Abstract
African swine fever (ASF) is a highly contagious and severe viral hemorrhagic disease in domestic and wild pigs. ASF seriously affects the global swine industry as the mortality rate can reach 100% with highly virulent strains. In 2007, ASF was introduced into the Caucasus and spread to Russia and later into other European and Asian countries. This study reported the first whole-genome sequence (WGS) of the ASF virus (ASFV) that was detected in a Mongolian wild boar. This sequence was then compared to other WGS samples from Asia and Europe. Results show that the ASFV Genotype II from Mongolia is similar to the Asian Genotype II WGS. However, there were three nucleotide differences found between the Asian and European genome sequences, two of which were non-synonymous. It was also observed that the European Genotype II ASFV WGS was more diverse than that of the Asian counterparts. The study demonstrates that the ASFV Genotype II variants found in wild boars and domestic pigs are highly similar, suggesting these animals might have had direct or indirect contact, potentially through outdoor animal breeding. In conclusion, this study provides a WGS and mutation spectrum of the ASFV Genotype II WGS in Asia and Europe and thus provides important insights into the origin and spread of ASFV in Mongolia.
1. Introduction
African swine fever (ASF) is a highly contagious and severe haemorrhagic viral disease in domestic pigs and wild boars, having serious effects on the global pig industry in more than 50 countries [1,2]. Virulent strains have a mortality rate of up to 100%, and there is no current control measure other than the culling of infected pig herds, which leads to enormous economic losses [3]. Since its first description in Kenya in 1921, the ASFV has spread from wild African Suidae to European domestic pigs brought to the African continent [4,5]. ASF was confined to Africa until the virus was diagnosed in Portugal in 1957, probably via a single-source introduction from Angola. This occurrence led to the ASFV epidemic in Europe, the Caribbean, and Brazil in 1960, caused by ASFVs belonging to Genotype I. This first incursion was eradicated from the affected European countries, apart from Sardinia, by the mid-1990s [6,7].
In 2007, however, ASFV Genotype II was introduced into the Caucasus and spread to Russia and later to other European countries [8]. Finally, in 2018, ASFV reached China and was further spread to South and Southwest Asia [5,9].
The first ASFV case in Mongolia was reported in 2019 and led to 105 affected farms and holdings and the elimination of more than 10% of the total pig population in the country [10,11]. In the fall of 2020, ASF-related mortality in wild boars was observed by local nut pickers and hunters in the Minjiin Khangai mountain area, covering the Tuv and Selenge province areas near the Russian border.
In Russia, cases of infected wild boars and domestic pigs were reported 60 km from the Mongolian border in the summer of 2020, just before the first carcasses were found in Mongolia (WOAH WAHIS, visited online 5 September 2020). These incidences led to passive and active surveillance in the Tuv and Selenge provinces of Mongolia in 2020, through which carcasses of wild boars were sampled for laboratory analysis.
This study reports the first whole-genome sequence (WGS) of the ASFV that was detected in a Mongolian wild boar in 2020. The sample was genotyped with phylogenetic analysis based on P54 and P72 gene segments. In addition, we compared the wild boar ASFV genome and that of a domestic pig from the index case in 2019 [10] that was generated in this study. Finally, the phylogenetic relationships with other WGSs of ASFVs from Europe and Asia was assessed.
2. Materials and Methods
2.1. Sample Collection and Rapid ASFV Detection
The field team, consisting of rangers, hunters, local veterinarians, and experts from the General Authority of Veterinary Service (GAVS), investigated the cases in Mongolia’s Tuv and Selenge provinces. They conducted a questionnaire survey with nut pickers and local hunters to locate grazing fields of wild boars and collected feces and tissue samples from three wild boar carcasses (Table 1). The first animal was hunted in the Selenge province while the second and third were found dead. The second animal was discovered deceased in the forest of Minjiin Khangai, which is approximately 80 km from the Russian border (49.15271 N, 108.280836 E), whereas the third animal was found dead near the Eluur river in the Tuv province, located at (48.850119 N, 108.880305 E) (Table 1, Supplementary Material Figure S1).

Table 1.
Tissue samples collected from three wild boars.
