Emergence of Carbapenem-Resistant Uropathogenic Escherichia coli (ST405 and ST167) Strains Carrying blaCTX-M-15, blaNDM-5 and Diverse Virulence Factors in Hospitalized Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Specimen Collection and Bacterial Identification
2.3. Molecular Confirmation
2.4. Antibiotic Susceptibility Pattern of CR-UPEC
2.5. Screening for Antimicrobial Resistance Determinants in UPEC
2.6. Virulence Profiling of CR-UPEC Isolates
2.7. Multi-Locus Sequence Typing (MLST)
2.8. Statistical Analysis
3. Results
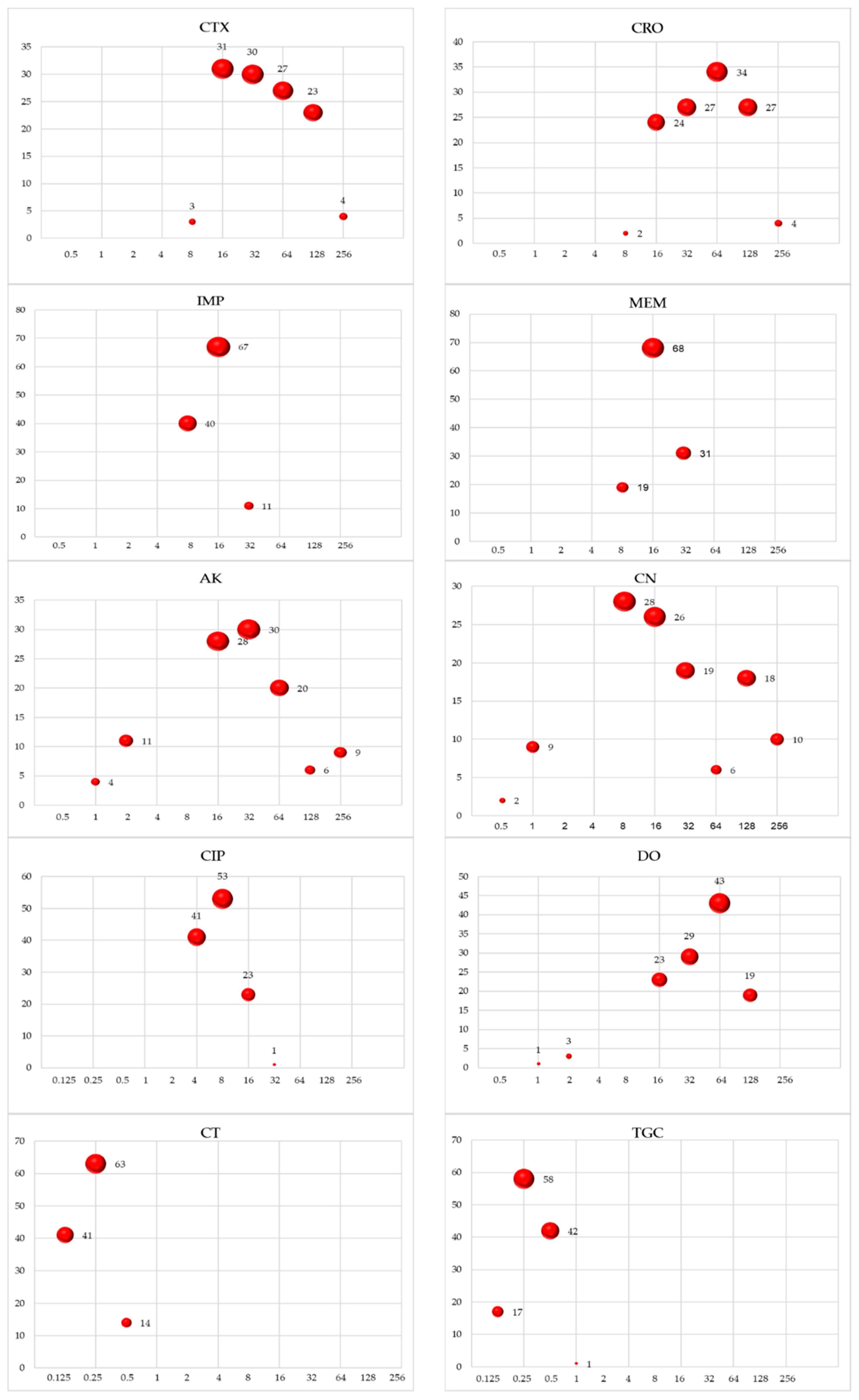
3.1. Antimicrobial Resistance
Characterization of Antibiotic Resistance Genes (ARGs)
3.2. Prevalence of Virulence Genes
3.3. Sequence Types of the Carb-R EPEC Isolates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sohail, M.; Khurshid, M.; Saleem, H.G.; Javed, H.; Khan, A.A. Characteristics and Antibiotic Resistance of Urinary Tract Pathogens Isolated from Punjab, Pakistan. Jundishapur J. Microbiol. 2015, 8, e19272. [Google Scholar] [CrossRef] [PubMed]
- Toval, F.; Köhler, C.D.; Vogel, U.; Wagenlehner, F.; Mellmann, A.; Fruth, A.; Schmidt, M.A.; Karch, H.; Bielaszewska, M.; Dobrindt, U. Characterization of Escherichia coli isolates from hospital inpatients or outpatients with urinary tract infection. J. Clin. Microbiol. 2014, 52, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Abd El Ghany, M.; Sharaf, H.; Al-Agamy, M.H.; Shibl, A.; Hill-Cawthorne, G.A.; Hong, P.Y. Genomic characterization of NDM-1 and 5, and OXA-181 carbapenemases in uropathogenic Escherichia coli isolates from Riyadh, Saudi Arabia. PLoS ONE 2018, 13, e0201613. [Google Scholar] [CrossRef]
- Terlizzi, M.E.; Gribaudo, G.; Maffei, M.E. UroPathogenic Escherichia coli (UPEC) Infections: Virulence Factors, Bladder Responses, Antibiotic, and Non-antibiotic Antimicrobial Strategies. Front. Microbiol. 2017, 8, 1566. [Google Scholar] [CrossRef]
- Whelan, S.; Lucey, B.; Finn, K. Uropathogenic Escherichia coli (UPEC)-Associated Urinary Tract Infections: The Molecular Basis for Challenges to Effective Treatment. Microorganisms 2023, 11, 2169. [Google Scholar] [CrossRef]
- Lee, D.S.; Lee, S.J.; Choe, H.S. Community-Acquired Urinary Tract Infection by Escherichia coli in the Era of Antibiotic Resistance. BioMed Res. Int. 2018, 2018, 7656752. [Google Scholar] [CrossRef]
- Byarugaba, D.K.; Erima, B.; Wokorach, G.; Alafi, S.; Kibuuka, H.; Mworozi, E.; Musinguzi, A.K.; Kiyengo, J.; Najjuka, F.; Wabwire-Mangen, F. Resistome and virulome of high-risk pandemic clones of multidrug-resistant extra-intestinal pathogenic Escherichia coli (ExPEC) isolated from tertiary healthcare settings in Uganda. PLoS ONE 2023, 18, e0294424. [Google Scholar] [CrossRef] [PubMed]
- Pitout, J.D.D.; Peirano, G.; Chen, L.; DeVinney, R.; Matsumura, Y. Escherichia coli ST1193: Following in the Footsteps of E. coli ST131. Antimicrob. Agents Chemother. 2022, 66, e0051122. [Google Scholar] [CrossRef]
