Identification of New Single Nucleotide Polymorphisms Potentially Related to Small Ruminant Lentivirus Infection Susceptibility in Goats Based on Data Selected from High-Throughput Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Blood Samples
2.2. Serological Testing of Serum Samples
2.3. Quantification of SRLV Proviral DNA
2.4. SNPs Identification Based on RNA-Seq Data
2.5. Polymorphism Selection and Genotyping
2.6. Statistical Analysis
3. Results
3.1. Serological Status of Goats
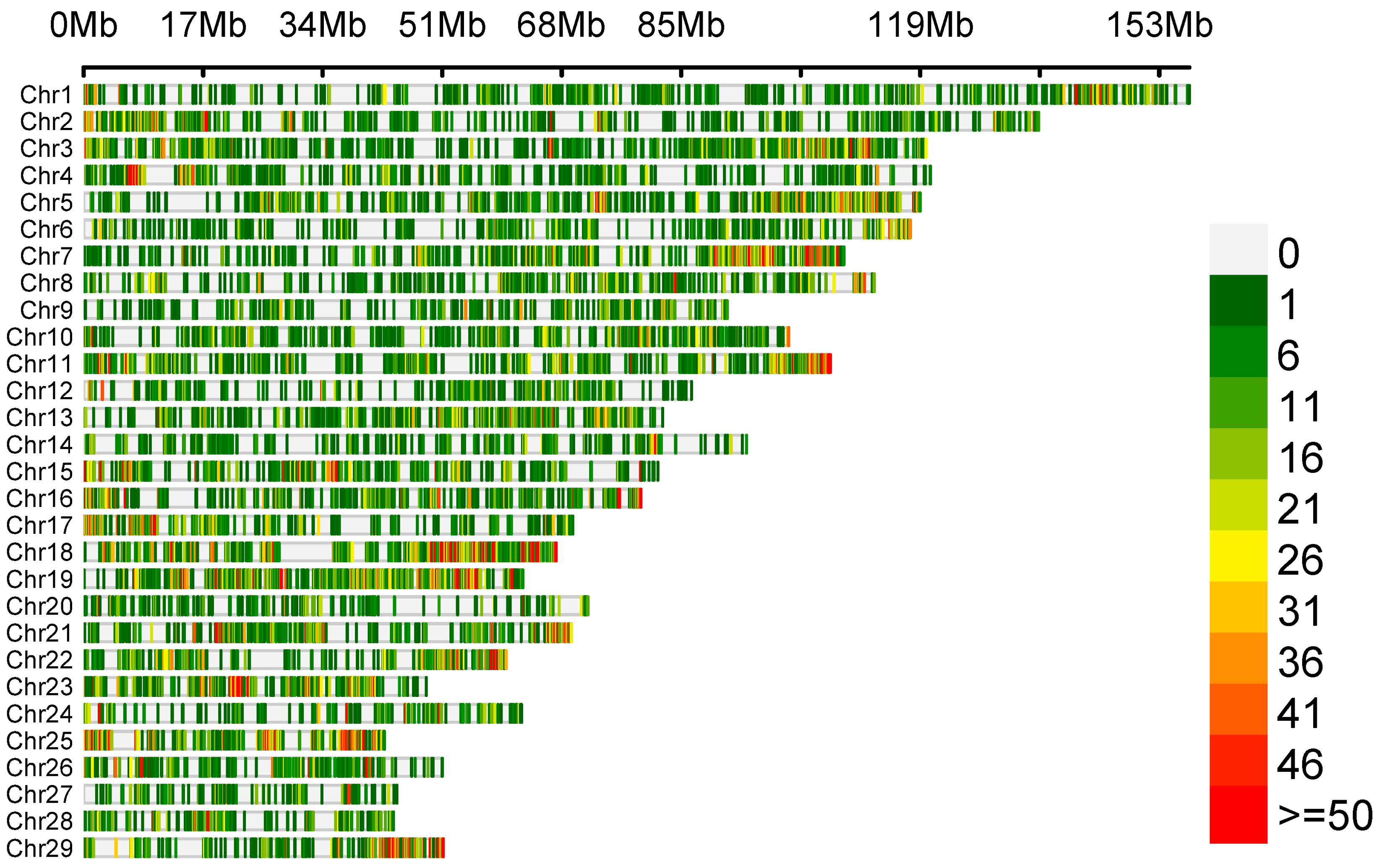
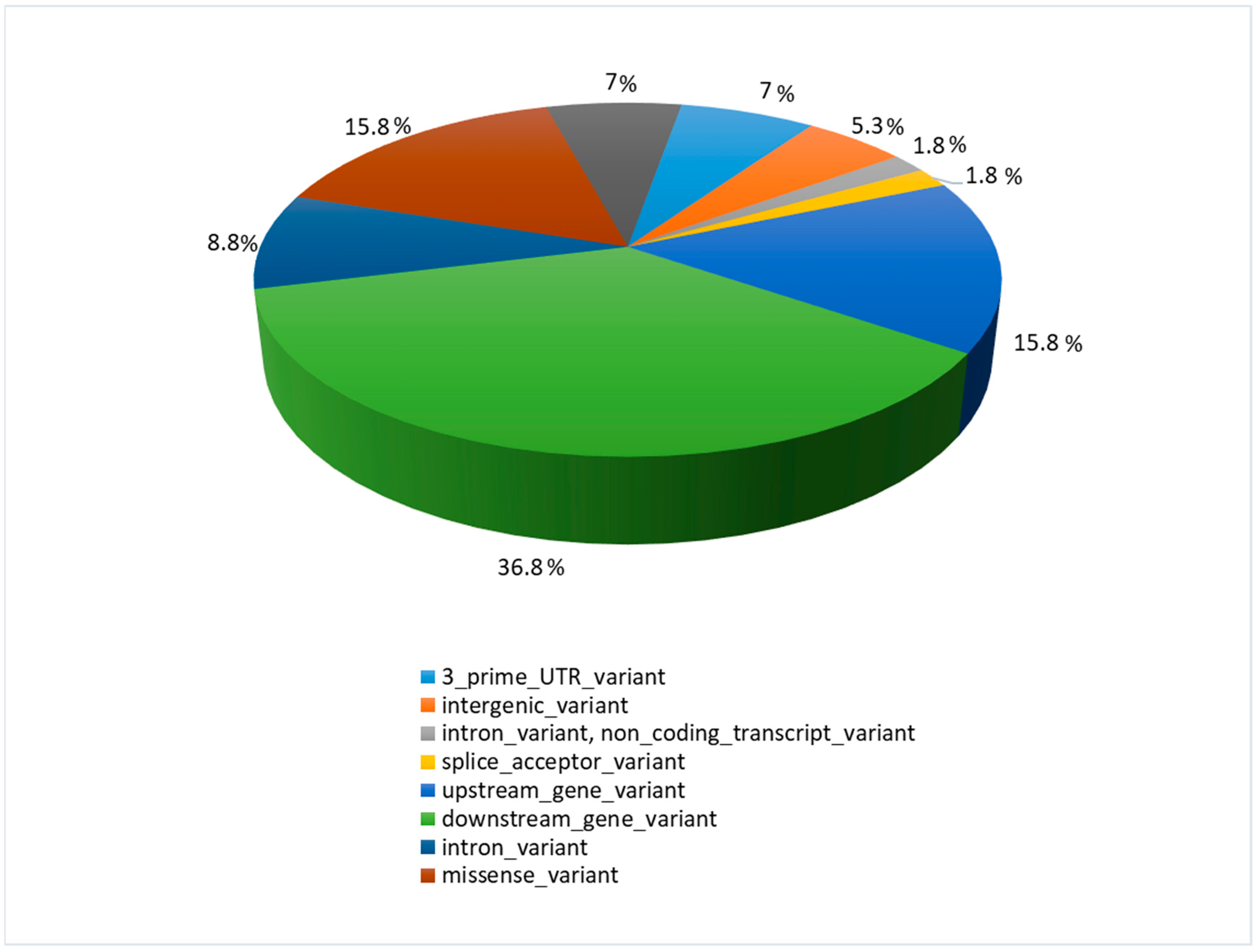
3.2. Whole-Transcriptome SNP Identification Results
3.3. Detected SNPs, Allele and Genotype Frequency
3.4. Association between SNPs and Provirus Copy Number
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Colitti, B.; Coradduzza, E.; Puggioni, G.; Capucchio, M.T.; Reina, R.; Bertolotti, L.; Rosati, S. A new approach for Small Ruminant Lentivirus full genome characterization revealed the circulation of divergent strains. PLoS ONE 2019, 14, e0212585. [Google Scholar] [CrossRef]
- Minguijón, E.; Reina, R.; Pérez, M.; Polledo, L.; Villoria, M.; Ramírez, H.; Leginagoikoa, I.; Badiola, J.J.; García-Marín, J.F.; de Andrés, D.; et al. Small ruminant lentivirus infections and diseases. Vet. Microbiol. 2015, 181, 75–89. [Google Scholar] [CrossRef]
- Blacklaws, B.A.; Berriatua, E.; Torsteinsdottir, S.; Watt, N.J.; de Andres, D.; Klein, D.; Harkiss, G.D. Transmission of small ruminant lentiviruses. Vet. Microbiol. 2004, 101, 199–208. [Google Scholar] [CrossRef]
- Peterhans, E.; Greenland, T.; Badiola, J.; Harkiss, G.; Bertoni, G.; Amorena, B.; Eliaszewicz, M.; Juste, R.A.; Krassnig, R.; Lafont, J.-P.; et al. Routes of transmission and consequences of small ruminant lentiviruses (SRLVs) infection and eradication schemes. Vet. Res. 2004, 35, 257–274. [Google Scholar] [CrossRef]
- Furtado Araújo, J.; Andrioli, A.; Pinheiro, R.R.; Sider, L.H.; de Sousa, A.L.M.; de Azevedo, D.A.A.; Peixoto, R.M.; Lima, A.M.C.; Damasceno, E.M.; Souza, S.C.R.; et al. Vertical transmissibility of small ruminant lentivirus. PLoS ONE 2020, 15, e0239916. [Google Scholar] [CrossRef]
