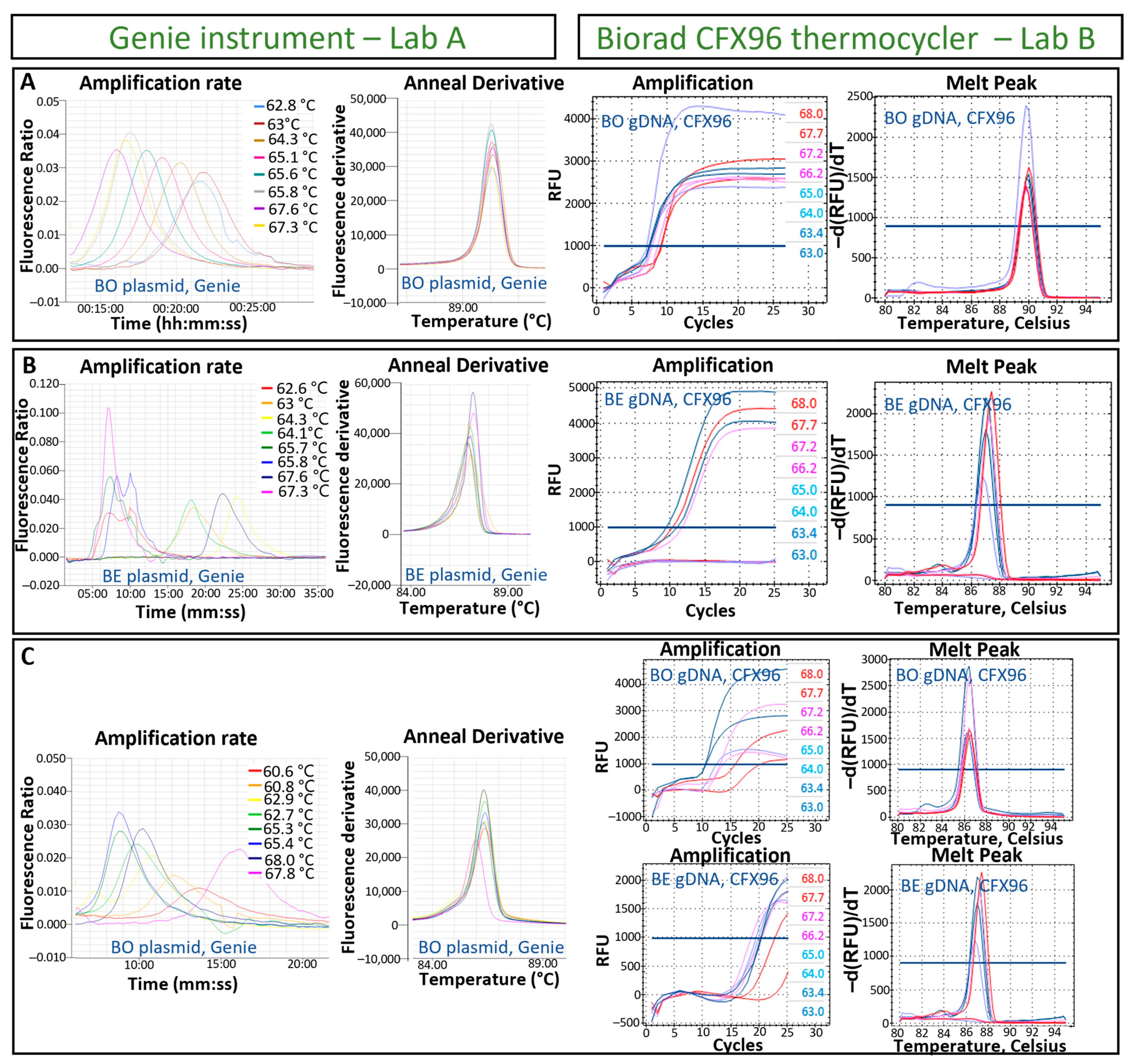
2.5. Optimization of the LAMP Assay Temperature
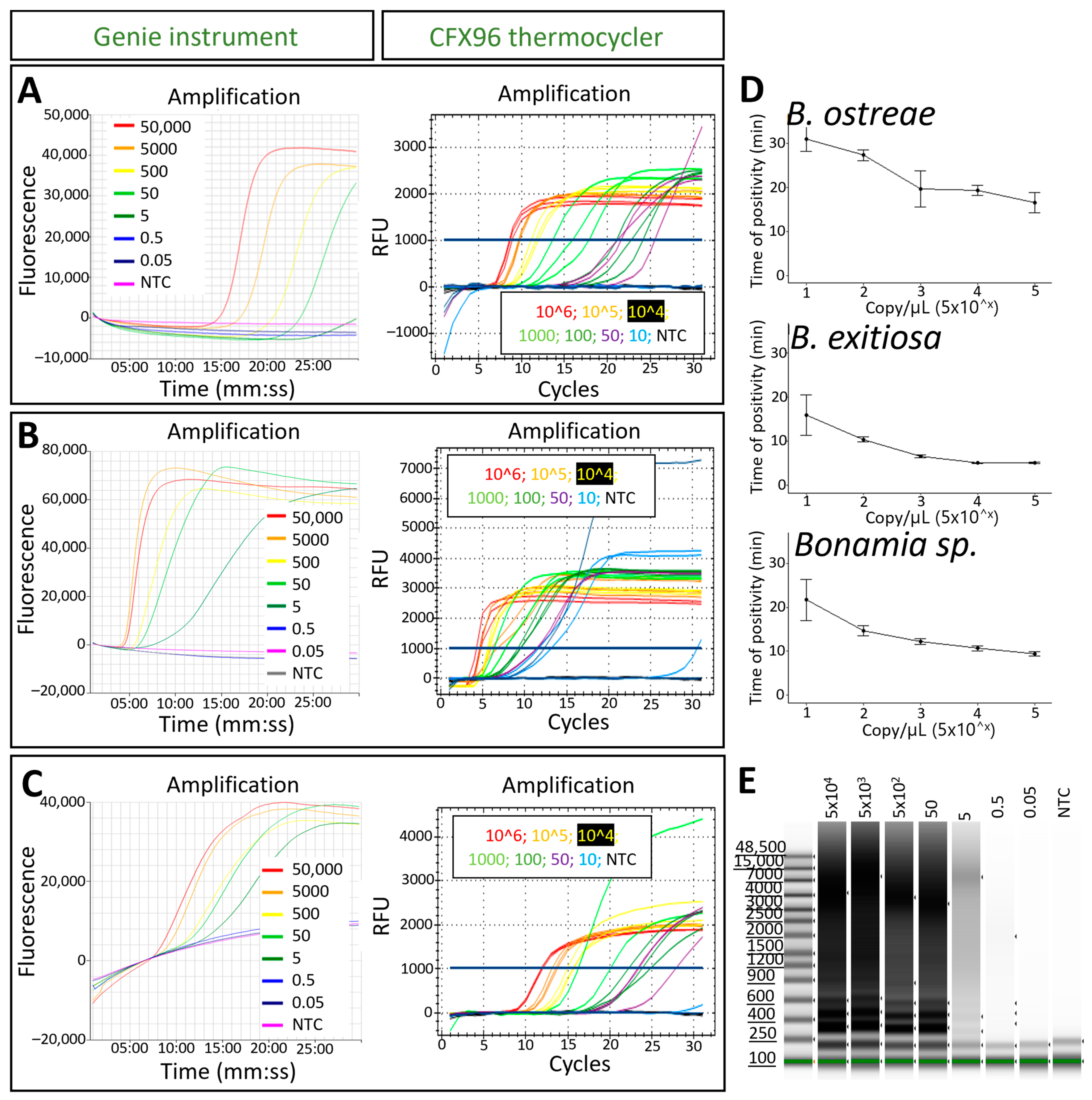
2.6. LAMP Analytical Sensitivity
2.7. LAMP Analytical Specificity
2.8. Diagnostic Accuracy of LAMP Assays for the Detection of Bonamiosis
3. Results
3.1. Designing of LAMP Tests for the Detection of B. ostreae and B. exitiosa and Optimisation of the Isothermal Temperature Reaction
3.2. LAMP Analytical Sensitivity
3.3. LAMP Analytical Specificity
3.4. LAMP Diagnostic Accuracy for the Detection of Bonamiosis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Infection with Bonamia Ostreae. In Aquatic Animal Health Code; World Oragnisation for Animal Health (WOAH): Paris, France, 2023; Chapter 11.3; Available online: https://www.woah.org/en/what-we-do/standards/codes-and-manuals/aquatic-code-online-access/?id=169&L=1&htmfile=chapitre_bonamia_ostreae.htm (accessed on 4 September 2023).
- Infection with Bonamia exitiosa. In Aquatic Animal Health Code; World Organisation for Animal Health (WOAH): Paris, France, 2023; Chapter 11.2; Available online: https://www.woah.org/en/what-we-do/standards/codes-and-manuals/aquatic-code-online-access/?id=169&L=1&htmfile=chapitre_bonamia_exitiosa.htm (accessed on 4 September 2023).