Blood samples from the first animal and feces collected from the wild boar grazing fields were tested on-site using the Ingezim® ASF CROM Ag test (Prod Ref: 11.ASF.K42). Additionally, tissue samples from the kidney, spleen, heart, liver, and lung samples (n = 5) of the third animal were subjected to the rapid test as per the manufacturer’s instructions. Upon arrival at Mongolia’s State Central Veterinary Laboratory (SCVL), tissue samples from the first and second animals were also analyzed using the rapid antigen test.
The domestic pig sample (Mon_Dom) was obtained during the initial outbreaks of ASFV, as reported by Ankhanbaatar et al., 2021 [10].
2.2. DNA Extraction, ASFV Detection, and Genotyping
Fifteen tissue samples (n = 15) were taken from three animals, as detailed in Table 1. At the Mongolian State Central Veterinary Laboratory (SCVL), these tissue samples were processed for nucleic acid extraction. Each sample was individually homogenized, then lysed with the addition of RLT plus lysis buffer. Total nucleic acid was extracted using the Qiagen RNeasy Mini Kit, excluding the on-column DNAse digestion step. The DNA was then amplified using the protocol described by King et al., 2003 [12] and Gallardo et al., 2009 [13]. In addition, clinical samples were sent to the Animal Production and Health Laboratory (APHL) of the Joint FAO/IAEA Center, Seibersdorf, Austria.
2.3. ASFV Whole-Genome Sequence and Assembly
ASFV DNA samples extracted from a domestic pig (archived DNA) and wild boar tissues were processed for whole-genome sequencing. Approximately 50–100 ng of the DNA was used to construct barcoded libraries using Ion Xpress™ Plus Fragment Library Kit and Ion Xpress barcode adapters (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s instructions. Initially, DNA was enzymatically fragmented into 200 bp lengths using Ion shear Plus reagents, followed by ligation of adapters and barcodes to the fragmented DNA and size selection using Pippin Prep (Sage Science, Inc, Beverly, MA, USA). Next, the size-selected libraries were further amplified for eight cycles using Platinum™ PCR SuperMix high-fidelity and library amplification primer mix provided with Ion Plus Fragment Library Kit. Finally, the amplified barcoded libraries at a concentration of 100 pM were pooled in equal volumes to be loaded onto the Ion Chef™ Instrument (Thermo Fisher Scientific, Waltham, MA, USA) for automated template preparation and chip loading using Ion 540™ Kit-Chef (Thermo Fisher Scientific, Waltham, MA, USA). Then, using the Ion Chef™ Instrument, the pooled libraries were clonally amplified on the Ion spheres (ISPs) by emulsion PCR, followed by automated loading template-enriched ISPs onto an Ion 540 chip. The sequencing was performed on an Ion S5TM next-generation sequencing system (Thermo Fisher Scientific, Waltham, MA, USA) with 500 flows to generate 200 bp reads.
The run was pre-processed using the torrent suite software to remove adapters. Then, the raw reads were quality-filtered using fastq-mcf v1.04.676 (ea-utils), and their quality was assessed with FastQC (v. 011.5). A Phred score of 20 was used and reads with read lengths between 50 bp and 250 bp were selected. Using the Denovo assembly’s contigs, BLAST searches identified ASFV_China/2018/AnhuiXCGQ (MK128995) as the most relevant reference for reference-guided assembly. After mapping the cleaned raw reads against the reference sequence using Bowtie 2 (v.2.3.5.1) [14,15], Mpileup files were generated using SAMtools (v1.11) [16]. Variant calling using the consensus caller method of BCFtools (v1.9) [17] was then performed. The consensus sequences were produced with vcfutils.pl (VCFtools v0.1.16) [18] and seqtk (v1.3.106) using a Phred score 20 and compared to the de novo assemblies after aligning with Mafft (v7475) [19,20]. The assemblies were also visualised using IGV.
2.4. Comparative Analysis of ASFV Genomes and Phylogenetic Analysis
Multiple-sequence alignment was performed and visualized using BioEdit (version 7.2) [21] and nucleotide changes were analyzed using RStudio (version 2022.12.0+353) [22]. ASFV genome sequences were aligned using MAFFT software (version 7.475) [19,20].
Partial genome sequence analysis: The p72 tree included 109 sequences retrieved from GenBank [23]. A neighbor-joining tree of the partial p72 gene and the evolutionary distances were computed using the Maximum Composite Likelihood method. In addition, a Minimum Evolution tree was depicted from 80 complete sequences of the P54 gene using the Kimura 2-parameter method on MegaX [24].