- Reina, R.; Berriatua, E.; Luján, L.; Juste, R.; Sánchez, A.; de Andrés, D.; Amorena, B. Prevention strategies against small ruminant lentiviruses: An update. Vet. J. 2009, 182, 31–37. [Google Scholar] [CrossRef]
- Crespo, H.; Bertolotti, L.; Proffiti, M.; Cascio, P.; Cerruti, F.; Acutis, P.L.; de Andrés, D.; Reina, R.; Rosati, S. Low proviral small ruminant lentivirus load as biomarker of natural restriction in goats. Vet. Microbiol. 2016, 192, 152–162. [Google Scholar] [CrossRef]
- White, S.N.; Mousel, M.R.; Reynolds, J.O.; Lewis, G.S.; Herrmann-Hoesing, L.M. Common promoter deletion is associated with 3.9-fold differential transcription of ovine CCR5 and reduced proviral level of ovine progressive pneumonia virus. Anim. Genet. 2009, 40, 583–589. [Google Scholar] [CrossRef]
- Heaton, M.P.; Clawson, M.L.; Chitko-Mckown, C.G.; Leymaster, K.A.; Smith, T.P.L.; Harhay, G.P.; White, S.N.; Herrmann-Hoesing, L.M.; Mousel, M.R.; Lewis, G.S.; et al. Reduced lentivirus susceptibility in sheep with TMEM154 mutations. PLoS Genet. 2012, 8, e1002467. [Google Scholar] [CrossRef]
- Arcangeli, C.; Lucarelli, D.; Torricelli, M.; Sebastiani, C.; Ciullo, M.; Pellegrini, C.; Felici, A.; Costarelli, S.; Giammarioli, M.; Feliziani, F.; et al. First Survey of SNPs in TMEM154, TLR9, MYD88 and CCR5 Genes in Sheep Reared in Italy and Their Association with Resistance to SRLVs Infection. Viruses 2021, 13, 1290. [Google Scholar] [CrossRef]
- Olech, M.; Ropka-Molik, K.; Szmatoła, T.; Piórkowska, K.; Kuźmak, J. Single Nucleotide Polymorphisms in Genes Encoding Toll-Like Receptors 7 and 8 and Their Association with Proviral Load of SRLVs in Goats of Polish Carpathian Breed. Animals 2021, 11, 1908. [Google Scholar] [CrossRef]
- Olech, M.; Ropka-Molik, K.; Szmatoła, T.; Piórkowska, K.; Kuźmak, J. Transcriptome Analysis for Genes Associated with Small Ruminant Lentiviruses Infection in Goats of Carpathian Breed. Viruses 2021, 13, 2054. [Google Scholar] [CrossRef]