- Carnegie, R.B.; Cochennec-Laureau, N. Microcell Parasites of Oysters: Recent Insights and Future Trends. Aquat. Living Resour. 2004, 17, 519–528. [Google Scholar] [CrossRef]
- Hill, K.M.; Stokes, N.A.; Webb, S.C.; Hine, P.M.; Kroeck, M.A.; Moore, J.D.; Morley, M.S.; Reece, K.S.; Burreson, E.M.; Carnegie, R.B. Phylogenetics of Bonamia Parasites Based on Small Subunit and Internal Transcribed Spacer Region Ribosomal DNA Sequence Data. Dis. Aquat. Organ. 2014, 110, 33–54. [Google Scholar] [CrossRef]
- Laing, I.; Dunn, P.; Peeler, E.J.; Feist, S.W.; Longshaw, M. Epidemiology of Bonamia in the UK, 1982 to 2012. Dis. Aquat. Organ. 2014, 110, 101–111. [Google Scholar] [CrossRef]
- Pichot, Y.; Comps, M.; Tige, G.; Grizel, H.; Rabouin, M.-A. Recherches Sur Bonamia Ostreae Gen. n., Sp. n., Parasite Nouveau de l’huitre Plate Ostrea edulis L. Rev. Trav. L’institut Pêches Marit. 1979, 43, 131–140. [Google Scholar]
- Cigarria, J.; Elston, R. Independent Introduction of Bonamia Ostreae, a Parasite of Ostrea Edulis, to Spain. Dis. Aquat. Org. 1997, 29, 157–158. [Google Scholar] [CrossRef]
- Hudson, E.B.; Hill, B.J. Impact and Spread of Bonamiasis in the UK. Aquaculture 1991, 93, 279–285. [Google Scholar] [CrossRef]
- Elston, R.; Farley, C.; Kent, M. Occurence and Significance of Bonamiasis in European Flat Oysters Ostrea Edulis in North America. Dis. Aquat. Organ. 1986, 2, 49–54. [Google Scholar] [CrossRef]
- Engelsma, M.Y.; Culloty, S.C.; Lynch, S.A.; Arzul, I.; Carnegie, R.B. Bonamia Parasites: A Rapidly Changing Perspective on a Genus of Important Mollusc Pathogens. Dis. Aquat. Organ. 2014, 110, 5–23. [Google Scholar] [CrossRef] [PubMed]
- Abollo, E.; Ramilo, A.; Casas, S.M.; Comesaña, P.; Cao, A.; Carballal, M.J.; Villalba, A. First Detection of the Protozoan Parasite Bonamia Exitiosa (Haplosporidia) Infecting Flat Oyster Ostrea Edulis Grown in European Waters. Aquaculture 2008, 274, 201–207. [Google Scholar] [CrossRef]
- Longshaw, M.; Stone, D.M.; Wood, G.; Green, M.J.; White, P. Detection of Bonamia Exitiosa (Haplosporidia) in European Flat Oysters Ostrea Edulis Cultivated in Mainland Britain. Dis. Aquat. Organ. 2013, 106, 173–179. [Google Scholar] [CrossRef]
- Montes, J.; Anadón, R.; Azevedo, C. A Possible Life Cycle for Bonamia Ostreae on the Basis of Electron Microscopy Studies. J. Invertebr. Pathol. 1994, 63, 1–6. [Google Scholar] [CrossRef]
- Balouet, G.; Poder, M.; Cahour, A. Haemocytic Parasitosis: Morphology and Pathology of Lesions in the French Flat Oyster, Ostrea edulis L. Aquaculture 1983, 34, 1–14. [Google Scholar] [CrossRef]
- Ramilo, A.; Villalba, A.; Abollo, E. Species-Specific Oligonucleotide Probe for Detection of Bonamia Exitiosa (Haplosporidia) Using in Situ Hybridisation Assay. Dis. Aquat. Organ. 2014, 110, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Engelsma, M.Y.; Kerkhoff, S.; Roozenburg, I.; Haenen, O.L.M.; Van Gool, A.; Sistermans, W.; Wijnhoven, S.; Hummel, H. Epidemiology of Bonamia Ostreae Infecting European Flat Oysters Ostrea Edulis from Lake Grevelingen, The Netherlands. Mar. Ecol. Prog. Ser. 2010, 409, 131–142. [Google Scholar] [CrossRef]
- Bradley, T.L.; Mercer, J.A.; Humphrey, J.D.; Moody, N.J.G.; Hunnam, J.C. Bonamia Exitiosa in Farmed Native Oysters Ostrea Angasi in Australia: Optimal Epidemiological QPCR Cut-Point and Clinical Disease Risk Factors. Dis. Aquat. Organ. 2020, 140, 151–165. [Google Scholar] [CrossRef] [PubMed]