Whole-genome sequence analysis: The total dataset consisted of 33 WGS covering genotypes II, III, IV, V, VII, IX, and X and the WGSs from a Mongolian wild boar and domestic pig. ASFVs from Genotypes III and VII were collected in South Africa, Genotype X in Kenya, Genotype IX in Uganda, Genotype V in Malawi, and Genotype IV in Namibia [6,25]. Out of about 70 ASFV Genotype II WGSs (>160,000 bp) deposited on GenBank, 25 showed complete metadata sets and were selected for comparison analysis. Other sequences showed missing information, including on host, country, and year of sample collection, or had incomplete sequences compared to the reference genome sequence collected in Georgia in 2007 (NCBI reference: NC 044959). The sequences were visualized, aligned, and analyzed using BioEdit and MAFFT. Any sequences retrieved from NCBI with unidentified nucleotides (represented by ‘N’) or incomplete sequences were eliminated. The final dataset included Genotype II ASFVs from China (6), Poland (6), Russia (3), Ukraine (1), Germany (1), Georgia (1), Timor-Leste (1), Korea (1), Lithuania (1), Italy (1), Belgium (1), Malawi (1), and India (1) (Table 2). While selecting Genotype II WGSs, we concentrated on sequences representative of different geographical regions in Europe and Asia since samples from Mongolia had been collected within these regions. Sequences from the Asian continent were derived from 2018–2022, whereas sequences from Europe were from 2007 to 2022. The two WGSs from Mongolia are included in Table 2 as Mon_Dom and Mon_WB. Sample Mon_Dom belongs to Genotype II, and Mon_WB was genotyped in this study. WGS data for Mon_WB and Mon_Dom have been submitted to Genbank. The Sequence Read Archive (SRA) identifiers for these sequences are SAMN36269616 for Mon_WB and SAMN36269617 for Mon_Dom. The Genbank accession numbers are OR271565 and OR271566 for Mon_WB and Mon_Dom, respectively.

Table 2.
Metadata of whole-genome sequence dataset.
Maximum likelihood (ML) phylogenetic trees were estimated by using IQ-TREE (version 1.6.12) [26] using the HKY nucleotide substitution model. Bootstrapping was performed to assess the robustness of tree topologies. The final tree was midpoint-rooted by iTOL version 6.6 [27]. In addition, to visualize ASFV Genotype II WGS, the software GrapeTree was applied for further comparative assessment. This open-source software tool uses a hierarchical clustering algorithm to group closely related strains [28].
3. Results
The rapid ASFV test conducted on-site using blood samples from the hunted wild boar (animal no. 1, Table 1) and feces collected from the fields yielded negative results. However, tissue samples obtained from wild boars found dead (animals no. 2 and 3, Table 1) and analyzed at the SCVL using the rapid ASFV test produced positive results. Real-time PCR was used to detect the ASFV genome on tissue samples from dead wild boars (animal 2 and 3, Table 1) using the method of King et al., 2003 [12]. In addition, the PCR amplification of p54 and p72 gene fragments was conducted following the protocol described in Gallardo et al. 2009. All six samples were positive (Supplementary Table S1). The sequencing coverage for the Mongolian wild boar (Mon_WB) WGS was 660 ± 200, and that for the Mongolian domestic pig was 133 ± 64. From the WGSs, P72 and P54 sequence segments were chosen and analyzed in a phylogenetic tree (Figure 1 and Figure 2). The analysis of both segments in the phylogenetic tree revealed that both Mon_WB and Mon_Dom belong to Genotype II with 99.99% similarity to other Genotype II sequences available in public databases. In addition, the sample Mon_WB showed high similarity to the index case Mon_Dom from 2019.
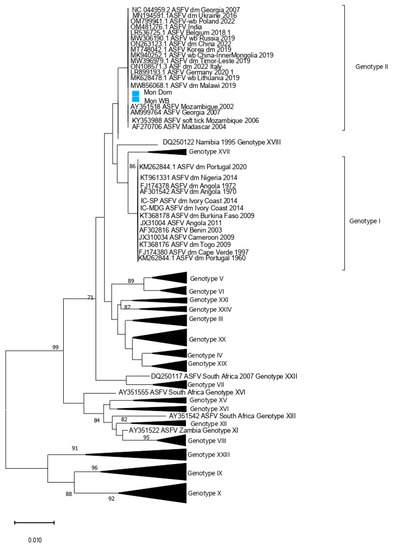
Figure 1.