- García-Meniño, I.; García, V.; Lumbreras-Iglesias, P.; Fernández, J.; Mora, A. Fluoroquinolone resistance in complicated urinary tract infections: Association with the increased occurrence and diversity of Escherichia coli of clonal complex 131, together with ST1193. Front. Cell. Infect. Microbiol. 2024, 14, 1351618. [Google Scholar] [CrossRef]
- Huang, J.; Lv, C.; Li, M.; Rahman, T.; Chang, Y.F.; Guo, X.; Song, Z.; Zhao, Y.; Li, Q.; Ni, P.; et al. Carbapenem-resistant Escherichia coli exhibit diverse spatiotemporal epidemiological characteristics across the globe. Commun. Biol. 2024, 7, 51. [Google Scholar] [CrossRef]
- Pöntinen, A.K.; Gladstone, R.A.; Pesonen, H.; Pesonen, M.; Cléon, F.; Parcell, B.J.; Kallonen, T.; Simonsen, G.S.; Croucher, N.J.; McNally, A.; et al. Modulation of multidrug-resistant clone success in Escherichia coli populations: A longitudinal, multi-country, genomic and antibiotic usage cohort study. Lancet Microbe 2024, 5, e142–e150. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Fernandez, A.; Villa, L.; Bibbolino, G.; Bressan, A.; Trancassini, M.; Pietropaolo, V.; Venditti, M.; Antonelli, G.; Carattoli, A. Novel Insights and Features of the NDM-5-Producing Escherichia coli Sequence Type 167 High-Risk Clone. mSphere 2020, 5, e00269-20. [Google Scholar] [CrossRef] [PubMed]
- Grönthal, T.; Österblad, M.; Eklund, M.; Jalava, J.; Nykäsenoja, S.; Pekkanen, K.; Rantala, M. Sharing more than friendship—Transmission of NDM-5 ST167 and CTX-M-9 ST69 Escherichia coli between dogs and humans in a family, Finland, 2015. Euro Surveill 2018, 23, 1700497. [Google Scholar] [CrossRef]
- Dwivedi, A.; Kumar, C.B.; Kumar, A.; Soni, M.; Sahu, V.; Awasthi, A.; Rathore, G. Molecular characterization of carbapenem resistant E. coli of fish origin reveals the dissemination of NDM-5 in freshwater aquaculture environment by the high risk clone ST167 and ST361. Environ. Sci. Pollut. Res. Int. 2023, 30, 49314–49326. [Google Scholar] [CrossRef] [PubMed]
- Alghoribi, M.F.; Gibreel, T.M.; Farnham, G.; Al Johani, S.M.; Balkhy, H.H.; Upton, M. Antibiotic-resistant ST38, ST131 and ST405 strains are the leading uropathogenic Escherichia coli clones in Riyadh, Saudi Arabia. J. Antimicrob. Chemother. 2015, 70, 2757–2762. [Google Scholar] [CrossRef]
- Chowdhury, P.R.; McKinnon, J.; Liu, M.; Djordjevic, S.P. Multidrug Resistant Uropathogenic Escherichia coli ST405 with a Novel, Composite IS26 Transposon in a Unique Chromosomal Location. Front. Microbiol. 2018, 9, 3212. [Google Scholar] [CrossRef]
- Gondal, A.J.; Choudhry, N.; Bukhari, H.; Rizvi, Z.; Yasmin, N. Characterization of Genomic Diversity among Carbapenem-Resistant Escherichia coli Clinical Isolates and Antibacterial Efficacy of Silver Nanoparticles from Pakistan. Microorganisms 2022, 10, 2283. [Google Scholar] [CrossRef]
- Habib, A.; Lo, S.; Villageois-Tran, K.; Petitjean, M.; Malik, S.A.; Armand-Lefèvre, L.; Ruppé, E.; Zahra, R. Dissemination of carbapenemase-producing Enterobacterales in the community of Rawalpindi, Pakistan. PLoS ONE 2022, 17, e0270707. [Google Scholar] [CrossRef]
