Gram-Negative ESKAPE Bacteria Surveillance in COVID-19 Pandemic Exposes High-Risk Sequence Types of Acinetobacter baumannii MDR in a Tertiary Care Hospital
Abstract
1. Introduction
2. Materials and Methods
2.1. Origin of the Gram-Negative ESKAPE Strains, Bacterial Identification, and Controls
2.2. MDR, XDR, and PDR Classification of Gram-Negative ESKAPE Bacteria
2.3. Carbapenemase Production and Their Relationship with Genotypes
2.3.1. Carbapenemase Detection by mCIM Assay
2.3.2. Screening to Confirm Carbapenemases Production in Gram-Negative ESKAPE Bacteria
2.4. adeABC Operon and Regulator Gene Detection adeRS in A. baumannii
2.5. Detection of High-Risk Sequence Types in Acinetobacter baumannii
2.6. Statistical Analysis
3. Results
3.1. Gram-Negative ESKAPE Bacterial Population Included in This Study
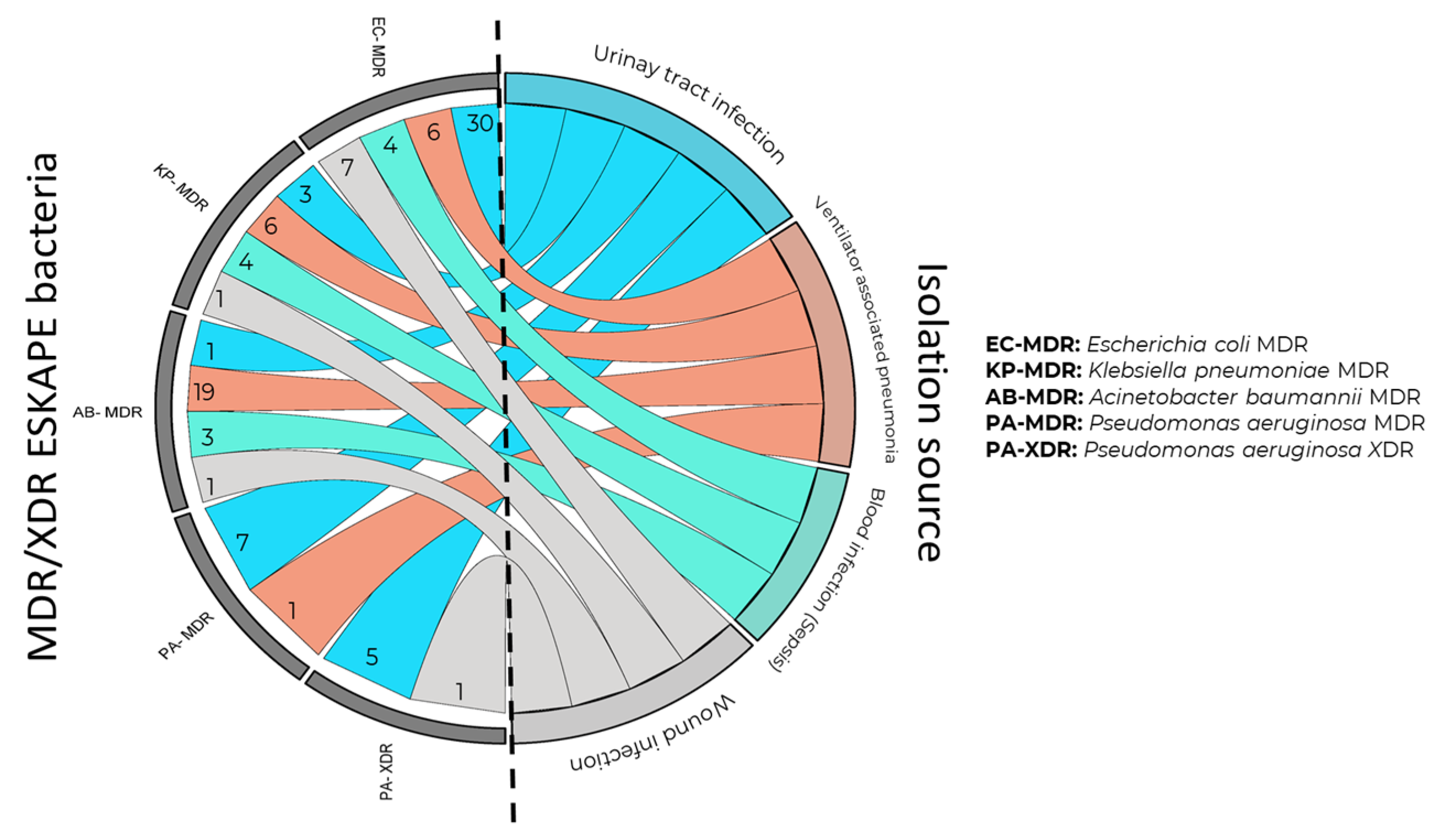
3.2. Clinical Origin of Gram-Negative ESKAPE Bacteria
3.3. MDR, XDR, and PDR Classification of Gram-Negative ESKAPE Bacteria
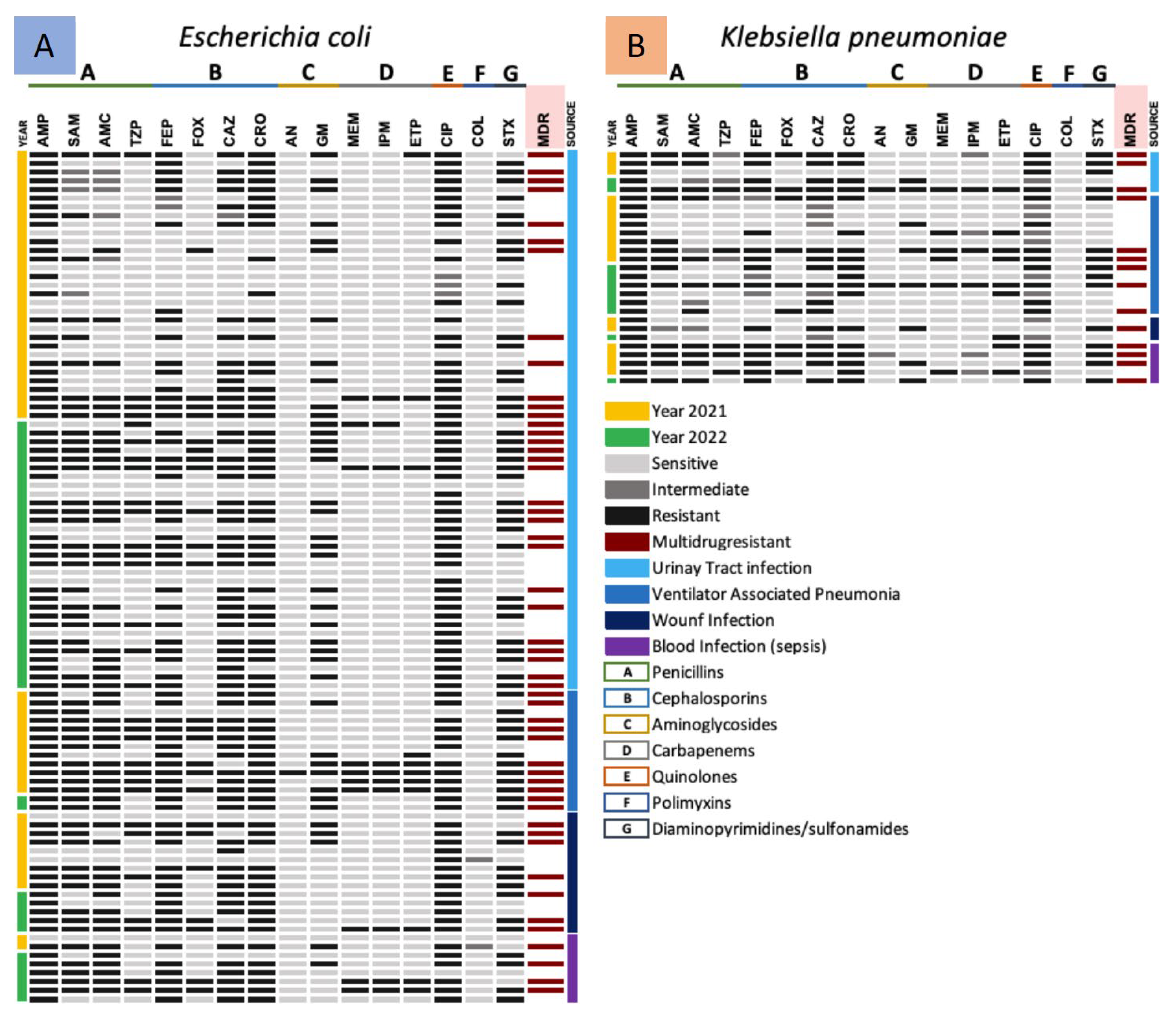
3.3.1. Antimicrobial Resistance Gram-Negative ESKAPE Bacteria (Enterobacterales)
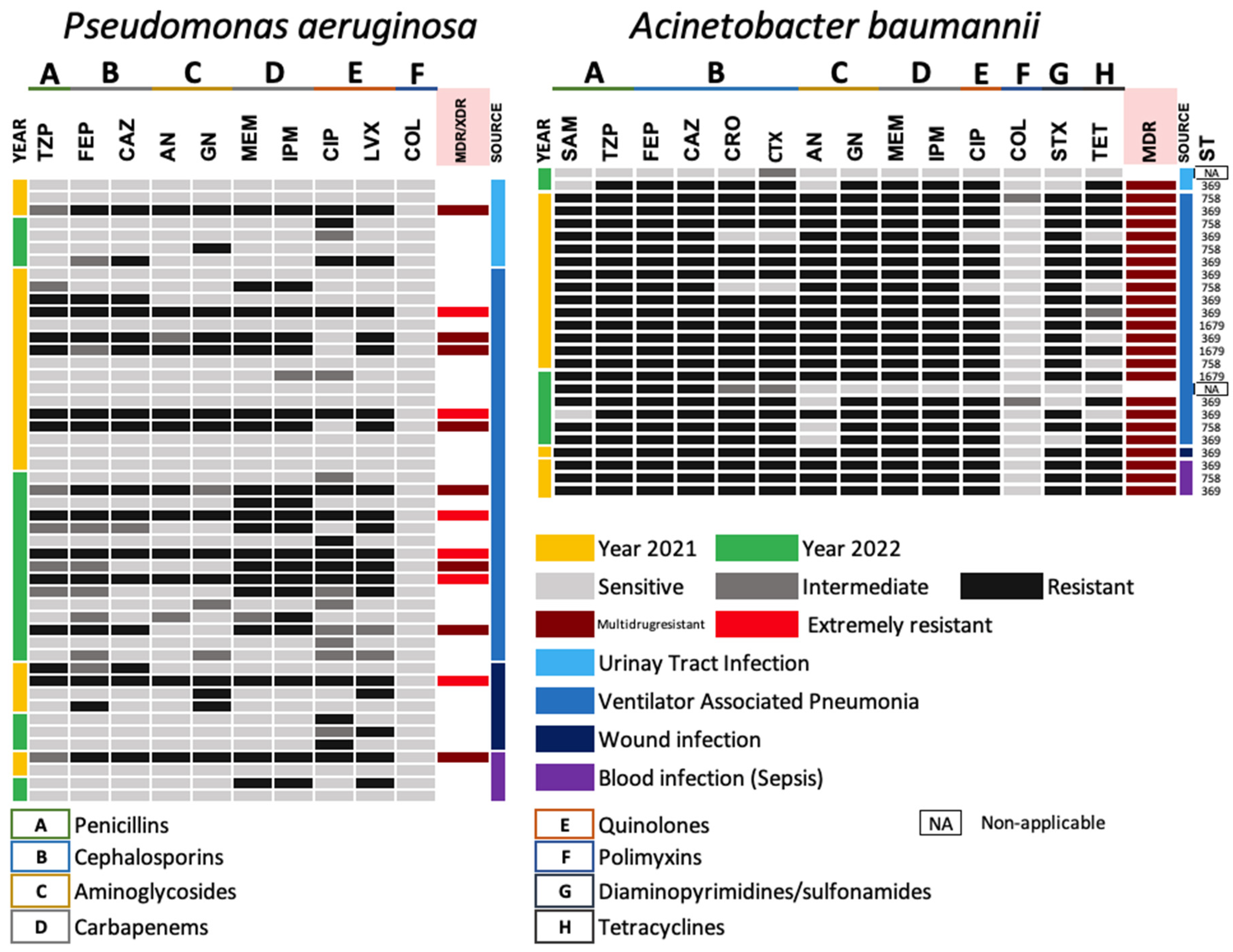
3.3.2. Antimicrobial Resistance Gram-Negative ESKAPE Bacteria (Non-Lactose Fermenting)
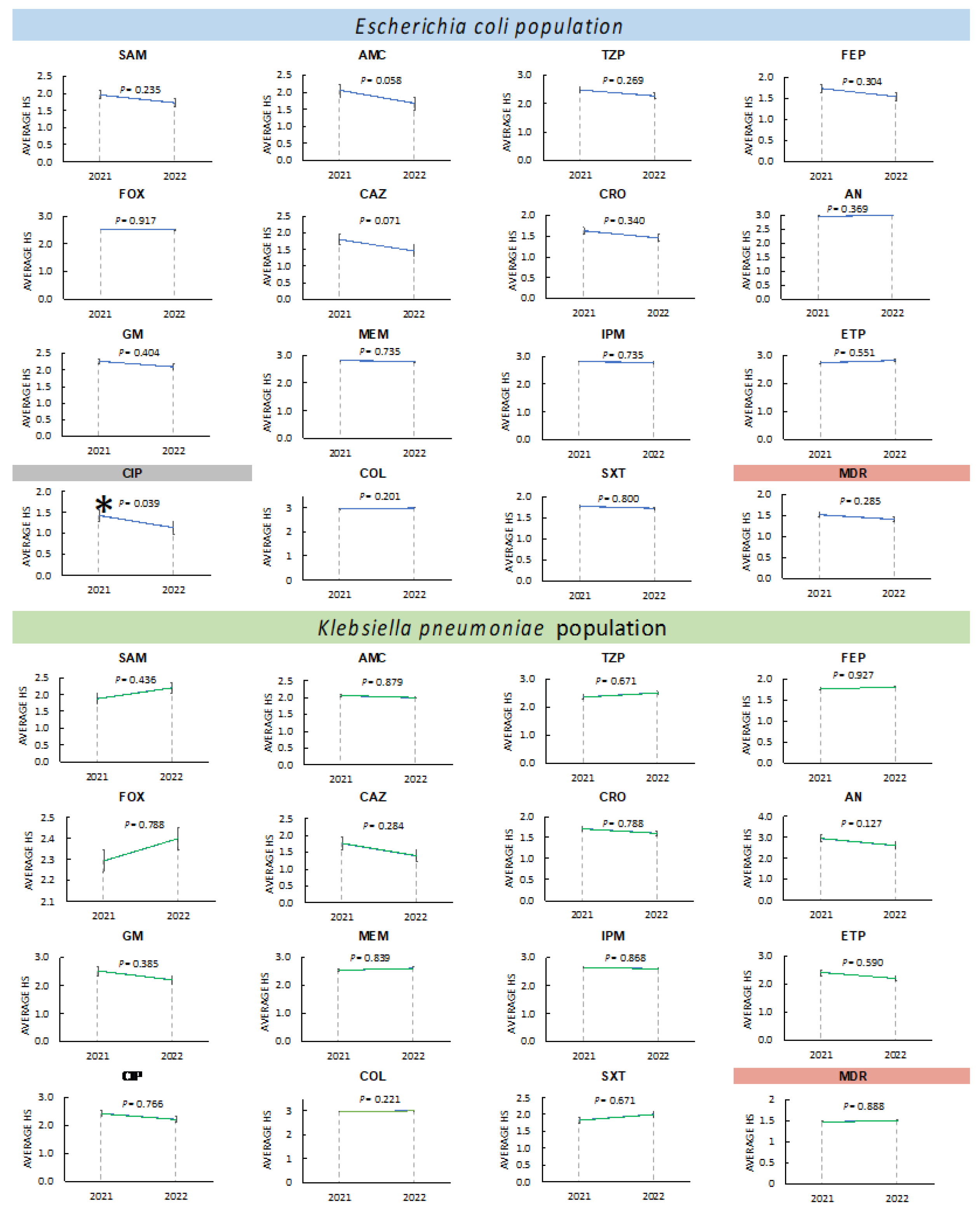
3.3.3. Changes in Antimicrobial Resistance in Enterobacterales during 2021 and 2022
3.3.4. Changes in Antimicrobial Resistance in Non-Fermenters during 2021 and 2022
3.4. Carbapenemase Production and Their Relationship with Genotypes
3.5. Detection of High-Risk Sequence Types in Acinetobacter baumannii
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef] [PubMed]
- Loyola-Cruz, M.Á.; Gonzalez-Avila, L.U.; Martínez-Trejo, A.; Saldaña-Padilla, A.; Hernández-Cortez, C.; Bello-López, J.M.; Castro-Escarpulli, G. ESKAPE and Beyond: The Burden of Coinfections in the COVID-19 Pandemic. Pathogens 2023, 12, 743. [Google Scholar] [CrossRef] [PubMed]
- Rödenbeck, M.; Ayobami, O.; Eckmanns, T.; Pletz, M.W.; Bleidorn, J.; Markwart, R. Clinical epidemiology and case fatality due to antimicrobial resistance in Germany: A systematic review and meta-analysis, 1 January 2010 to 31 December 2021. Euro Surveill. 2023, 28, 2200672. [Google Scholar] [CrossRef] [PubMed]
- Mouanga-Ndzime, Y.; Onanga, R.; Longo-Pendy, N.M.; Bignoumba, M.; Bisseye, C. Epidemiology of community origin of major multidrug-resistant ESKAPE uropathogens in a paediatric population in South-East Gabon. Antimicrob. Resist. Infect. Control 2023, 12, 47. [Google Scholar] [CrossRef] [PubMed]
- Durán-Manuel, E.M.; Cruz-Cruz, C.; Ibáñez-Cervantes, G.; Bravata-Alcantara, J.C.; Sosa-Hernández, O.; Delgado-Balbuena, L.; León-García, G.; Cortés-Ortíz, I.A.; Cureño-Díaz, M.A.; Castro-Escarpulli, G.; et al. Clonal dispersion of Acinetobacter baumannii in an intensive care unit designed to patients COVID-19. J. Infect. Dev. Ctries 2021, 15, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Alcántar-Curiel, M.D.; Huerta-Cedeño, M.; Jarillo-Quijada, M.D.; Gayosso-Vázquez, C.; Fernández-Vázquez, J.L.; Hernández-Medel, M.L.; Zavala-Pineda, M.; Morales-Gil, M.Á.; Hernández-Guzmán, V.A.; Bolaños-Hernández, M.I.; et al. Gram-negative ESKAPE bacteria bloodstream infections in patients during the COVID-19 pandemic. PeerJ. 2023, 11, e15007. [Google Scholar] [CrossRef] [PubMed]