- Carrasco, N.; Villalba, A.; Andree, K.B.; Engelsma, M.Y.; Lacuesta, B.; Ramilo, A.; Gairín, I.; Furones, M.D. Bonamia Exitiosa (Haplosporidia) Observed Infecting the European Flat Oyster Ostrea Edulis Cultured on the Spanish Mediterranean Coast. J. Invertebr. Pathol. 2012, 110, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Hine, P.M.; Cochennec-Laureau, N.; Berthe, F.C.J. Bonamia Exitiosus n.Sp. (Haplosporidia) Infecting Flat Oysters Ostrea Chilensis in New Zealand. Dis. Aquat. Organ. 2001, 47, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Carnegie, R.B.; Barber, B.J.; Culloty, S.C.; Figueras, A.J.; Distel, D.L. Development of a PCR Assay for Detection of the Oyster Pathogen Bonamia Ostreae and Support for Its Inclusion in the Haplosporidia. Dis. Aquat. Organ. 2000, 42, 199–206. [Google Scholar] [CrossRef]
- Cochennec, N.; Le Roux, F.; Berthe, F.; Gerard, A. Detection of Bonamia Ostreae Based on Small Subunit Ribosomal Probe. J. Invertebr. Pathol. 2000, 76, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Corbeil, S.; Arzul, I.; Diggles, B.; Heasman, M.; Chollet, B.; Berthe, F.C.J.; Crane, M.S.J. Development of a TaqMan PCR Assay for the Detection of Bonamia Species. Dis. Aquat. Organ. 2006, 11, 75–80. [Google Scholar] [CrossRef]
- Marty, G.D.; Bower, S.M.; Clarke, K.R.; Meyer, G.; Lowe, G.; Osborn, A.L.; Chow, E.P.; Hannah, H.; Byrne, S.; Sojonky, K.; et al. Histopathology and a Real-Time PCR Assay for Detection of Bonamia Ostreae in Ostrea Edulis Cultured in Western Canada. Aquaculture 2006, 261, 33–42. [Google Scholar] [CrossRef]
- Robert, M.; Garcia, C.; Chollet, B.; Lopez-Flores, I.; Ferrand, S.; François, C.; Joly, J.P.; Arzul, I. Molecular Detection and Quantification of the Protozoan Bonamia Ostreae in the Flat Oyster, Ostrea Edulis. Mol. Cell. Probes 2009, 23, 264–271. [Google Scholar] [CrossRef] [PubMed]
- Ramilo, A.; Navas, J.I.; Villalba, A.; Abollo, E. Species-Specific Diagnostic Assays for Bonamia Ostreae and B. Exitiosa in European Flat Oyster Ostrea Edulis: Conventional, Real-Time and Multiplex PCR. Dis. Aquat. Organ. 2013, 104, 149–161. [Google Scholar] [CrossRef] [PubMed]
- European Union Reference Laboratory (EURL) for Molluscs Diseases Bonamia Ostreae and Bonamia Exitiosa Detection by Taqman® Real Time Polymerase Chain Reaction, 2nd ed.; La Temblade: Ifremer, France, 2023; Available online: https://www.eurl-mollusc.eu/content/download/137231/file/B.ostreae%26B.exitiosa%20_TaqmanRealTimePCR_editionN%C2%B02.pdf (accessed on 4 September 2023).
- Mérou, N.; Lecadet, C.; Pouvreau, S.; Arzul, I. An EDNA/ERNA-Based Approach to Investigate the Life Cycle of Non-Cultivable Shellfish Micro-Parasites: The Case of Bonamia Ostreae, a Parasite of the European Flat Oyster Ostrea Edulis. Microb. Biotechnol. 2020, 13, 1807–1818. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, L.v.G.; Nielsen, J.W.; Villadsen, M.K.; Vismann, B.; Dalvin, S.; Mathiessen, H.; Madsen, L.; Kania, P.W.; Buchmann, K. A Non-Lethal Method for Detection of Bonamia Ostreae in Flat Oyster (Ostrea Edulis) Using Environmental DNA. Sci. Rep. 2020, 10, 16143. [Google Scholar] [CrossRef]