- Khan, A.Y.; Ahmad, S.S.; Avais, M.; Ashraf, K. Molecular prevalence with associated risk factors and haemato-serum electrolyte analysis of E. coli O157: H7 in Canine pups with diarrhoea. Pak. Vet. J. 2022, 42, 161–166. [Google Scholar]
- Li, X.; Zhu, X.; Xue, Y. Drug resistance and genetic relatedness of Escherichia coli from mink in Northeast China. Pak. Vet. J. 2023, 43, 824–827. [Google Scholar]
- Tian, X.; Zheng, X.; Sun, Y.; Fang, R.; Zhang, S.; Zhang, X.; Lin, J.; Cao, J.; Zhou, T. Molecular Mechanisms and Epidemiology of Carbapenem-Resistant Escherichia coli Isolated from Chinese Patients During 2002–2017. Infect. Drug Resist. 2020, 13, 501–512. [Google Scholar] [CrossRef] [PubMed]
- von Wintersdorff, C.J.; Penders, J.; van Niekerk, J.M.; Mills, N.D.; Majumder, S.; van Alphen, L.B.; Savelkoul, P.H.; Wolffs, P.F. Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer. Front. Microbiol. 2016, 7, 173. [Google Scholar] [CrossRef] [PubMed]
- Coque, T.M.; Novais, A.; Carattoli, A.; Poirel, L.; Pitout, J.; Peixe, L.; Baquero, F.; Cantón, R.; Nordmann, P. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase CTX-M-15. Emerg. Infect. Dis. 2008, 14, 195–200. [Google Scholar] [CrossRef]
- Tian, G.B.; Rivera, J.I.; Park, Y.S.; Johnson, L.E.; Hingwe, A.; Adams-Haduch, J.M.; Doi, Y. Sequence type ST405 Escherichia coli isolate producing QepA1, CTX-M-15, and RmtB from Detroit, Michigan. Antimicrob. Agents Chemother. 2011, 55, 3966–3967. [Google Scholar] [CrossRef]
- Shin, J.; Kim, D.H.; Ko, K.S. Comparison of CTX-M-14- and CTX-M-15-producing Escherichia coli and Klebsiella pneumoniae isolates from patients with bacteremia. J. Infect. 2011, 63, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Ye, K.; Li, X.; Ye, L.; Guo, L.; Wang, L.; Yang, J. Genetic characterization of Carbapenem-Resistant Escherichia coli from China, 2015–2017. BMC Microbiol. 2021, 21, 248. [Google Scholar] [CrossRef]
- Peirano, G.; Pitout, J.D.D. Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae: Update on Molecular Epidemiology and Treatment Options. Drugs 2019, 79, 1529–1541. [Google Scholar] [CrossRef]
- Bitar, I.; Piazza, A.; Gaiarsa, S.; Villa, L.; Pedroni, P.; Oliva, E.; Nucleo, E.; Pagani, L.; Carattoli, A.; Migliavacca, R. ST405 NDM-5 producing Escherichia coli in Northern Italy: The first two clinical cases. Clin. Microbiol. Infect. 2017, 23, 489–490. [Google Scholar] [CrossRef]
- Sumbana, J.J.; Santona, A.; Fiamma, M.; Taviani, E.; Deligios, M.; Zimba, T.; Lucas, G.; Sacarlal, J.; Rubino, S.; Paglietti, B. Extraintestinal Pathogenic Escherichia coli ST405 Isolate Coharboring blaNDM-5 and blaCTXM-15: A New Threat in Mozambique. Microb. Drug Resist. 2021, 27, 1633–1640. [Google Scholar] [CrossRef]
- Corbellini, S.; Scaltriti, E.; Piccinelli, G.; Gurrieri, F.; Mascherpa, M.; Boroni, G.; Amolini, C.; Caruso, A.; De Francesco, M.A. Genomic characterisation of Escherichia coli isolates co-producing NDM-5 and OXA-1 from hospitalised patients with invasive infections. J. Glob. Antimicrob. Resist. 2022, 28, 136–139. [Google Scholar] [CrossRef]