Phylogenetic tree based on partial P72 gene inferred using neighbor-joining method. The Mongolia ASFV samples from wild boars and domestic pigs are highlighted in blue.
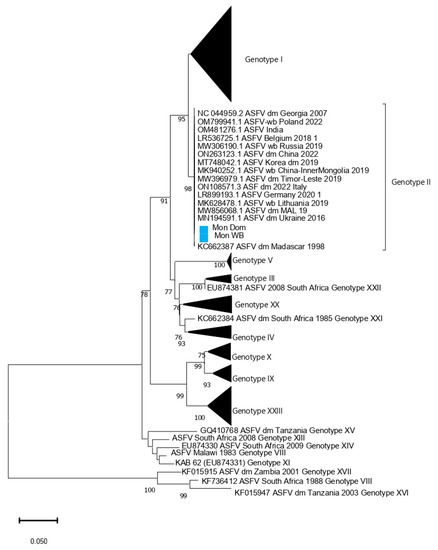
Figure 2.
Minimum Evolution tree based on full P54 gene sequences. The Mongolia ASFV samples from wild boars and domestic pigs are highlighted in blue.
The maximum likelihood phylogenetic tree of ASFV whole genomes showed two primary clades with 100% bootstrap support (Figure 3). Primary clade I included two subclades, comprising the previously identified Genotypes III, IV, V, VII, IX, and X, all from Africa. Primary clade II included Genotype II viruses from Africa, Europe, and Asia.
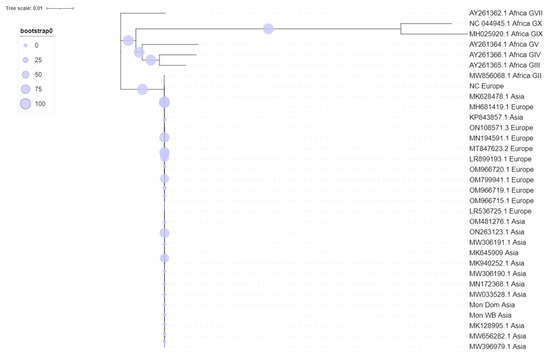
Figure 3.
Maximum likelihood phylogenetic midpoint-rooted tree of ASFV genome sequences.
Groupwise comparison between the Asian and European sequences using Mega (version 11) revealed a similarity of 99.99% within sequences and 409 mutations, of which 207 were deletions. There were three region-specific nucleotide changes between the ASFV Genotype II sequences from Asia and Europe (Table 3). The first distinct nucleotide change was on gene ASFV G ACD 00190 CDS, the second was on the gene MGF 360-10L CDS, and the third mutation was on a non-translation region between B602L and B385R. The first two nucleotide changes were non-synonymous (M/F and N/S, respectively).

Table 3.
Genotype II differences in nucleotide.
Within the Asian and European ASFV Genotype II sequences, there were 74 and 119 nucleotide changes, respectively. Among the fourteen sequences from Asia in Table 3, thirteen had a gap at position 9114 in the gene ASFV G ACD 00190 CDS, while the sequence from India in 2022 (OM481276) had an “a” at that position. In contrast, each of two sequences from Europe (MH681419, MK628478) had a gap at the same position, representing outbreaks in Poland in 2015 and Lithuania in 2019. Thirteen out of fourteen Asian sequences had a “c” at position 22996 within the MGF 360-10L CDS (Table 3), while all European sequences had a “t”, including the sequence from Russia (KP843857). All European sequences and two from Asia (KP843857 and OM481276) had a gap at the third mutation within the intergenic region between B602L and B385R, whereas 12 out of 14 Asian sequences had a “g” (Table 3). The unique sequence from Africa followed the same pattern as the European sequences, with an “a” at the first, a “t” at the second, and a gap at the third nucleotide position (Table 3).
Five mutation sites were identified by comparing sequences from Mongolia with the reference genome of ASFV Genotype II from Georgia in 2007. Among these, two were minor mutations, and three major changes were located in the MGF 110 region, resulting in amino acid changes.