- Colussi, S.; Desiato, R.; Beltramo, C.; Peletto, S.; Modesto, P.; Maniaci, M.G.; Campia, V.; Quasso, A.; Rosati, S.; Bertolotti, L.; et al. A single nucleotide variant in the promoter region of the CCR5 gene increases susceptibility to arthritis encephalitis virus in goats. BMC Vet. Res. 2019, 15, 230. [Google Scholar] [CrossRef]
- Olech, M.; Kuźmak, J. Molecular characterization of small ruminant lentiviruses of subtype A5 detected in naturally infected but clinically healthy goats of carpathian breed. Pathogens 2020, 9, 992. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Bioinformatics: Cambridge, UK, 2010. [Google Scholar]
- Dodt, M.; Roehr, J.T.; Ahmed, R.; Dieterich, C. FLEXBAR-flexible barcode and adapter processing for next-generation sequencing platforms. Biology 2012, 1, 895–905. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Picard Toolkit. Broad Institute, GitHub Repos. 2019. Available online: http://broadinstitute.github.io/picard (accessed on 10 April 2022).
- Van der Auwera, G.; O’Connor, B. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra; O’Reilly Media: Sebastopol, CA, USA, 2020; ISBN 9781491975190. [Google Scholar]
- Teitell, M.A. OCA-B regulation of B-cell development and function. Trends Immunol. 2003, 24, 546–553. [Google Scholar] [CrossRef]
- Luo, Y.; Roeder, R.G. B-cell-specific coactivator OCA-B: Biochemical aspects, role in B-cell development and beyond. Cold Spring Harb. Symp. Quant. Biol. 1999, 64, 119–131. [Google Scholar] [CrossRef]
- Kim, U.; Qin, X.F.; Gong, S.; Stevens, S.; Luo, Y.; Nussenzweig, M.; Roeder, R.G. The B-cell-specific transcription coactivator OCA-B/OBF-1/Bob-1 is essential for normal production of immunoglobulin isotypes. Nature 1996, 383, 542–547. [Google Scholar] [CrossRef]
- Nielsen, P.J.; Georgiev, O.; Lorenz, B.; Schaffner, W. B lymphocytes are impaired in mice lacking the transcriptional co-activator Bob1/OCA-B/OBF1. Eur. J. Immunol. 1996, 26, 3214–3218. [Google Scholar] [CrossRef]
- Zhou, H.; Brekman, A.; Zuo, W.-L.; Ou, X.; Shaykhiev, R.; Agosto-Perez, F.J.; Wang, R.; Walters, M.S.; Salit, J.; Strulovici-Barel, Y.; et al. POU2AF1 Functions in the Human Airway Epithelium To Regulate Expression of Host Defense Genes. J. Immunol. 2016, 196, 3159–3167. [Google Scholar] [CrossRef]