- Slown, S.; Walas, N.; Amato, H.K.; Lloyd, T.; Varghese, V.; Bender, M.; Pandori, M.; Graham, J. Clonal Lineages and Virulence Factors of Carbapenem Resistant E. coli in Alameda County, California, 2017–2019. Antibiotics 2022, 11, 1794. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Ortiz de la Rosa, J.M.; Sakaoglu, Z.; Kusaksizoglu, A.; Sadek, M.; Nordmann, P. NDM-35-Producing ST167 Escherichia coli Highly Resistant to β-Lactams Including Cefiderocol. Antimicrob. Agents Chemother. 2022, 66, e0031122. [Google Scholar] [CrossRef] [PubMed]
- Peterhans, S.; Stevens, M.J.A.; Nüesch-Inderbinen, M.; Schmitt, S.; Stephan, R.; Zurfluh, K. First report of a bla(NDM-5)-harbouring Escherichia coli ST167 isolated from a wound infection in a dog in Switzerland. J. Glob. Antimicrob. Resist. 2018, 15, 226–227. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Wang, P.; Cheng, J.; Qin, S.; Xie, W. Characterization of a novel bla (NDM-5)-harboring IncFII plasmid and an mcr-1-bearing IncI2 plasmid in a single Escherichia coli ST167 clinical isolate. Infect. Drug Resist. 2019, 12, 511–519. [Google Scholar] [CrossRef]
- Manyahi, J.; Moyo, S.J.; Kibwana, U.; Goodman, R.N.; Allman, E.; Hubbard, A.T.M.; Blomberg, B.; Langeland, N.; Roberts, A.P. First identification of bla (NDM-5) producing Escherichia coli from neonates and a HIV infected adult in Tanzania. J. Med. Microbiol. 2022, 71, 001513. [Google Scholar] [CrossRef]
- Ragupathi, N.K.D.; Veeraraghavan, B.; Sethuvel, D.P.M.; Anandan, S.; Vasudevan, K.; Neeravi, A.R.; Daniel, J.L.K.; Sathyendra, S.; Iyadurai, R.; Mutreja, A. First Indian report on genome-wide comparison of multidrug-resistant Escherichia coli from blood stream infections. PLoS ONE 2020, 15, e0220428. [Google Scholar]
- Fuga, B.; Sellera, F.P.; Cerdeira, L.; Esposito, F.; Cardoso, B.; Fontana, H.; Moura, Q.; Cardenas-Arias, A.; Sano, E.; Ribas, R.M.; et al. WHO Critical Priority Escherichia coli as One Health Challenge for a Post-Pandemic Scenario: Genomic Surveillance and Analysis of Current Trends in Brazil. Microbiol. Spectr. 2022, 10, e0125621. [Google Scholar] [CrossRef]
- Bojesen, A.M.; Ahmed, U.; Skaarup, H.; Espinosa-Gongora, C. Recurring outbreaks by the same Escherichia coli ST10 clone in a broiler unit during 18 months. Vet. Res. 2022, 53, 2. [Google Scholar] [CrossRef]
- Kudinha, T.; Kong, F. Possible step-up in prevalence for Escherichia coli ST131 from fecal to clinical isolates: Inferred virulence potential comparative studies within phylogenetic group B2. J. Biomed. Sci. 2022, 29, 78. [Google Scholar] [CrossRef]
- Linkevicius, M.; Bonnin, R.A.; Alm, E.; Svartström, O.; Apfalter, P.; Hartl, R.; Hasman, H.; Roer, L.; Räisänen, K.; Dortet, L.; et al. Rapid cross-border emergence of NDM-5-producing Escherichia coli in the European Union/European Economic Area, 2012 to June 2022. Eurosurveillance 2023, 28, 2300209. [Google Scholar] [CrossRef]

| Resistance Traits | Total Isolates n = 118 n (%) | Hospital A n = 59 n (%) | Hospital B n = 33 n (%) | Hospital C n = 26 n (%) | p-Value |