- Loyola-Cruz, M.Á.; Durán-Manuel, E.M.; Cruz-Cruz, C.; Márquez-Valdelamar, L.M.; Bravata-Alcántara, J.C.; Cortés-Ortíz, I.A.; Cureño-Díaz, M.A.; Ibáñez-Cervantes, G.; Fernández-Sánchez, V.; Castro-Escarpulli, G.; et al. ESKAPE bacteria characterization reveals the presence of Acinetobacter baumannii and Pseudomonas aeruginosa outbreaks in COVID-19/VAP patients. Am. J. Infect. Control 2023, 51, 729–737. [Google Scholar] [CrossRef] [PubMed]
- Luyt, C.E.; Sahnoun, T.; Gautier, M.; Vidal, P.; Burrel, S.; Pineton de Chambrun, M.; Chommeloux, J.; Desnos, C.; Arzoine, J.; Nieszkowska, A.; et al. Ventilator-associated pneumonia in patients with SARS-CoV-2-associated acute respiratory distress syndrome requiring ECMO: A retrospective cohort study. Ann. Intensive Care 2020, 10, 158. [Google Scholar] [CrossRef]
- Lima, W.G.; Brito, J.C.M.; da Cruz Nizer, W.S. Ventilator-associated pneumonia (VAP) caused by carbapenem-resistant Acinetobacter baumannii in patients with COVID-19: Two problems, one solution? Med. Hypotheses 2020, 144, 110139. [Google Scholar] [CrossRef]
- Póvoa, H.C.C.; Chianca, G.C.; Iorio, N.L.P.P. COVID-19: An Alert to Ventilator-Associated Bacterial Pneumonia. Infect. Dis. Ther. 2020, 9, 417–420. [Google Scholar] [CrossRef]
- Masoud, S.S.; Kovacevich, A.; Gangji, R.; Nyawale, H.; Nyange, M.; Ntukula, A. Extent and Resistance Patterns of ESKAPE Pathogens Isolated in Pus Swabs from Hospitalized Patients. Can. J. Infect. Dis. Med. Microbiol. 2022, 2022, 3511306. [Google Scholar] [CrossRef] [PubMed]
- Marturano, J.E.; Lowery, T.J. ESKAPE Pathogens in Bloodstream Infections Are Associated With Higher Cost and Mortality but Can Be Predicted Using Diagnoses Upon Admission. Open Forum Infect. Dis. 2019, 6, 12. [Google Scholar] [CrossRef] [PubMed]
- Zhen, X.; Lundborg, C.S.; Sun, X.; Hu, X.; Dong, H. Economic burden of antibiotic resistance in ESKAPE organisms: A systematic review. Antimicrob. Resist. Infect. Control 2019, 8, 137. [Google Scholar] [CrossRef] [PubMed]
- Sosa-Hernández, O.; Matías-Téllez, B.; Estrada-Hernández, A.; Cureño-Díaz, M.A.; Bello-López, J.M. Incidence and costs of ventilator-associated pneumonia in the adult intensive care unit of a tertiary referral hospital in Mexico. Am. J. Infect. Control 2019, 9, e21–e25. [Google Scholar] [CrossRef] [PubMed]
- Kaier, K.; Heister, T.; Motschall, E.; Hehn, P.; Bluhmki, T.; Wolkewitz, M. Impact of mechanical ventilation on the daily costs of ICU care: A systematic review and meta regression. Epidemiol Infect. 2019, 147, e314. [Google Scholar] [CrossRef] [PubMed]
- Luckraz, H.; Manga, N.; Senanayake, E.L.; Abdelaziz, M.; Gopal, S.; Charman, S.C.; Giri, R.; Oppong, R.; Andronis, L. Cost of treating ventilator-associated pneumonia post cardiac surgery in the National Health Service: Results from a propensity-matched cohort study. J. Intensive Care Soc. 2018, 19, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Ladbrook, E.; Khaw, D.; Bouchoucha, S.; Hutchinson, A. A systematic scoping review of the cost-impact of ventilator-associated pneumonia (VAP) intervention bundles in intensive care. Am. J. Infect. Control 2021, 7, 928–936. [Google Scholar] [CrossRef] [PubMed]