- Feng, C.; Wang, C.; Lin, X.; Zhang, Y.; Lv, J.; Deng, J.; Yuan, X.; Mei, L.; Wu, S. Development of a Loop-Mediated Isothermal Amplification Method for Detection of Perkinsus Spp. in Mollusks. Dis. Aquat. Organ. 2013, 104, 141–148. [Google Scholar] [CrossRef]
- Wu, L.; Ye, L.; Wang, Z.; Cui, Y.; Wang, J. Utilization of Recombinase Polymerase Amplification Combined with a Lateral Flow Strip for Detection of Perkinsus Beihaiensis in the Oyster Crassostrea Hongkongensis. Parasit. Vectors 2019, 11, 360. [Google Scholar] [CrossRef]
- Chen, M.H.; Kuo, S.T.; Renault, T.; Chang, P.H. The Development of a Loop-Mediated Isothermal Amplification Assay for Rapid and Sensitive Detection of Abalone Herpesvirus DNA. J. Virol. Methods 2014, 196, 199–203. [Google Scholar] [CrossRef]
- Ren, W.; Renault, T.; Cai, Y.; Wang, C. Development of a Loop-Mediated Isothermal Amplification Assay for Rapid and Sensitive Detection of Ostreid Herpesvirus 1 DNA. J. Virol. Methods 2010, 32, 993–996. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Xie, Z.; Pang, Y.; Deng, X.; Xie, Z.; Liu, J. Development of a Loop-Mediated Isothermal Amplification Assay for Visual Detection of Marteilia Refringens in Shellfish. Chin. J. Vet. Sci. 2012, 32, 993–996. [Google Scholar] [CrossRef]
- Zaczek-Moczydłowska, M.A.; Mohamed-Smith, L.; Toldrà, A.; Hooper, C.; Campàs, M.; Dolors Furones, M.; Bean, T.P.; Campbell, K. A Single-Tube Hnb-Based Loop-Mediated Isothermal Amplification for the Robust Detection of the Ostreid Herpesvirus 1. Int. J. Mol. Sci. 2020, 21, 6605. [Google Scholar] [CrossRef] [PubMed]
- Biswas, G.; Sakai, M. Loop-Mediated Isothermal Amplification (LAMP) Assays for Detection and Identification of Aquaculture Pathogens: Current State and Perspectives. Appl. Microbiol. Biotechnol. 2014, 98, 2881–2895. [Google Scholar] [CrossRef]
- Tomlinson, J.A.; Dickinson, M.; Hobden, E.; Robinson, S.; Giltrap, P.M.; Boonham, N. A Five-Minute DNA Extraction Method for Expedited Detection of Phytophthora Ramorum Following Prescreening Using Phytophthora Spp. Lateral Flow Devices. J. Microbiol. Methods 2010, 81, 116–120. [Google Scholar] [CrossRef]
- Lau, H.Y.; Botella, J.R. Advanced DNA-Based Point-of-Care Diagnostic Methods for Plant Diseases Detection. Front. Plant Sci. 2017, 8, 2016. [Google Scholar] [CrossRef]
- Ali, N.; Rampazzo, R.d.C.P.; Costa, A.D.T.; Krieger, M.A. Current Nucleic Acid Extraction Methods and Their Implications to Point-of-Care Diagnostics. BioMed Res. Int. 2017, 2017, 9306564. [Google Scholar] [CrossRef]
- Hartikainen, H.; Stentiford, G.D.; Bateman, K.S.; Berney, C.; Feist, S.W.; Longshaw, M.; Okamura, B.; Stone, D.; Ward, G.; Wood, C.; et al. Mikrocytids Are a Broadly Distributed and Divergent Radiation of Parasites in Aquatic Invertebrates. Curr. Biol. 2014, 24, 807–812. [Google Scholar] [CrossRef] [PubMed]
- Arzul, I.; Garcia, C.; Chollet, B.; Serpin, D.; Lupo, C.; Noyer, M.; Tourbiez, D.; Berland, C.; Dégremont, L.; Travers, M.-A. First Characterization of the Parasite Haplosporidium Costale in France and Development of a Real-Time PCR Assay for Its Rapid Detection in the Pacific Oyster, Crassostrea Gigas. Transbound. Emerg. Dis. 2022, 69, e2041–e2058. [Google Scholar] [CrossRef] [PubMed]