|---|---|---|---|---|---|
| Antimicrobial agents | |||||
| Amoxicillin-clavulanate | 118 (100) | 59 (100) | 33 (100) | 26 (100) | - |
| Piperacillin-tazobactam | 118 (100) | 59 (100) | 33 (100) | 26 (100) | - |
| Cefotaxime | 118 (100) | 59 (100) | 33 (100) | 26 (100) | - |
| Ceftriaxone | 118 (100) | 59 (100) | 33 (100) | 26 (100) | - |
| Imipenem | 118 (100) | 59 (100) | 33 (100) | 26 (100) | - |
| Meropenem | 118 (100) | 59 (100) | 33 (100) | 26 (100) | - |
| Amikacin | 103 (87.3) | 51 (86.4) | 30 (90.9) | 22 (84.6) | 0.742 |
| Gentamicin | 107 (90.7) | 51 (86.4) | 30 (90.9) | 26 (100) | 0.14 |
| Doxycycline | 114 (96.6) | 56 (94.9) | 33 (100) | 25 (96.2) | 0.429 |
| Nalidixic acid | 118 (100) | 59 (100) | 33 (100) | 26 (100) | - |
| Ciprofloxacin | 118 (100) | 59 (100) | 33 (100) | 26 (100) | - |
| Sulfamethoxazole-trimethoprim | 114 (96.6) | 59 (100) | 30 (90.9) | 25 (96.2) | 0.069 |
| Nitrofurantoin | 15 (12.7) | 7 (11.9) | 3 (9.1) | 5 (19.2) | 0.491 |
| Fosfomycin | 9 (7.6) | - | - | 9 (34.6) | <0.001 |
| Colistin | - | - | - | - | - |
| Tigecycline | - | - | - | - | - |
| Resistance genes | |||||
| blaSHV | 15 (12.7) | 7 (11.9) | - | 8 (30.8) | 0.002 |
| blaTEM | 47 (39.8) | 25 (42.4) | 12 (36.4) | 10 (38.5) | 0.842 |
| blaCTXM-1 | 12 (10.2) | 8 (13.6) | - | 4 (15.4) | 0.072 |
| blaCTXM-15 | 97 (82.2) | 44 (74.6) | 28 (84.8) | 25 (96.2) | 0.051 |
| blaOXA-48 | 14 (11.9) | 3 (5.1) | - | 11 (42.3) | <0.001 |
| blaNDM-1 | 32 (27.1) | 13 (22.0) | 6 (18.2) | 13 (50) | 0.011 |
| blaNDM-5 | 86 (72.9) | 46 (78) | 27 (81.8) | 13 (50) | 0.011 |
| aac(6′)-Ib | 91 (77.1) | 45 (76.3) | 25 (75.8) | 21 (80.8) | 0.88 |
| aph(3″)-Ib | 89 (75.4) | 43 (72.9) | 22 (66.7) | 24 (92.3) | 0.062 |
| ant(2″)-Ia | 9 (7.6) | - | - | 9 (34.6) | <0.001 |
| rmtB | 22 (18.6) | 12 (20.3) | 7 (21.2) | 3 (11.5) | 0.571 |
| tetA | 45 (38.1) | 22 (37.3) | 7 (21.2) | 16 (61.5) | 0.007 |
| tetB | 90 (76.3) | 42 (71.2) | 28 (84.8) | 20 (76.9) | 0.334 |
| qnrB | 8 (6.8) | 5 (8.5) | 3 (9.1) | - | 0.296 |
| qnrS | 10 (8.5) | 9 (15.3) | - | 1 (3.8) | 0.026 |
| qepA | 62 (52.5) | 29 (49.2) | 18 (54.5) | 15 (57.7) | 0.74 |
| sul1 | 104 (88.1) | 58 (98.3) | 30 (90.9) | 16 (61.5) | <0.001 |
| sul2 | 25 (21.2) | 13 (22) | 3 (9.1) | 9 (34.6) | 0.057 |
| Virulence genes | |||||
| traT | 110 (93.2) | 54 (91.5) | 30 (90.9) | 26 (100) | 0.296 |
| iutA | 105 (89) | 51 (86.4) | 28 (84.8) | 26 (100) | 0.123 |
| irp2 | 81 (68.6) | 43 (72.9) | 25 (75.8) | 13 (50) | 0.065 |
| hylA | 54 (45.8) | 25 (42.4) | 11 (33.3) | 18 (69.2) | 0.017 |
| fyuA | 98 (83.1) | 51 (86.4) | 30 (90.9) | 17 (65.4) | 0.021 |
| papC | 94 (79.7) | 51 (86.4) | 30 (90.9) | 13 (50) | <0.001 |
| papG | 80 (67.8) | 45 (76.3) | 27 (81.8) | 8 (30.8) | <0.001 |
| capU | 34 (28.8) | 13 (22) | 6 (18.2) | 15 (57.7) | 0.001 |
| fimH | 114 (96.6) | 56 (94.9) | 33 (100) | 25 (96.2) | 0.429 |
| kpsMTII | 93 (78.8) | 51 (86.4) | 30 (90.9) | 12 (46.2) | <0.001 |