- Venkateswaran, P.; Vasudevan, S.; David, H.; Shaktivel, A.; Shanmugam, K.; Neelakantan, P.; Solomon, A.P. Revisiting ESKAPE Pathogens: Virulence, resistance, and combating strategies focusing on quorum sensing. Front. Cell. Infect. Microbiol. 2023, 13, 1159798. [Google Scholar] [CrossRef]
- Peterson, L.R. Bad bugs, no drugs: No ESCAPE revisited. Clin. Infect. Dis. 2009, 6, 992–993. [Google Scholar] [CrossRef]
- Pépin, J.; Valiquette, L.; Cossette, B. Mortality attributable to nosocomial Clostridium difficile-associated disease during an epidemic caused by a hypervirulent strain in Quebec. CMAJ. 2005, 9, 1037–1042. [Google Scholar] [CrossRef]
- Sosa-Hernández, O.; Matías-Téllez, B.; González-Martínez, J.; Juárez-Vargas, R.; González-González, N.E.; Estrada-Hernández, A.; Ruíz-Santana, M.; Bravata-Alcántara, J.C.; Bello-López, J.M. Implementation of control measures against an outbreak due to Clostridioides difficile producing toxin B in a tertiary hospital in Mexico. J. Prev. Med. Hyg. 2021, 2, E508–E513. [Google Scholar]
- Gajdács, M.; Albericio, F. Antibiotic Resistance: From the Bench to Patients. Antibiotics 2019, 3, 129. [Google Scholar] [CrossRef]
- Cruz-Córdova, A.; Mancilla-Rojano, J.; Luna-Pineda, V.M.; Escalona-Venegas, G.; Cázares-Domínguez, V.; Ormsby, C.; Franco-Hernández, I.; Zavala-Vega, S.; Hernández, M.A.; Medina-Pelcastre, M.; et al. Molecular Epidemiology, Antibiotic Resistance, and Virulence Traits of Stenotrophomonas maltophilia Strains Associated with an Outbreak in a Mexican Tertiary Care Hospital. Front. Cell. Infect. Microbiol. 2020, 10, 50. [Google Scholar] [CrossRef] [PubMed]
- Benkő, R.; Gajdács, M.; Matuz, M.; Bodó, G.; Lázár, A.; Hajdú, E.; Papfalvi, E.; Hannauer, P.; Erdélyi, P.; Pető, Z. Prevalence and Antibiotic Resistance of ESKAPE Pathogens Isolated in the Emergency Department of a Tertiary Care Teaching Hospital in Hungary: A 5-Year Retrospective Survey. Antibiotics 2020, 9, 624. [Google Scholar] [CrossRef] [PubMed]
- Cortés-Ortíz, I.A.; Juárez-Gómez, J.C.; Cu-Quijano, C.; Flores-Paz, R.; Durán-Manuel, E.M.; Cruz-Cruz, C.; Gutiérrez-Muñoz, V.H.; Sosa-Hernández, O.; Escobar-Escamilla, N.; Bravata-Alcántara, J.C.; et al. Klebsiella pneumoniae blaNDM-1 carrying a class 1 integron causing a hospital outbreak in a Mexican attention center. J. Infect. Dev. Ctries 2021, 1, 657–664. [Google Scholar] [CrossRef] [PubMed]
- Durán-Manuel, E.M.; Loyola-Cruz, M.Á.; Cruz-Cruz, C.; Ibáñez-Cervantes, G.; Gaytán-Cervantes, J.; González-Torres, C.; Quiroga-Vargas, E.; Calzada-Mendoza, C.C.; Cureño-Díaz, M.A.; Fernández-Sánchez, V.; et al. Massive sequencing of the V3-V4 hypervariable region of bronchoalveolar lavage from patients with COVID-19 and VAP reveals the collapse of the pulmonary microbiota. J. Med. Microbiol. 2022, 71, 001634. [Google Scholar] [CrossRef] [PubMed]