- Clewley, J.P.; Arnold, C. MEGALIGN. The Multiple Alignment Module of LASERGENE. Methods Mol. Biol. 1997, 70, 119–129. [Google Scholar] [PubMed]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for Inferring Very Large Phylogenies by Using the Neighbor-Joining Method. Proc. Natl. Acad. Sci. USA 2004, 101, 30. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Culloty, S.C.; Mulcahy, M.F. Bonamia Ostreae in the Native Oyster Ostrea Edulis: A Review. Mar. Environ. Health Ser. 2007, 29, 1–36. [Google Scholar]
- Ramilo, A.; González, M.; Carballal, M.J.; Darriba, S.; Abollo, E.; Villalba, A. Oyster Parasites Bonamia Ostreae and B. Exitiosa Co-Occur in Galicia (NW Spain): Spatial Distribution and Infection Dynamics. Dis. Aquat. Organ. 2014, 110, 123–133. [Google Scholar] [CrossRef]
- Hill, B.; Reese, A.; Dixon, P.; Oidtmann, B.; Paley, R.; Peeler, E.; Stentiford, G.; Stone, D.; Way, K.; Hine, M.; et al. Epidemiology of Different Agents Causing Disease in Aquatic Animals. EFSA Support. Publ. 2010, 7, 37E. [Google Scholar] [CrossRef]
- Prado-Alvarez, M.; Couraleau, Y.; Chollet, B.; Tourbiez, D.; Arzul, I. Whole-Genome Amplification: A Useful Approach to Characterize New Genes in Unculturable Protozoan Parasites Such as Bonamia Exitiosa. Parasitology 2015, 142, 1523–1534. [Google Scholar] [CrossRef]
- Chou, P.H.; Lin, Y.C.; Teng, P.H.; Chen, C.L.; Lee, P.Y. Real-Time Target-Specific Detection of Loop-Mediated Isothermal Amplification for White Spot Syndrome Virus Using Fluorescence Energy Transfer-Based Probes. J. Virol. Methods 2011, 173, 67–74. [Google Scholar] [CrossRef]
- Hardinge, P.; Murray, J.A.H. Reduced False Positives and Improved Reporting of Loop-Mediated Isothermal Amplification Using Quenched Fluorescent Primers. Sci. Rep. 2019, 9, 7400. [Google Scholar] [CrossRef]
- Canier, L.; Noyer, M.; Nadeau, D.; Chollet, B.; Garcia, C.; Arzul, I. PCR Diagnosis of Regulated Mollusc Diseases Present in Europe. In Proceedings of the 21st International Conference on Diseases of Fish and Shellfish, Aberdeen, UK, 11–14 September 2023; EAFP: Aberdeen, UK, 2023. [Google Scholar]
- Heywood, J.L.; Sieracki, M.E.; Bellows, W.; Poulton, N.J.; Stepanauskas, R. Capturing Diversity of Marine Heterotrophic Protists: One Cell at a Time. ISME J. 2011, 5, 674–684. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.J.; Lu, J.F.; Su, X.R.; Jin, J.L.; Li, S.Y.; Zhou, Y.; Wang, L.; Shao, X.B.; Wang, Y.H.; Yan, M.C.; et al. Simultaneous Detection of Multiple Bacterial and Viral Aquatic Pathogens Using a Fluorogenic Loop-Mediated Isothermal Amplification-Based Dual-Sample Microfluidic Chip. J. Fish Dis. 2021, 44, 401–413. [Google Scholar] [CrossRef]
- Kaneko, H.; Kawana, T.; Fukushima, E.; Suzutani, T. Tolerance of Loop-Mediated Isothermal Amplification to a Culture Medium and Biological Substances. J. Biochem. Biophys. Methods 2007, 70, 499–501. [Google Scholar] [CrossRef]
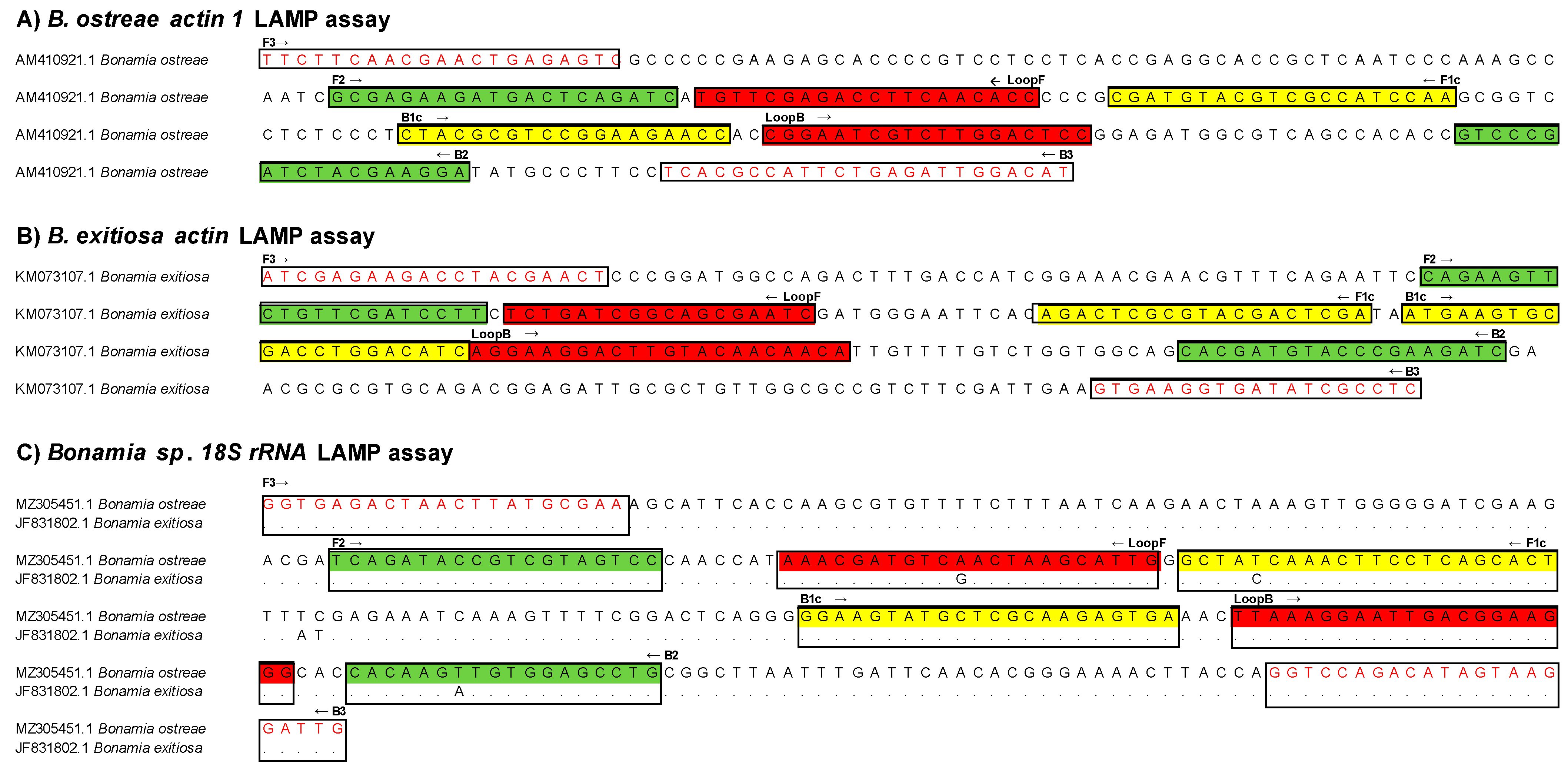
| LAMP Assay | Primer | Primer Sequence (5′-3′) | Position | Tm °C | GC% | Amplicon * |
|---|---|---|---|---|---|---|
| B. ostreae actin-1 | BO_F3 | TTCTTCAACGAACTGAGAGTC | 37 | 59.9 | 42.9 | 158 bp |
| BO_B3 | ATGTCCAATCTCAGAATGGC | 308 | 60 | 45 | ||
| BO_FIP (F1c + F2) | TTGGATGGCGACGTACATCGGCGAGAAGATGACTCAGATC | |||||
| BO_BIP (B1c + B2) | CTACGCGTCCGGAAGAACCTCCTTCGTAGATCGGGAC | |||||