| Resistance Traits | ST405 (n = 42) | ST167 (n = 25) | ST10 (n = 11) | ST101 (n = 9) | ST131 (n = 8) | ST940 (n = 6) | ST648 (n = 5) | ST410 (n = 4) | ST1702 (n = 4) | ST2851 (n = 4) | p-Value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | ||
| Amoxicillin-clavulanate | 42 (100) | 25 (100) | 11 (100) | 9 (100) | 8 (100) | 6 (100) | 5 (100) | 4 (100) | 4 (100) | 4 (100) | - |
| Piperacillin-tazobactam | 42 (100) | 25 (100) | 11 (100) | 9 (100) | 8 (100) | 6 (100) | 5 (100) | 4 (100) | 4 (100) | 4 (100) | - |
| Cefotaxime | 42 (100) | 25 (100) | 11 (100) | 9 (100) | 8 (100) | 6 (100) | 5 (100) | 4 (100) | 4 (100) | 4 (100) | - |
| Ceftriaxone | 42 (100) | 25 (100) | 11 (100) | 9 (100) | 8 (100) | 6 (100) | 5 (100) | 4 (100) | 4 (100) | 4 (100) | - |
| Imipenem | 42 (100) | 25 (100) | 11 (100) | 9 (100) | 8 (100) | 6 (100) | 5 (100) | 4 (100) | 4 (100) | 4 (100) | - |
| Meropenem | 42 (100) | 25 (100) | 11 (100) | 9 (100) | 8 (100) | 6 (100) | 5 (100) | 4 (100) | 4 (100) | 4 (100) | - |
| Amikacin | 42 (100) | 25 (100) | - | 9 (100) | 8 (100) | 6 (100) | 5 (100) | 4 (100) | - | 4 (100) | <0.001 |
| Gentamicin | 42 (100) | 25 (100) | - | 9 (100) | 8 (100) | 6 (100) | 5 (100) | 4 (100) | 4 (100) | 4 (100) | <0.001 |
| Doxycycline | 42 (100) | 25 (100) | 11 (100) | 9 (100) | 8 (100) | 6 (100) | 5 (100) | - | 4 (100) | 4 (100) | <0.001 |
| Nalidixic acid | 42 (100) | 25 (100) | 11 (100) | 9 (100) | 8 (100) | 6 (100) | 5 (100) | 4 (100) | 4 (100) | 4 (100) | - |
| Ciprofloxacin | 42 (100) | 25 (100) | 11 (100) | 9 (100) | 8 (100) | 6 (100) | 5 (100) | 4 (100) | 4 (100) | 4 (100) | - |
| Sulfamethoxazole-trimethoprim | 42 (100) | 25 (100) | 11 (100) | 9 (100) | 8 (100) | 2 (33.3) | 5 (100) | 4 (100) | 4 (100) | 4 (100) | <0.001 |
| Nitrofurantoin | - | - | 10 (90.9) | 5 (55.6) | - | - | - | - | - | - | <0.001 |
| Fosfomycin | - | - | - | 9 (100) | - | - | - | - | - | - | <0.001 |
| Colistin | - | - | - | - | - | - | - | - | - | - | - |
| Tigecycline | - | - | - | - | - | - | - | - | - | - | - |
| Resistance genes | |||||||||||
| blaSHV | - | - | 7 (63.6) | 8 (88.9) | - | - | - | - | - | - | <0.001 |
| blaTEM | 6 (14.3) | 4 (16) | 11 (100.0) | 6 (66.7) | 5 (62.5) | 6 (100) | 5 (100) | 4 (100) | - | - | <0.001 |
| blaCTXM-1 | - | - | 8 (72.7) | - | - | - | - | - | 4 (100) | - | <0.001 |
| blaCTXM-15 | 42 (100) | 21 (84) | 3 (27.3) | 9 (100) | 8 (100) | - | 2 (40) | 4 (100) | 4 (100) | 4 (100) | <0.001 |
| blaOXA-48 | - | 4 (16) | 1 (9.1) | 9 (100) | - | - | - | - | - | - | <0.001 |
| blaNDM-1 | - | - | 11 (100) | 9 (100) | 8 (100) | - | - | - | 4 (100) | - | <0.001 |
| blaNDM-5 | 42 (100) | 25 (100) | - | - | - | 6 (100) | 5 (100) | 4 (100) | - | 4 (100) | <0.001 |
| aac(6′)-Ib | 42 (100) | 25 (100) | - | 9 (100) | 8 (100) | 6 (100) | - | - | - | 1 (25) | <0.001 |
| aph(3″)-Ib | 42 (100) | 25 (100) | - | 9 (100) | 8 (100) | - | - | - | 4 (100) | 1 (25) | <0.001 |
| ant(2″)-Ia | - | - | - | 9 (100) | - | - | - | - | - | - | <0.001 |
| rmtB | 10 (23.8) | - | - | - | - | - | 5 (100) | 4 (100) | - | 3 (75) | <0.001 |
| tetA | 16 (38.1) | 14 (56) | - | 9 (100) | - | - | 2 (40) | - | - | 4 (100) | <0.001 |