In Figure 4, a GrapeTree visualization of 27 WGS shows that the European and Asian sequences are separated into two distinct clusters, with the European sequences being more dispersed than the Asian sequences. The tree is based on asymmetric Hamming-like distances in which the directionality is from more complete to less complete profiles. Within the GrapeTree visualization, the Asian sequences have the closest connection to sequence LR536725 from Belgium, which was collected in 2018. The sequence from Malawi, dating from 2019, is connected to the Georgian sequence from 2007 (NC044959). The GrapeTree image visualizes data from Table 3 and shows that the European and Asian sequences form two separate groups. Sample Mon_WB is placed next to the index case Mon_Dom, further indicating a high similarity between these samples.
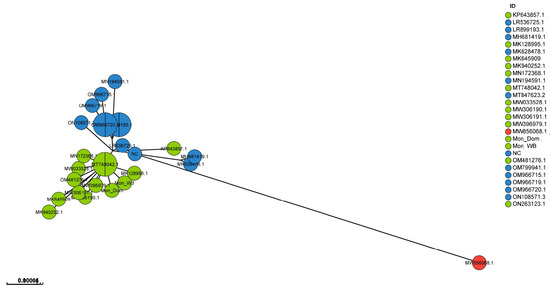
Figure 4.
GrapeTree interface exemplified with a precalculated Newick tree based on 27 WGSs of ASFV Genotype II. Nodes in blue include European sequences, nodes in green show Asian sequences, and the sequence from Africa is shown in red.
4. Discussion
The World Organisation of Animal Health ranks ASF as the most important cause of mortality in domestic pigs globally [3].
The comparative analysis of ASFV Genotype II genomes indicates that ASFVs from Mongolia have genetic similarities to Asian Genotype II ASFVs. Interestingly, three nucleotide changes between Asian and European WGS were identified, including two non-synonymous within the MGF360 gene and the gene ACD 00190 CDS. MFG 360 gene products have been reported to be related to the suppression of a type I interferon response [11]. In contrast, the gene ACD 00190 CDS is only associated with hypothetical proteins and its function is currently unknown [29]. The WGSs obtained from GenBank (NCBI) were produced using a range of sequencing platforms, such as Illumina NovaSeq and IonTorrent, indicating that the observed mutations are not exclusively caused by errors inherent to a single technology. Furthermore, recent studies have reported mutations in genes MFG 360 and ACD 00190 CDS [30].
Due to its large genome size (>190 kb), the ASFV, like many other large DNA viruses, mutates relatively slowly. Despite the occurrence of 409 nucleotide substitutions between the Asian and European sequences, these groups still exhibit a homogeneity of over 99.99%. Thus, the evolutionary trend of the ASFV is generally constant [31]. The number of substitutions within the European sequences is 119, which is 61% higher than that within the Asian group, which has 74 changes. The greater diversity within the European group is also better visualized in the GrapeTree image, which displays the genetic relationships calculated from the minimum spanning tree algorithm (MSTree V2) [28]. One possible explanation for the greater diversity among the European ASFVs could be the longer timespan covered by these sequences, which ranges from 2007 to 2022, compared to the Asian sequences from isolates dated from 2018. The sequence from Korea is placed the closest to European sequences, indicating a higher similarity between MT748042 and sequences from Europe. Similar findings have been reported by Kim et al., 2023, showing that the Korea/YC1/2019 strain shared the highest similarity with the Georgia 2007, Belgium 2018/1, and ASFV-wbBS01 strains [32]. However, comparison with sequences from domestic pigs was not investigated.
Interestingly, the sequence of the sample collected in Malawi in 2019 is connected to the reference sequence from Georgia from 2007 based on asymmetric Hamming-like distances (Figure 4). The Malawian sequence was compared in a previous study to other African sequences by Hakizimana et al., 2021 [33]. They concluded that the sequence from Malawi collected in 2019 resembled sequences from Tanzania in 2017 with 99.7% nucleotide identity. In addition, the Malawian ASFV strain had more than 99% nucleotide identity with ASFV Genotype II viruses previously described in Europe and Asia, suggesting a possible common ancestor [34].