- Cureño-Díaz, M.A.; Durán-Manuel, E.M.; Cruz-Cruz, C.; Ibáñez-Cervantes, G.; Rojo-Gutiérrez, M.I.; Moncayo-Coello, C.V.; Loyola-Cruz, M.Á.; Castro-Escarpulli, G.; Hernández, D.M.R.; Bello-López, J.M. Impact of the modification of a cleaning and disinfection method of mechanical ventilators of COVID-19 patients and ventilator-associated pneumonia: One year of experience. Am. J. Infect. Control 2021, 49, 1474–1480. [Google Scholar] [CrossRef] [PubMed]
- Ibáñez-Cervantes, G.; Cruz-Cruz, C.; Durán-Manuel, E.M.; Loyola-Cruz, M.Á.; Cureño-Díaz, M.A.; Castro-Escarpulli, G.; Lugo-Zamudio, G.E.; Rojo-Gutiérrez, M.I.; Razo-Blanco Hernández, D.M.; López-Ornelas, A.; et al. Disinfection efficacy of ozone on ESKAPE bacteria biofilms: Potential use in difficult-to-access medical devices. Am. J. Infect. Control 2023, 51, 11–17. [Google Scholar] [CrossRef]
- López-Jácome, L.E.; Fernández-Rodríguez, D.; Franco-Cendejas, R.; Camacho-Ortiz, A.; Morfin-Otero, M.D.R.; Rodríguez-Noriega, E.; Ponce-de-León, A.; Ortiz-Brizuela, E.; Rojas-Larios, F.; Velázquez-Acosta, M.D.C.; et al. Increment Antimicrobial Resistance During the COVID-19 Pandemic: Results from the Invifar Network. Microb. Drug. Resist. 2022, 28, 338–345. [Google Scholar]
- Langford, B.J.; Soucy, J.R.; Leung, V.; So, M.; Kwan, A.T.H.; Portnoff, J.S.; Bertagnolio, S.; Raybardhan, S.; MacFadden, D.R.; Daneman, N. Antibiotic resistance associated with the COVID-19 pandemic: A systematic review and meta-analysis. Clin. Microbiol. Infect. 2023, 29, 302–309. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 32nd ed.; CLSI Suppl. M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2022. [Google Scholar]
- Jiménez Pearson, M.A.; Galas, M.; Corso, A.; Hormazábal, J.C.; Duarte; Valderrama, C.; Salgado Marcano, N.; Ramón-Pardo, P.; Melano, R.G. Consenso latinoamericano para definir, categorizar y notificar patógenos multirresistentes, con resistencia extendida o panresistentes [Latin American consensus to define, categorize, and report multidrug-resistant, extensively drug-resistant, or pandrug-resistant pathogensConsenso latino-americano para definição, categorização e notificação de patógenos multirresistentes, com resistência ampliada ou panresistentes]. Rev. Panam. Salud. Publica. 2019, 43, e65. [Google Scholar] [PubMed]
- Yu, Y.; Ouyang, Y.; Yao, W. shinyCircos: An R/Shiny application for interactive creation of Circos plot. Bioinformatics 2018, 34, 1229–1231. [Google Scholar] [CrossRef] [PubMed]
- Pierce, V.M.; Simner, P.J.; Lonsway, D.R.; Roe-Carpenter, D.E.; Johnson, J.K.; Brasso, W.B.; Bobenchik, A.M.; Lockett, Z.C.; Charnot-Katsikas, A.; Ferraro, M.J.; et al. Modified carbapenem inactivation method for phenotypic detection of carbapenemase production among Enterobacteriaceae. J. Clin. Microbiol. 2017, 55, 2321–2333. [Google Scholar] [CrossRef] [PubMed]
- Dallenne, C.; Da Costa, A.; Decré, D.; Favier, C.; Arlet, G. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Poirel, L.; Carrër, A.; Toleman, M.; Walsh, T. How to detect NDM-1 producers. J. Clin. Microbiol. 2011, 49, 718–721. [Google Scholar] [CrossRef] [PubMed]
- Hujer, K.M.; Hujer, A.M.; Hulten, E.A.; Bajaksouzian, S.; Adams, J.M.; Donskey, C.J.; Ecker, D.J.; Massire, C.; Eshoo, M.W.; Sampath, R.; et al. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob. Agents Chemother. 2006, 50, 4114–4123. [Google Scholar] [CrossRef] [PubMed]