| tetB | 36 (85.7) | 11 (44) | 11 (100) | 9 (100) | 8 (100) | 6 (100) | 5 (100) | - | 4 (100) | - | <0.001 |
| qnrB | - | - | - | - | 8 (100) | - | - | - | - | - | <0.001 |
| qnrS | - | 4 (16) | - | - | - | 2 (33.3) | - | 4 (100) | - | - | <0.001 |
| qepA | 42 (100) | 11 (44) | - | 9 (100) | - | - | - | - | - | - | <0.001 |
| sul1 | 42 (100) | 25 (100) | 10 (90.9) | - | 8 (100) | 2 (33.3) | 5 (100) | 4 (100) | 4 (100) | 4 (100) | <0.001 |
| sul2 | - | - | 8 (72.7) | 9 (100) | 8 (100) | - | - | - | - | - | <0.001 |
| Virulence factors | |||||||||||
| traT | 42 (100) | 25 (100) | 11 (100) | 9 (100) | - | 6 (100) | 5 (100) | 4 (100) | 4 (100) | 4 (100) | <0.001 |
| iutA | 42 (100) | 25 (100) | 11 (100) | 9 (100) | - | 6 (100) | - | 4 (100) | 4 (100) | 4 (100) | <0.001 |
| irp2 | 42 (100) | 25 (100) | - | - | - | 6 (100) | - | 4 (100) | - | 4 (100) | <0.001 |
| shylA | - | 25 (100) | 11 (100) | 9 (100) | - | - | 5 (100) | - | 4 (100) | - | <0.001 |
| fyuA | 42 (100) | 25 (100) | - | - | - | 6 (100) | 5 (100) | 4 (100) | 4 (100) | 4 (100) | <0.001 |
| papC | 42 (100) | 25 (100) | - | - | 8 (100) | 6 (100) | 5 (100) | 4 (100) | - | 4 (100) | <0.001 |
| papG | 42 (100) | - | 11 (100) | - | 8 (100) | 6 (100) | 5 (100) | 4 (100) | - | 4 (100) | <0.001 |
| capU | - | - | 11 (100) | 9 (100) | - | 6 (100) | - | 4 (100) | 4 (100) | - | <0.001 |
| fimH | 42 (100) | 25 (100) | 11 (100) | 9 (100) | 8 (100) | 6 (100) | 5 (100) | - | 4 (100) | 4 (100) | <0.001 |
| kpsMTII | 42 (100) | 25 (100) | 11 (100) | - | - | 6 (100) | 5 (100) | - | - | 4 (100) | <0.001 |
| STs | Co-Occurrence of Resistance Genes (n) | Antimicrobial Resistant Phenotype (n) | Co-Occurrence of Virulence Genes |
|---|---|---|---|
| ST405 (n = 42) | blaTEM, blaCTX-M-15, blaNDM-5, aac(6′)-Ib, aph(3″)-Ib, tetA, qepA, sul1 (06) | AMC, TZP, CTX, CRO, IPM, MEM, AK, CN, DO, NA, CIP, SXT (42) | traT, iutA, irp2, fyuA, papC, papG, fimH, kpsMTII |
| blaCTX-M-15, blaNDM-5, aac(6′)-Ib, aph(3″)-Ib, rmtB, tetA, tetB, qepA, sul1 (10) | |||
| blaCTX-M-15, blaNDM-5, aac(6′)-Ib, aph(3″)-Ib, tetB, qepA, sul1 (26) | |||
| ST167 (n = 25) | blaCTX-M-15, blaNDM-5, aac(6′)-Ib, aph(3″)-Ib, tetB, qepA, sul1 (11) | AMC, TZP, CTX, CRO, IPM, MEM, AK, CN, DO, NA, CIP, SXT (25) | traT, iutA, irp2, hylA, fyuA, papC, fimH, kpsMTII |
| blaCTX-M-15, blaNDM-5, aac(6′)-Ib, aph(3″)-Ib, tetA, sul1 (06) | |||
| blaTEM, blaCTX-M-15, blaOXA-48, blaNDM-5, aac(6′)-Ib, aph(3″)-Ib, tetA, sul1 (04) | |||
| blaNDM-5, aac(6′)-Ib, aph(3″)-Ib, tetA, qnrS, sul1 (04) | |||
| ST10 (n = 11) | blaSHV, blaTEM, blaCTX-M-1, blaNDM-1, tetB, sul1, sul2 (07) | AMC, TZP, CTX, CRO, IPM, MEM, DO, NA, CIP, SXT, F (10) | traT, iutA, hylA, papG, capU, fimH, kpsMTII |
| blaTEM, blaCTX-M-15, blaNDM-1, tetB, sul1 (03) | |||
| blaTEM, blaCTX-M-1, blaOXA-48, blaNDM-1, tetB, sul2 (01) | AMC, TZP, CTX, CRO, IPM, MEM, DO, NA, CIP, SXT (01) | ||
| ST101 (n = 9) | blaSHV, blaTEM, blaCTX-M-15, blaOXA-48, blaNDM-1, aac(6′)-Ib, aph(3″)-Ib, ant(2″)-Ia, tetA, tetB, qepA, sul2 (05) | AMC, TZP, CTX, CRO, IPM, MEM, AK, CN, DO, NA, CIP, SXT, F, FOS (05) | traT, iutA, hylA, capU, FimH |