- Brady, P.N.; Goel, A.; Johnson, M.A. Poly(ADP-Ribose) Polymerases in Host-Pathogen Interactions, Inflammation, and Immunity. Microbiol. Mol. Biol. Rev. 2019, 83, 10–1128. [Google Scholar] [CrossRef]
- Mehrotra, P.; Riley, J.P.; Patel, R.; Li, F.; Voss, L.; Goenka, S. PARP-14 functions as a transcriptional switch for Stat6-dependent gene activation. J. Biol. Chem. 2011, 286, 1767–1776. [Google Scholar] [CrossRef]
- Caprara, G.; Prosperini, E.; Piccolo, V.; Sigismondo, G.; Melacarne, A.; Cuomo, A.; Boothby, M.; Rescigno, M.; Bonaldi, T.; Natoli, G. PARP14 Controls the Nuclear Accumulation of a Subset of Type I IFN-Inducible Proteins. J. Immunol. 2018, 200, 2439–2454. [Google Scholar] [CrossRef]
- Grunewald, M.E.; Chen, Y.; Kuny, C.; Maejima, T.; Lease, R.; Ferraris, D.; Aikawa, M.; Sullivan, C.S.; Perlman, S.; Fehr, A.R. The coronavirus macrodomain is required to prevent PARP-mediated inhibition of virus replication and enhancement of IFN expression. PLoS Pathog. 2019, 15, e1007756. [Google Scholar] [CrossRef]
- Atasheva, S.; Akhrymuk, M.; Frolova, E.I.; Frolov, I. New PARP gene with an anti-alphavirus function. J. Virol. 2012, 86, 8147–8160. [Google Scholar] [CrossRef]
- Atasheva, S.; Frolova, E.I.; Frolov, I. Interferon-stimulated poly(ADP-Ribose) polymerases are potent inhibitors of cellular translation and virus replication. J. Virol. 2014, 88, 2116–2130. [Google Scholar] [CrossRef]
- Goodier, J.L.; Pereira, G.C.; Cheung, L.E.; Rose, R.J.; Kazazian, H.H.J. The Broad-Spectrum Antiviral Protein ZAP Restricts Human Retrotransposition. PLoS Genet. 2015, 11, e1005252. [Google Scholar] [CrossRef]
- Kerns, J.A.; Emerman, M.; Malik, H.S. Positive selection and increased antiviral activity associated with the PARP-containing isoform of human zinc-finger antiviral protein. PLoS Genet. 2008, 4, e21. [Google Scholar] [CrossRef]
- Daugherty, M.D.; Young, J.M.; Kerns, J.A.; Malik, H.S. Rapid evolution of PARP genes suggests a broad role for ADP-ribosylation in host-virus conflicts. PLoS Genet. 2014, 10, e1004403. [Google Scholar] [CrossRef]