- Alcántar-Curiel, M.D.; Rosales-Reyes, R.; Jarillo-Quijada, M.D.; Gayosso-Vázquez, C.; Fernández-Vázquez, J.L.; Toledano-Tableros, J.E.; Giono-Cerezo, S.; Garza-Villafuerte, P.; López-Huerta, A.; Vences-Vences, D.; et al. Carbapenem-Resistant Acinetobacter baumannii in Three Tertiary Care Hospitals in Mexico: Virulence Profiles, Innate Immune Response and Clonal Dissemination. Front. Microbiol. 2019, 10, 2116. [Google Scholar] [CrossRef] [PubMed]
- Bartual, S.G.; Seifert, H.; Hippler, C.; Domínguez, M.A.; Wisplinghoff, H.; Rodríguez-Valera, F. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 2005, 43, 4382–4390. [Google Scholar] [CrossRef]
- Rouzé, A.; Martin-Loeches, I.; Povoa, P.; Makris, D.; Artigas, A.; Bouchereau, M.; Lambiotte, F.; Metzelard, M.; Cuchet, P.; Boulle Geronimi, C.; et al. Relationship between SARS-CoV-2 infection and the incidence of ventilator-associated lower respiratory tract infections: A European multicenter cohort study. Intensive Care Med. 2021, 47, 188–198. [Google Scholar] [CrossRef]
- Nseir, S.; Martin-Loeches, I.; Povoa, P.; Metzelard, M.; Du Cheyron, D.; Lambiotte, F.; Tamion, F.; Labruyere, M.; Makris, D.; Boulle Geronimi, C.; et al. Relationship between ventilator-associated pneumonia and mortality in COVID-19 patients: A planned ancillary analysis of the coVAPid cohort. Crit. Care 2021, 25, 177. [Google Scholar] [CrossRef]
- Meawed, T.E.; Ahmed, S.M.; Mowafy, S.M.S.; Samir, G.M.; Anis, R.H. Bacterial and fungal ventilator associated pneumonia in critically ill COVID-19 patients during the second wave. J. Infect. Public Health 2021, 14, 1375–1380. [Google Scholar] [CrossRef]
- Alali, W.Q.; Abdo, N.M.; AlFouzan, W.; Dhar, R. Antimicrobial resistance pattern in clinical Escherichia coli and Pseudomonas aeruginosa isolates obtained from a secondary-care hospital prior to and during the COVID-19 pandemic in Kuwait. Germs 2022, 12, 372–383. [Google Scholar] [CrossRef] [PubMed]
- Wardoyo, E.H.; Suardana, I.W.; Yasa, I.W.P.S.; Sukrama, I.D.M. Antibiotics susceptibility of Escherichia coli isolates from clinical specimens before and during COVID-19 pandemic. Iran J. Microbiol. 2021, 2, 156–160. [Google Scholar]
- Chatterjee, N.; Nirwan, P.K.; Srivastava, S.; Rati, R.; Sharma, L.; Sharma, P.; Dwivedi, P.; Jaggi, N. Trends in carbapenem resistance in Pre-COVID and COVID times in a tertiary care hospital in North India. Ann. Clin. Microbiol. Antimicrob. 2023, 22, 1. [Google Scholar] [CrossRef] [PubMed]
- Royer, S.; Faria, A.L.; Seki, L.M.; Chagas, T.P.; Campos, P.A.; Batistão, D.W.; Asensi, M.D.; Gontijo, P.P.; Ribas, R.M. Spread of multidrug-resistant Acinetobacter baumannii and Pseudomonas aeruginosa clones in patients with ventilator-associated pneumonia in an adult intensive care unit at a university hospital. Braz. J. Infect. Dis. 2015, 4, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Jamnani, A.N.; Montazeri, M.; Mirzakhani, M.; Moosazadeh, M.; Haghighi, M. Evaluation of Bacterial Coinfection and Antibiotic Resistance in Patients with COVID-19 Under Mechanical Ventilation. SN Compr. Clin. Med. 2022, 4, 19. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Chávez, L.A.; Esteban-Dionicio, M.L.; Rodriguez-Mendoza, C.R.E. Microbiological profile of bacteria causing ventilator-associated pneumonia in the intensive care unit of a high-complexity hospital. Rev. Peru. Med. Exp. Salud Publica 2023, 40, 115–117. [Google Scholar] [CrossRef] [PubMed]