This study analyzed the phylogenetic relationship between ASFVs collected in wild boars and domestic pigs in Mongolia and other Genotype II ASFVs from Eurasia, including the ASFV reference strain from Georgia in 2007 (NCBI reference: NC 044959). The results suggest a likely introduction of the virus through cross-border spread due to close geographic proximity to Russia and animal movements from North to Southeast Asia [11]. However, there is also a possibility that the virus might have already circulated in Mongolia and spread to neighboring countries before its detection. The first reported case of the ASFV in Russia was in 2007, leading to subsequent reported cases over the years until the virus was detected in China in 2018, followed by its detection in Mongolia, Vietnam, Cambodia, Korea, and Timor-Leste [11,34]. Limitations in using the WGS of the ASFV for molecular epidemiology have been highlighted in a study by Forth et al. (2020). The study compared 29 WGSs of corresponding Genotype II strains from 10 affected countries in Asia, Europe, and Africa. Forth et al. reported a high genetic similarity among circulating ASFV strains and the absence of a clear pattern or phenotype change [35]. However, in our study, we were able to find patterns that distinguished European and Asian ASFV Genotype II WGSs using our curated dataset with complete metadata and sequences as compared to the reference genome sequence collected in Georgia in 2007 (NCBI reference: NC_044959). In addition, the study revealed a high similarity between the ASFV whole-genome sequences of Genotype II in domestic pigs and wild boars. This suggests that ASFV strains obtained from infected domestic pigs could have impacted wild boars and vice versa. This result is consistent with previous research conducted in Europe [5]. Direct contact between infectious domestic pigs and wild boars and the consumption of infected raw and frozen pork, blood, offal, and carcasses of dead wild boars and domestic pigs have been reported as the main transmission routes of the ASFV to domestic pigs and likely occur between wild boars and domestic pigs [36,37]. In the Caucasus, ASFV spread is attributed to wild boars’ movements [38]. The infected wild boar population and the low adherence to on-farm biosecurity in Eastern European and Asian countries may explain the tremendous spread of this disease, e.g., free-range pig management systems have been observed in this region [39]. In addition to carrier animals, several other mechanisms can lead to the long-term circulation of the ASFV in pig or wild boar populations. The most important are human-induced factors such as the illegal movement of infected pork, swill feeding, and insects [40].
5. Conclusions
In this study, we showed that the ASFV Genotype II sequences in wild boars and domestic pigs are similar and that the evolutionary trend of the ASFV is generally constant. In conclusion, this study expands our knowledge of the genetic diversity of ASFVs and supports published data on current epidemiological studies on ASFV circulation in Europe and Asia.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pathogens12091143/s1, Figure S1: Carcasses of Wild Boars Found in Tuv and Selenge Provinces.; Table S1: PCR Analysis of p54 and p72 Gene Fragments using Extracted DNA from Six Types of Tissue Samples (Animals 2 and 3).
Author Contributions
Conceptualization, all authors; methodology, C.E.L., A.A., T.B.K.S., U.A., D.G.-O., E.-O.T.-O. and H.O.A.; formal analysis, A.A., T.T. and C.E.L.; resources, G.C.; writing—original draft preparation, A.A. and U.A.; writing—review and editing, all authors. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by VETLAB network initiative of the Joint FAO/IAEA Centre through the IAEA Peaceful Uses Initiative Project funded by the Government of the Unites States of America.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are available upon reasonable request.
Acknowledgments
We would like to thank the Laboratory Unit—Emergency Prevention System (EMPRES), Animal Health Service, Food and Agriculture Organization of the United Nations (FAO-UN), Rome, Italy, for their support (GCP/GLO/074/USA). We also thank the General Authority of Veterinary Service, Institute of Biology, and rangers of the Khan Khentii Strictly Protected Area in Mongolia for their assistance during field sampling.
Conflicts of Interest
The authors declare no conflict of interest.
Disclaimer
The views expressed in this publication are those of the author(s) and do not necessarily reflect the views or policies of the Food and Agriculture Organization of the United Nations.