- Wattenhofer, M.; Shibuya, K.; Kudoh, J.; Lyle, R.; Michaud, J.; Rossier, C.; Kawasaki, K.; Asakawa, S.; Minoshima, S.; Berry, A.; et al. Isolation and characterization of the UBASH3A gene on 21q22.3 encoding a potential nuclear protein with a novel combination of domains. Hum. Genet. 2001, 108, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Paisie, T.K.; Chen, S.; Concannon, P. UBASH3A Regulates the Synthesis and Dynamics of TCR–CD3 Complexes. J. Immunol. 2019, 203, 2827–2836. [Google Scholar] [CrossRef] [PubMed]
- Yamagata, K.; Nakayamada, S.; Zhang, T.; Nguyen, A.P.; Ohkubo, N.; Iwata, S.; Kato, S.; Tanaka, Y. IL-6 production through repression of UBASH3A gene via epigenetic dysregulation of super-enhancer in CD4+ T cells in rheumatoid arthritis. Inflamm. Regen. 2022, 42, 46. [Google Scholar] [CrossRef] [PubMed]
- Vukojević, K.; Šoljić, V.; Martinović, V.; Raguž, F.; Filipović, N. The Ubiquitin-Associated and SH3 Domain-Containing Proteins (UBASH3) Family in Mammalian Development and Immune Response. Int. J. Mol. Sci. 2024, 25, 1932. [Google Scholar] [CrossRef]
- Mayers, J.R.; Torrence, M.E.; Danai, L.V.; Papagiannakopoulos, T.; Davidson, S.M.; Bauer, M.R.; Lau, A.N.; Ji, B.W.; Dixit, P.D.; Hosios, A.M.; et al. Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science 2016, 353, 1161–1165. [Google Scholar] [CrossRef]
- Jiang, J.; Li, Z. Metabolomics Strategy Assisted by Transcriptomics Analysis to Identify Potential Biomarkers Associated with Tuberculosis. Infect. Drug Resist. 2021, 14, 4795–4807. [Google Scholar] [CrossRef]
- Li, J.-T.; Yin, M.; Wang, D.; Wang, J.; Lei, M.-Z.; Zhang, Y.; Liu, Y.; Zhang, L.; Zou, S.-W.; Hu, L.-P.; et al. BCAT2-mediated BCAA catabolism is critical for development of pancreatic ductal adenocarcinoma. Nat. Cell Biol. 2020, 22, 167–174. [Google Scholar] [CrossRef]
- Cai, Z.; Chen, J.; Yu, Z.; Li, H.; Liu, Z.; Deng, D.; Liu, J.; Chen, C.; Zhang, C.; Ou, Z.; et al. BCAT2 Shapes a Noninflamed Tumor Microenvironment and Induces Resistance to Anti-PD-1/PD-L1 Immunotherapy by Negatively Regulating Proinflammatory Chemokines and Anticancer Immunity. Adv. Sci. 2023, 10, 2207155. [Google Scholar] [CrossRef]