| BO_LoopF | GGTGTTGAAGGTCTCGAACA | 156 | 62.3 | 50 | ||
| BO_LoopB | CGGAATCGTCTTGGACTCC | 216 | 62.2 | 57.9 | ||
| B. exitiosa actin | BE_F3 | ATCGAGAAGACCTACGAACT | 472 | 60 | 45 | 155 bp |
| BE_B3 | GAGGCGATATCACCTTCAC | 763 | 59.8 | 52.6 | ||
| BE_FIB (F1C + F2) | TCGAGTCGTACGCGAGTCTCAGAAGTTCTGTTCGATCCTT | |||||
| BE_BIP (B1c + B2) | ATGAAGTGCGACCTGGACATCGATCTTCGGGTACATCGTG | |||||
| BE_LoopF | GATTCGCTGCCGATCAGA | 578 | 62 | 55.6 | ||
| BE_LoopR | AGGAAGGACTTGTACAACAACA | 634 | 61.9 | 40.9 | ||
| B. ostreae 18S | B_F3 | GGTGAGACTAACTTATGCGAA | 881 | 59.8 | 42.9 | 169 bp |
| B_B3 | CAATCCTTACTATGTCTGGACC | 1185 | 60 | 45.5 | ||
| B_FIB (F1C + F2) | AGTGCTGAGGAAGTTTGRTAGCTCAGATACCGTCGTAGTCC | |||||
| B_BIP (B1c + B2) | GGAAGTATGCTCGCAAGAGTGACAGGCTCCACAWCTTGTG | |||||
| B_LoopF | CAATGCTTAGTYGACATCGTTT | 1007 | 61.6 | 40.9 | ||
| B_LoopR | CTTAAAGGAATTGACGGAAGGG | 1086 | 61.3 | 45.5 |
| LAMP Assay | Species-Specific B. ostreae | Species-Specific B. exitiosa | Generic Bonamia | |||
|---|---|---|---|---|---|---|
| Copies/µL | A | B | A | B | A | B |
| 106 | 4/4 | 6/6 | 4/4 | 6/6 | 4/4 | 6/6 |
| 105 | 4/4 | 6/6 | 4/4 | 6/6 | 4/4 | 6/6 |
| 104 | 4/4 | 6/6 | 4/4 | 6/6 | 4/4 | 6/6 |
| 103 | 4/4 | 6/6 | 4/4 | 6/6 | 4/4 | 6/6 |
| 102 | 4/4 | 6/6 | 4/4 | 6/6 | 4/4 | 6/6 |
| 50 | 4/4 | 6/6 | 4/4 | 6/6 | 4/4 | 6/6 |
| 10 | 2/4 | 2/6 | 4/4 | 4/6 | 3/4 | 2/6 |
| 1 | 0/4 | 0/6 | 0/4 | 1/6 | 0/4 | 0/6 |
| NTC | 0/4 | 0/6 | 0/4 | 0/6 | 0/4 | 0/6 |
| B. ostreae | LAMP B. ostreae | LAMP Generic Bonamia | Taqman qPCR | ||
| gDNA Dilution | Positive/Total | TM °C | Positive/Total | TM °C | Positive/Total |
| 1/10 | 9/9 | 90 | 9/9 | 86.4–86.6 | 9/9 |
| 1/100 | 9/9 | 90 | 9/9 | 86.4 | 9/9 |
| 1/1000 | 9/9 (1 doubtful) | 90 | 9/9 | 86.4–87 | 9/9 |
| 1/10,000 | 0/9 | none | 8/9 (1 doubtful) | 86.2–86.4 | 6/9 |
| 1/100,000 | 0/9 | none | 1/9 | 86.2 | 0/9 |
| 1/1,000,000 | 0/9 | none | 0/9 | none | 0/9 |
| NTC | 0/9 | none | 0/9 | none | 0/9 |
| B. exitiosa | LAMP B. exitiosa | LAMP Generic Bonamia | Taqman qPCR | ||
| gDNA Dilution | Positive/Total | TM °C | Positive/Total | TM °C | Positive/Total |
| 1/10 | 9/9 | 87.2–87.6 | 9/9 | 86.0–86.6 | 9/9 |
| 1/100 | 9/9 (1 doubtful) | 86.2–87.0 | 9/9 | 86.2–86.4 | 9/9 |
| 1/1000 | 5/9 (1 doubtful) | 86.4–88.0 | 9/9 | 86.0–86.2 | 9/9 |
| 1/10,000 | 1/9 | 87.6 | 2/9 (1 doubtful) | 86.0–86.2 | 8/9 |
| 1/100,000 | 0/9 | none | 2/9 (1 doubtful) | 86.0–86.2 | 1/9 |
| 1/1,000,000 | 0/9 | none | 0/9 | none | 0/9 |
| NTC | 0/9 | none | 0/9 | none | 0/9 |
| LAMP B. ostreae | LAMP B. exitiosa | LAMP Bonamia sp. | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Pathogen | Host | Origin | Results | TM | Results | TM | Results | Tm | |
| Exclusivity | Marteilia type O | Ostrea edulis | France (Ifremer) | − | None | − | None | − | None |
| Mikrocytos veneroïdes | Donax trunculus | France (Ifremer) | − | 87.00 * | − | None | − | None | |
| Haplosporidium costale | Magallana (ex Crassostrea) gigas | France (Ifremer) | − | None | − | None | +/− | 86.80 ** | |
| Haplosporidium nelsoni & H. costale | Crassostrea virginica | USA (VIMS) | − | None | − | None | − | None | |
| Mikrocytos mackini | Magallana (ex Crassostrea) gigas | Canada (DFO) | − | None | − | None | − | None | |