References
- Bao, J.; Zhang, Y.; Shi, C.; Wang, Q.; Wang, S.; Wu, X.; Cao, S.; Xu, F.; Wang, Z. Genome-Wide Diversity Analysis of African Swine Fever Virus Based on a Curated Dataset. Animals 2022, 12, 2446. [Google Scholar] [CrossRef] [PubMed]
- Schulz, K.; Boklund, A. The Epidemiology of African Swine Fever, Its Complexity and the Requirement for Multiple Solution Approaches. Animals 2020, 10, 1900. [Google Scholar] [CrossRef] [PubMed]
- Njau, E.P.; Domelevo Entfellner, J.B.; Machuka, E.M.; Bochere, E.N.; Cleaveland, S.; Shirima, G.M.; Kusiluka, L.J.; Upton, C.; Bishop, R.P.; Pelle, R.; et al. The first genotype II African swine fever virus isolated in Africa provides insight into the current Eurasian pandemic. Sci. Rep. 2021, 11, 13081. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, R.E. On a form of swine fever occurring in British east-Africa (Kenya Colony). J. Comp. Pathol. Ther. 1921, 34, 159–191. [Google Scholar] [CrossRef]
- Giammarioli, M.; Alessandro, D.; Cammà, C.; Masoero, L.; Torresi, C.; Marcacci, M.; Zoppi, S.; Curini, V.; Rinaldi, A.; Rossi, E.; et al. Molecular Characterization of the First African Swine Fever Virus Genotype II Strains Identified from Mainland Italy, 2022. Pathogens 2023, 12, 372. [Google Scholar] [CrossRef] [PubMed]
- Portugal, R.; Coelho, J.; Hoeper, D.; Little, N.; Smithson, C.; Upton, C.; Martins, C.; Leitão, A.; Keil, G. Related strains of African swine fever virus with different virulence: Genome comparison and analysis. J. Gen. Virol. 2014, 96, 408–419. [Google Scholar] [CrossRef]
- Fiori, M.S.; Sanna, D.; Scarpa, F.; Floris, M.; Di Nardo, A.; Ferretti, L.; Loi, F.; Cappai, S.; Sechi, A.M.; Angioi, P.P.; et al. A Deeper Insight into Evolutionary Patterns and Phylogenetic History of ASFV Epidemics in Sardinia (Italy) through Extensive Genomic Sequencing. Viruses 2021, 13, 1994. [Google Scholar] [CrossRef] [PubMed]
- Beltrán-Alcrudo, D.; Lubroth, J.; Depner, K.; De La Rocque, S. African swine fever in the Caucasus. FAO Empres. Watch. 2008, 1, 1–8. [Google Scholar]
- Zhou, X.; Li, N.; Luo, Y.; Liu, Y.; Miao, F.; Chen, T.; Zhang, S.; Cao, P.; Li, X.; Tian, K.; et al. Emergence of African Swine Fever in China, 2018. Transbound. Emerg. Dis. 2018, 65, 1482–1484. [Google Scholar] [CrossRef]
- Ankhanbaatar, U.; Sainnokhoi, T.; Khanui, B.; Ulziibat, G.; Jargalsaikhan, T.; Dulam, P.; Settypalli, B.K.; Flannery, J.; Dundon, W.; Basan, G.; et al. African Swine Fever Virus Genotype II in Mongolia, 2019. Transbound. Emerg. Dis. 2021, 68, 2787–2794. [Google Scholar] [CrossRef]
- Afonso, C.L.; Piccone, M.E.; Zaffuto, K.M.; Neilan, J.; Kutish, G.F.; Lu, Z.; Balinsky, C.A.; Gibb, T.R.; Bean, T.J.; Zsak, L.; et al. African swine fever virus multigene family 360 and 530 genes affect host interferon response. J. Virol. 2004, 78, 1858–1864. [Google Scholar] [CrossRef] [PubMed]
- King, D.P.; Reid, S.M.; Hutchings, G.H.; Grierson, S.S.; Wilkinson, P.J.; Dixon, L.K.; Bastos, A.D.; Drew, T.W. Development of a TaqMan PCR assay with internal amplification control for the detection of African swine fever virus. J. Virol. Methods 2003, 107, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, C.; Mwaengo, D.M.; Macharia, J.M.; Arias, M.; Taracha, E.A.; Soler, A.; Okoth, E.; Martín, E.; Kasiti, J.; Bishop, R.P. Enhanced discrimination of African swine fever virus isolates through nucleotide sequencing of the p54, p72, and pB602L (CVR) genes. Virus Genes 2009, 38, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Wilks, C.; Antonescu, V.; Charles, R. Scaling read aligners to hundreds of threads on general-purpose processors. Bioinformatics 2018, 35, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.E.; Handsaker, R.; Lunter, G.; Marth, G.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef]
- Nakamura, T.; Yamada, K.D.; Tomii, K.; Katoh, K. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics 2018, 34, 2490–2492. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- RStudio Team. RStudio: Integrated Development for R. Rstudio; PBC: Boston, MA, USA, 2020; Available online: http://www.rstudio.com/ (accessed on 1 September 2022).