- Saleem, M.; Syed, K.A.S.; Hossain, A.; Alenazi, F.; Said, K.B.; Moursi, S.A.; Almalaq, H.A.; Mohamed, H.; Rakha, E.; Alharbi, M.S.; et al. Pathogen Burden Among ICU Patients in a Tertiary Care Hospital in Hail Saudi Arabia with Particular Reference to β-Lactamases Profile. Infect. Drug Resist. 2023, 16, 769–778. [Google Scholar] [CrossRef]
- Jones, N.I.; Harmer, C.J.; Hamidian, M.; Hall, R.M. Evolution of Acinetobacter baumannii plasmids carrying the oxa58 carbapenemase resistance gene via plasmid fusion, IS26-mediated events and dif module shuffling. Plasmid 2022, 121, 102628. [Google Scholar] [CrossRef]
- Hamidian, M.; Kenyon, J.J.; Holt, K.E.; Pickard, D.; Hall, R.M. A conjugative plasmid carrying the carbapenem resistance gene blaOXA-23 in AbaR4 in an extensively resistant GC1 Acinetobacter baumannii isolate. J. Antimicrob. Chemother. 2014, 10, 625–628. [Google Scholar] [CrossRef]
- Zhang, L.; Fu, Y.; Zhang, L.; Xu, Q.; Yang, Y.; He, J.; Leptihn, S.; Loh, B.; Moran, R.A.; Van-Schaik, W.; et al. Co-evolutionary adaptations of Acinetobacter baumannii and a clinical carbapenemase-encoding plasmid during carbapenem exposure. Evol. Appl. 2022, 7, 1045–1061. [Google Scholar] [CrossRef] [PubMed]
- Levy-Blitchtein, S.; Roca, I.; Plasencia-Rebata, S.; Vicente-Taboada, W.; Velásquez-Pomar, J.; Muñoz, L.; Moreno-Morales, J.; Pons, M.J.; Del Valle-Mendoza, J.; Vila, J. Emergence and spread of carbapenem-resistant Acinetobacter baumannii international clones II and III in Lima, Peru. Emerg. Microbes Infect. 2018, 7, 119. [Google Scholar] [CrossRef] [PubMed]
- Ploy, M.C.; Denis, F.; Courvalin, P.; Lambert, T. Molecular characterization of integrons in Acinetobacter baumannii: Description of a hybrid class 2 integron. Antimicrob. Agents Chemother. 2000, 44, 2684–2688. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Meletis, G.; Exindari, M.; Vavatsi, N.; Sofianou, D.; Diza, E. Mechanisms responsible for the emergence of carbapenem resistance in Pseudomonas aeruginosa. Hippokratia 2012, 6, 303–307. [Google Scholar]
- Słoczyńska, A.; Wand, M.E.; Bock, L.J.; Tyski, S.; Laudy, A.E. Efflux-Related Carbapenem Resistance in Acinetobacter baumannii Is Associated with Two-Component Regulatory Efflux Systems’ Alteration and Insertion of ΔAbaR25-Type Island Fragment. Int. J. Mol. Sci. 2023, 24, 9525. [Google Scholar] [CrossRef] [PubMed]
- Shafigh, N.; Roodneshin, F.; Shafigh, E.; Kermany, M.P.Z.N.; Ziaie, S.; Miller, J.; Gharehbeglou, M.; Masoumzadeh, N.; Hasheminik, M.; Avval, J.O. Rapid spread and high mortality due to community-acquired pneumonia caused by Acinetobacter baumannii (CAP-AB) in a COVID-19 intensive care unit. J. Family Med. Prim. Care 2022, 12, 7957–7959. [Google Scholar]