| blaSHV, blaCTX-M-15, blaOXA-48, blaNDM-1, aac(6′)-Ib, aph(3″)-Ib, ant(2″)-Ia, tetA, tetB, qepA, sul2 (03) | AMC, TZP, CTX, CRO, IPM, MEM, AK, CN, DO, NA, CIP, SXT, FOS (04) | ||
| blaTEM, blaCTX-M-15, blaOXA-48, blaNDM-1, aac(6′)-Ib, aph(3″)-Ib, ant(2″)-Ia, tetA, tetB, qepA, sul2 (01) | |||
| ST131 (n = 8) | blaTEM, blaCTX-M-15, blaNDM-1, aac(6′)-Ib, aph(3″)-Ib, tetB, qnrB, sul1 (05) | AMC, TZP, CTX, CRO, IPM, MEM, AK, CN, DO, NA, CIP, SXT (08) | fyuA, papC, papG, capU, FimH |
| blaCTX-M-15, blaNDM-1, aac(6′)-Ib, aph(3″)-Ib, tetB, qnrB, sul1, sul2 (03) | |||
| ST940 (n = 6) | blaTEM, aac(6′)-Ib, blaNDM-5, tetB (04) | AMC, TZP, CTX, CRO, IPM, MEM, AK, CN, DO, NA, CIP (06) | traT, iutA, irp2, fyuA, papC, papG, fimH, kpsMTII |
| blaNDM-5, aac(6′)-Ib, tetB,qnrS, sul1 (02) | |||
| ST648 (n = 5) | blaTEM, blaNDM-5, rmtB, tetB, sul1 (03) | AMC, TZP, CTX, CRO, IPM, MEM, AK, CN, DO, NA, CIP, SXT (05) | traT, hylA, fyuA, papC, papG, fimH, kpsMTII |
| blaTEM, blaCTX-M-15, blaNDM-5, rmtB tetA, tetB, sul1 (02) | |||
| ST410 (n = 4) | blaCTX-M-15, blaNDM-5, rmtB, sul1 (04) | AMC, TZP, CTX, CRO, IPM, MEM, AK, CN, NA, CIP, SXT (04) | traT, iutA, irp2, fyuA, papC, papG, capU |
| ST1702 (n = 4) | blaCTX-M-1, blaCTX-M-15, blaNDM-1, aph(3″)-Ib, tetB, sul1 (04) | AMC, TZP, CTX, CRO, IPM, MEM, CN, DO, NA, CIP, SXT (04) | traT, iutA, hylA, fyuA, capU, FimH |
| ST2851 (n = 4) | blaCTX-M-15, blaNDM-5, rmtB, tetA, sul1 (03) | AMC, TZP, CTX, CRO, IPM, MEM, AK, CN, DO, NA, CIP, SXT (04) | traT, iutA, irp2, fyuA, papC, papG, fimH, kpsMTII |
| blaCTX-M-15, blaNDM-5, aac(6′)-Ib, aph(3″)-Ib, tetA, sul1 (01) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mujahid, F.; Rasool, M.H.; Shafiq, M.; Aslam, B.; Khurshid, M. Emergence of Carbapenem-Resistant Uropathogenic Escherichia coli (ST405 and ST167) Strains Carrying blaCTX-M-15, blaNDM-5 and Diverse Virulence Factors in Hospitalized Patients. Pathogens 2024, 13, 964. https://doi.org/10.3390/pathogens13110964
Mujahid F, Rasool MH, Shafiq M, Aslam B, Khurshid M. Emergence of Carbapenem-Resistant Uropathogenic Escherichia coli (ST405 and ST167) Strains Carrying blaCTX-M-15, blaNDM-5 and Diverse Virulence Factors in Hospitalized Patients. Pathogens. 2024; 13(11):964. https://doi.org/10.3390/pathogens13110964
Chicago/Turabian StyleMujahid, Fatima, Muhammad Hidayat Rasool, Muhammad Shafiq, Bilal Aslam, and Mohsin Khurshid. 2024. "Emergence of Carbapenem-Resistant Uropathogenic Escherichia coli (ST405 and ST167) Strains Carrying blaCTX-M-15, blaNDM-5 and Diverse Virulence Factors in Hospitalized Patients" Pathogens 13, no. 11: 964. https://doi.org/10.3390/pathogens13110964
APA StyleMujahid, F., Rasool, M. H., Shafiq, M., Aslam, B., & Khurshid, M. (2024). Emergence of Carbapenem-Resistant Uropathogenic Escherichia coli (ST405 and ST167) Strains Carrying blaCTX-M-15, blaNDM-5 and Diverse Virulence Factors in Hospitalized Patients. Pathogens, 13(11), 964. https://doi.org/10.3390/pathogens13110964