- Nong, X.; Zhang, C.; Wang, J.; Ding, P.; Ji, G.; Wu, T. The mechanism of branched-chain amino acid transferases in different diseases: Research progress and future prospects. Front. Oncol. 2022, 12, 988290. [Google Scholar] [CrossRef]

| Reference Sequence | Sequences of Primers (5′–3′) | Orientation | Size | Identified SNP Based on RNA-Seq Data (HGVS Name) |
|---|---|---|---|---|
| POU2AF1 ENSCHIG00000020104 | TGGTATCTGCCTCAAGTCCC | F | 149 bp | rs666106235 (NC_030822.1:g.60734296G>A) |
| GACTCCCTTCTCTCTTGCCT | R | |||
| BCAT2 ENSCHIG00000008019 | GTACAGGATTTGGTGCACGG | F | 155 bp | rs644972268 (NC_030825.1:g.56430949T>C) |
| AGGTTATTTGTCTCGCCCCA | R | |||
| TMEM154 ENSCHIG00000026757 | ATTTCTCTGTCACCTGGCCA | F | 155 bp | rs654984114 (NC_030824.1:g.65805153T>C) |
| AGACAGCAAACAAAGCAAGTATT | R | |||
| UBASH3A ENSCHIG00000022525 | GAGGAAGGAAAATGGGAGTTGG | F | 150 bp | rs668378283 (NC_030808.1:g.142319001A>G) |
| TCTGCGGAGTCCCTTCTC | R | rs658091735 (NC_030808.1:g.142318972G>A) | ||
| PARP14 ENSCHIG00000018391 | CGGGTACTCACTGGATGCTA | F | 203 bp | rs664150071 (NC_030808.1:g.66713881C>T) |
| TCTGCAAAGGTTACCAAAATGTT | R |
| Gene Name | loc | Position | Genotyping Method | Variant Type | Restriction Enzyme | rs | HGVS Nomenclature | GERP |
|---|---|---|---|---|---|---|---|---|
| POU2AF1 | 15 | 60,734,296 | PCR-RFLP | ds | DraI | rs666106235 | NC_030822.1:g.60734296G>A | −6.03 |
| BCAT2 | 18 | 56,430,949 | ms | MnlI | rs644972268 | NC_030825.1:g.56430949T>C | 0.14 | |
| TMEM154 | 17 | 65,805,153 | ds | HgaI | rs654984114 | NC_030824.1:g.65805153T>C | −0.84 | |
| UBASH3A | 1 | 142,319,001 | Sanger sequencing | ms | _ | rs668378283 | NC_030808.1:g.142319001A>G | −6.05 |
| 142,318,972 | sv | rs658091735 | NC_030808.1:g.142318972G>A | −7.13 | ||||
| PARP14 | 1 | 66,713,881 | sv | rs664150071 | NC_030808.1:g.66713881C>T | −1.04 |
| Gene/Polymorphism | Genotype | Allele | HWE * (p-Value) | |||
|---|---|---|---|---|---|---|