- Hwang, S.M.; Cho, H.W.; Kim, T.Y.; Park, J.S.; Jung, J.; Song, K.H.; Lee, H.; Kim, E.S.; Kim, H.B.; Park, K.U. Whole-Genome Sequencing for Investigating a Health Care-Associated Outbreak of Carbapenem-Resistant Acinetobacter baumannii. Diagnostics 2021, 11, 201. [Google Scholar] [CrossRef]
- Vanegas, J.M.; Higuita, L.F.; Vargas, C.A.; Cienfuegos, A.V.; Rodríguez, E.A.; Roncancio, G.E.; Jiménez, J.N. Carbapenem-resistant Acinetobacter baumannii causing osteomyelitis and infections of skin and soft tissues in hospitals of Medellín, Colombia. Biomedica 2015, 35, 522–530. [Google Scholar]
- Mancilla-Rojano, J.; Castro-Jaimes, S.; Ochoa, S.A.; Bobadilla Del Valle, M.; Luna-Pineda, V.M.; Bustos, P.; Laris-González, A.; Arellano-Galindo, J.; Parra-Ortega, I.; Hernández-Castro, R.; et al. Whole-Genome Sequences of Five Acinetobacter baumannii strains from a child with leukemia M2. Front. Microbiol. 2019, 10, 132. [Google Scholar] [CrossRef]
- Public Databases for Molecular Typing and Microbial Genome Diversity (PubMLST). Available online: https://pubmlst.org/bigsdb?page=profileInfo&db=pubmlst_abaumannii_seqdef&scheme_id=1&profile_id=1679 (accessed on 21 November 2023).

| Primer | Molecular Target | Sequence (5′→3′) | Size (bp) | Reference |
|---|---|---|---|---|
| IMP-F | blaIMP | TTGACACTCCATTTACDG | 139 | [35] |
| IMP-R | GATYGAGAATTAAGCCACYCT | |||
| VIM-F | blaVIM | GATGGTGTTTGGTCGCATA | 390 | |
| VIM-R | CGAATGCGCAGCACCAG | |||
| KPC-F | blaKPC | CATTCAAGGGCTTTCTTGCTGC | 538 | |
| KPC-R | ACGACGGCATAGTCATTTGC | |||
| OXA-48F | blaOXA-48 | GCACTTCTTTTGTGATGGC | 281 | |
| OXA-48R | GAGCACTTCTTTTGTGATGGC | |||
| NDM-F | blaNDM | GGTTTGGCGAT CTGGTTTTC | 621 | [36] |
| NDM-R | CGGAATGGCTCATCACGATC | |||
| OXA-23F | blaOXA-23 | GATGTGTCATAGTATTCGTCG | 1065 | [37] |
| OXA-23R | TCACAACAACTAAAAGCACTG | |||
| OXA-40F | blaOXA-40 | TCTAGTTTCTCTCAGTGCATGTTCATC | 749 | [38] |
| OXA-40R | CATTACGAATAGAACCAGACATTCC | |||
| gltA-F | Citrate synthase | AATTTACAGTGGCACATTAGGTCCC | 722 | [39] |
| gltA-R | GCAGAGATACCAGCAGAGATACACG | |||
| gyrB-F | DNA gyrase subunit B | TGTAAAACGACGGCCAGTGCNGGRTCYTTYTCYTGRCA | 909 | |
| gyrB-R | CAGGAAACAGCTATGACCAYGSNGGNGGNAARTTYRA | |||
| gdhB-F | Glucose dehydrogenase B | GCTACTTTTATGCAACAGAGCC | 775 | |
| gdhB-R | GTTGAGTTGGCGTATGTTGTGC | |||
| recA-F | Homologous recombination factor | CCTGAATCTTCYGGTAAAAC | 425 | |
| recA-R | GTTTCTGGGCTGCCAAACATTAC | |||
| cpn60-F | 60 kDa chaperonin | ACTGTACTTGCTCAAGC | 479 | |
| cpn60-R | TTCAGCGATGATAAGAAGTGG | |||
| gpi-F | Glucose-6-phosphate isomerase | AATACCGTGGTGCTACGGG | 508 | |
| gpi-R | AACTTGATTTTCAGGAGC | |||
| rpoD-F | RNA polymerase sigma factor rpoD (Sigma-70) | ACGACTGACCCGGTACGCATGTAYATGMGNGARATCGC NACNCT | 492 | |
| rpoD-R | ATAGAAATAACCAGACGTAAGTTNGCYTCNACCATYTG YTTYTT |
| Gram-Negative ESKAPE Bacteria Causing Nosocomial Infections | Analyzed Strains by Year n (%) | Total n (%) | |
|---|---|---|---|
| 2021 | 2022 | ||
| Escherichia coli | 54 (47.4) | 44 (51.2) | 98 (49.0) |
| Klebsiella pneumoniae | 17 (14.9) | 10 (11.6) | 27 (13.5) |
| Pseudomonas aeruginosa | 25 (21.9) | 24 (27.9) | 49 (24.5) |
| Acinetobacter baumannii | 18 (15.8) | 8 (9.30) | 26 (13.0) |