| Inclusivity | Bonamia exitiosa | Ostrea edulis | France (Ifremer) | − | None | + | 87.40 | + | 86.2 |
| Bonamia exitiosa | Ostrea edulis | France (Ifremer) | − | None | + | 87.60 | + | 86.2 | |
| Bonamia exitiosa | Ostrea edulis | Turkey (VCRI) | − | None | −/+ | 87.40 | + | 86.2 | |
| Bonamia ostreae | Ostrea edulis | France (Ifremer) | + | 89.80 | − | None | + | 86.4 | |
| Bonamia. ostreae | Ostrea edulis | France (Ifremer) | + | 90.00 | − | None | + | 86.4 | |
| Bonamia ostreae | Ostrea edulis | France (Ifremer) | + | 89.80 | − | None | + | 86.4 | |
| Sample | Status | LAMP B. ostreae | LAMP B. exitiosa | LAMP Bonamia sp. | |||
|---|---|---|---|---|---|---|---|
| Lab A (Tp) | Lab B (TM) | Lab A (Tp) | Lab B (TM) | Lab A (Tp) | Lab B (TM) | ||
| 1 | Bo+ Be++ | +(12:00) | +(90.00) | +(11:00) | +(87.60) | +(11:59) | +(86.40) |
| 2 | negative | – | – | – | – | – | – |
| 3 | Be+ | – | – | +(17:30) | +(87.40) | +(12:14) | +(86.20) |
| 4 * | Be++ | – | – | – | +(87.40) | +(11:44) | +(86.20) |
| 5 | negative | – | – | – | – | – | – |
| 6 | Bo++ | +(12:00) | +(90.00) | – | – | +(9:59) | +(86.20) |
| 7 | negative | – | – | – | – | – | – |
| 8 | Be+ | – | – | +(12:45) | +(87.60) | +(13:15) | +(86.20) |
| 9 * | Be+ | – | – | +(17:30) | – | +(12:30) | +(86.20) |
| 10 | Bo+ | +(8:30) | +(90.00) | – | – | +(9:15) | +(86.40) |
| 11 | Be++ | – | – | +(15:15) | +(87.60) | +(10:00) | +(86.20) |
| 12 | negative | – | – | – | – | – | – |
| 13 | negative | – | – | – | – | – | – |
| 14 | Bo++ Be+ | +(16:45) | +(90.00) | +(12:00) | +(87.40) | +(10:15) | +(86.40) |
| 15 | Bo++ Be+ | +(9:00) | +(90.00) | +(13:40) | +(87.40) | +(10:00) | +(86.00) |
| 16 | Bo+ | +(8:30) | +(90.20) | – | – | +(10:30) | +(86.40) |
| 17 | Be++ | – | – | +(12:00) | +(87.60) | +(9:30) | +(86.20) |
| 18 | Bo+ Be++ | +(9:00) | +(90.00) | +(10:15) | +(87.60) | +(13:00) | +(86.40) |
| 19 | Bo++ | +(7:45) | +(90.00) | – | – | +(9:30) | +(86.40) |
| 20 | negative | – | – | – | – | – | – |
| 21 | negative | – | – | – | – | – | – |
| 22 | negative | – | – | – | – | – | – |
| 23 | Bo+ | +(8:15) | +(90.00) | – | – | +(10:15) | +(86.40) |
| 24 | Bo++ | +(9:00) | +(90.00) | – | – | +(9:15) | +(86.40) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cano, I.; Wood, G.; Stone, D.; Noyer, M.; Canier, L.; Arzul, I. Loop-Mediated Isothermal Amplification for the Fast Detection of Bonamia ostreae and Bonamia exitiosa in Flat Oysters. Pathogens 2024, 13, 132. https://doi.org/10.3390/pathogens13020132
Cano I, Wood G, Stone D, Noyer M, Canier L, Arzul I. Loop-Mediated Isothermal Amplification for the Fast Detection of Bonamia ostreae and Bonamia exitiosa in Flat Oysters. Pathogens. 2024; 13(2):132. https://doi.org/10.3390/pathogens13020132
Chicago/Turabian StyleCano, Irene, Gareth Wood, David Stone, Mathilde Noyer, Lydie Canier, and Isabelle Arzul. 2024. "Loop-Mediated Isothermal Amplification for the Fast Detection of Bonamia ostreae and Bonamia exitiosa in Flat Oysters" Pathogens 13, no. 2: 132. https://doi.org/10.3390/pathogens13020132
APA StyleCano, I., Wood, G., Stone, D., Noyer, M., Canier, L., & Arzul, I. (2024). Loop-Mediated Isothermal Amplification for the Fast Detection of Bonamia ostreae and Bonamia exitiosa in Flat Oysters. Pathogens, 13(2), 132. https://doi.org/10.3390/pathogens13020132