| ref hom | het | alt hom | ref | alt | ||
| POU2AF1_G>A | 0.22 (13) | 0.5 (30) | 0.28 (17) | 0.47 | 0.53 | 0.97 |
| BCAT2_T>C | 0.07 (4) | 0.40 (24) | 0.53 (32) | 0.27 | 0.73 | 0.86 |
| TMEM154_T>C | 0.15 (9) | 0.55 (33) | 0.30 (18) | 0.22 | 0.78 | 0.33 |
| UBASH3A_A>G | 0.68 (41) | 0.28 (17) | 0.04 (2) | 0.70 | 0.30 | 0.88 |
| UBASH3A_G>A | 0.56 (34) | 0.40 (24) | 0.04 (2) | 0.60 | 0.40 | 0.36 |
| PARP14_C>T | 0 (0) | 0.28 (19) | 0.68 (41) | 0.23 | 0.77 | 0.15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Materniak-Kornas, M.; Ropka-Molik, K.; Piórkowska, K.; Kowalik, J.; Szmatoła, T.; Sikora, J.; Kawęcka, A.; Kuźmak, J. Identification of New Single Nucleotide Polymorphisms Potentially Related to Small Ruminant Lentivirus Infection Susceptibility in Goats Based on Data Selected from High-Throughput Sequencing. Pathogens 2024, 13, 830. https://doi.org/10.3390/pathogens13100830
Materniak-Kornas M, Ropka-Molik K, Piórkowska K, Kowalik J, Szmatoła T, Sikora J, Kawęcka A, Kuźmak J. Identification of New Single Nucleotide Polymorphisms Potentially Related to Small Ruminant Lentivirus Infection Susceptibility in Goats Based on Data Selected from High-Throughput Sequencing. Pathogens. 2024; 13(10):830. https://doi.org/10.3390/pathogens13100830
Chicago/Turabian StyleMaterniak-Kornas, Magdalena, Katarzyna Ropka-Molik, Katarzyna Piórkowska, Joanna Kowalik, Tomasz Szmatoła, Jacek Sikora, Aldona Kawęcka, and Jacek Kuźmak. 2024. "Identification of New Single Nucleotide Polymorphisms Potentially Related to Small Ruminant Lentivirus Infection Susceptibility in Goats Based on Data Selected from High-Throughput Sequencing" Pathogens 13, no. 10: 830. https://doi.org/10.3390/pathogens13100830
APA StyleMaterniak-Kornas, M., Ropka-Molik, K., Piórkowska, K., Kowalik, J., Szmatoła, T., Sikora, J., Kawęcka, A., & Kuźmak, J. (2024). Identification of New Single Nucleotide Polymorphisms Potentially Related to Small Ruminant Lentivirus Infection Susceptibility in Goats Based on Data Selected from High-Throughput Sequencing. Pathogens, 13(10), 830. https://doi.org/10.3390/pathogens13100830