| Total | 114 (100) | 86 (100) | 200 (100) |
| Clinical Origin | Analyzed Strains by Year n (%) | Total | |
|---|---|---|---|
| 2021 | 2022 | ||
| Urinary tract infection (UTI) | 37 (32.4) | 39 (45.3) | 76 (38.0) |
| Ventilator-associated pneumonia (VAP) | 50 (43.9) | 29 (33.7) | 79 (39.5) |
| Wound infection | 16 (14.0) | 9 (10.5) | 25 (12.5) |
| Blood infection (Sepsis) | 11 (9.7) | 9 (10.5) | 20 (10.0) |
| Total | 114 (100) | 86 (100) | 200 (100) |
| ESKAPE | Carbapenemase Detection (n/%) | Efflux Pump adeABC | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phenotype n (%) by | Genotype by End-Point PCR | |||||||||
| Disc Diffusion | mCIM Assay | blaNDM | blaVIM | blaIMP | blaKPC | blaOXA-48 | blaOXA-40 | blaOXA-23 | ||
| E. coli | 12 (17.6) | 8 (25.85) | 4 (50) | 2 (25) | 0 (0) | 1 (12.5) | 1 (12.5) | NA* | NA | NA |
| K. pneumoniae | 11 (16.2) | 4 (12.9) | 1 (25) | 1 (25) | 0 (0) | 0 (0) | 2 (50) | NA | NA | NA |
| P. aeruginosa | 21 (30.8) | 2 (6.5) | 0 (0) | 2 (100) | 0 (0) | 0 (0) | 0 (0) | NA | NA | NA |
| A. baumannii | 24 (35.4) | 17 (54.8) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 17 (100) | 0 (0) | 24 (100) |
| Total | 68 (100) | 31 (100) | ||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cureño-Díaz, M.A.; Plascencia-Nieto, E.S.; Loyola-Cruz, M.Á.; Cruz-Cruz, C.; Nolasco-Rojas, A.E.; Durán-Manuel, E.M.; Ibáñez-Cervantes, G.; Gómez-Zamora, E.; Tamayo-Ordóñez, M.C.; Tamayo-Ordóñez, Y.d.J.; et al. Gram-Negative ESKAPE Bacteria Surveillance in COVID-19 Pandemic Exposes High-Risk Sequence Types of Acinetobacter baumannii MDR in a Tertiary Care Hospital. Pathogens 2024, 13, 50. https://doi.org/10.3390/pathogens13010050
Cureño-Díaz MA, Plascencia-Nieto ES, Loyola-Cruz MÁ, Cruz-Cruz C, Nolasco-Rojas AE, Durán-Manuel EM, Ibáñez-Cervantes G, Gómez-Zamora E, Tamayo-Ordóñez MC, Tamayo-Ordóñez YdJ, et al. Gram-Negative ESKAPE Bacteria Surveillance in COVID-19 Pandemic Exposes High-Risk Sequence Types of Acinetobacter baumannii MDR in a Tertiary Care Hospital. Pathogens. 2024; 13(1):50. https://doi.org/10.3390/pathogens13010050
Chicago/Turabian StyleCureño-Díaz, Mónica Alethia, Estibeyesbo Said Plascencia-Nieto, Miguel Ángel Loyola-Cruz, Clemente Cruz-Cruz, Andres Emmanuel Nolasco-Rojas, Emilio Mariano Durán-Manuel, Gabriela Ibáñez-Cervantes, Erika Gómez-Zamora, María Concepción Tamayo-Ordóñez, Yahaira de Jesús Tamayo-Ordóñez, and et al. 2024. "Gram-Negative ESKAPE Bacteria Surveillance in COVID-19 Pandemic Exposes High-Risk Sequence Types of Acinetobacter baumannii MDR in a Tertiary Care Hospital" Pathogens 13, no. 1: 50. https://doi.org/10.3390/pathogens13010050
APA StyleCureño-Díaz, M. A., Plascencia-Nieto, E. S., Loyola-Cruz, M. Á., Cruz-Cruz, C., Nolasco-Rojas, A. E., Durán-Manuel, E. M., Ibáñez-Cervantes, G., Gómez-Zamora, E., Tamayo-Ordóñez, M. C., Tamayo-Ordóñez, Y. d. J., Calzada-Mendoza, C. C., & Bello-López, J. M. (2024). Gram-Negative ESKAPE Bacteria Surveillance in COVID-19 Pandemic Exposes High-Risk Sequence Types of Acinetobacter baumannii MDR in a Tertiary Care Hospital. Pathogens, 13(1), 50. https://doi.org/10.3390/pathogens13010050