- NCBI-National Center for Biotechnology Information (NCBI) [Internet]. National Library of Medicine (US). National Center for Biotechnology Information: Bethesda, MD, USA. 1988. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 6 March 2023).
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms Molecular Biology and Evolution; Oxford University Press: Oxford, UK, 2018; Volume 35, p. 1547. [Google Scholar]
- Zhang, Y.; Ke, J.; Zhang, J.; Yang, J.; Yue, H.; Zhou, X.; Qi, Y.; Zhu, R.; Miao, F.; Li, Q.; et al. African Swine Fever Virus Bearing an I226R Gene Deletion Elicits Robust Immunity in Pigs to African Swine Fever. J. Virol. 2021, 95, e0119921. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Alikhan, N.F.; Sergeant, M.J.; Luhmann, N.; Vaz, C.; Francisco, A.P.; Carriço, J.A.; Achtman, M. GrapeTree: Visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018, 28, 1395–1404. [Google Scholar] [CrossRef] [PubMed]
- Olesen, A.S.; Kodama, M.; Lohse, L.; Accensi, F.; Rasmussen, T.B.; Lazov, C.M.; Limborg, M.T.; Gilbert, M.T.P.; Bøtner, A.; Belsham, G.J. Identification of African Swine Fever Virus Transcription within Peripheral Blood Mononuclear Cells of Acutely Infected Pigs. Viruses 2021, 13, 2333. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Núñez, D.; Castillo-Rosa, E.; Vigara-Astillero, G.; García-Belmonte, R.; Gallardo, C.; Revilla, Y. Identification and Isolation of Two Different Subpopulations within African Swine Fever Virus Arm/07 Stock. Vaccines 2020, 8, 625. [Google Scholar] [CrossRef] [PubMed]
- Rolesu, S.; Mandas, D.; Loi, F.; Oggiano, A.; Dei Giudici, S.; Franzoni, G.; Guberti, V.; Cappai, S. African swine fever in smallholder Sardinian farms: Last ten years of network transmission reconstruction and analysis. Front. Vet. Med. 2021, 8, 692448. [Google Scholar] [CrossRef]
- Kim, G.; Park, J.-E.; Kim, S.-J.; Kim, Y.; Kim, W.; Kim, Y.-K.; Jheong, W. Complete genome analysis of the African swine fever virus isolated from a wild boar responsible for the first viral outbreak in Korea, 2019. Front. Vet. Sci. 2023, 9, 1080397. [Google Scholar] [CrossRef]
- Hakizimana, J.N.; Ntirandekura, J.B.; Yona, C.; Nyabongo, L.; Kamwendo, G.; Chulu, J.L.C.; Ntakirutimana, D.; Kamana, O.; Nauwynck, H.; Misinzo, G. Complete genome analysis of African swine fever virus responsible for outbreaks in domestic pigs in 2018 in Burundi and 2019 in Malawi. Trop. Anim. Health Prod. 2021, 53, 438. [Google Scholar] [CrossRef]
- Mazur-Panasiuk, N.; Wozniakowski, G.; Niemczuk, K. The first complete genomic sequences of african swine fever virus isolated in Poland. Sci. Rep. 2019, 9, 4556. [Google Scholar] [CrossRef] [PubMed]
- Forth, J.H.; Forth, L.F.; Blome, S.; Höper, D.; Beer, M. African swine fever whole-genome sequencing—Quantity wanted but quality needed. PLoS Pathog. 2020, 16, e1008779. [Google Scholar] [CrossRef]
- Guinat, C.; Gogin, A.; Blome, S.; Keil, G.; Pollin, R.; Pfeiffer, D.U.; Dixon, L. Transmission routes of African swine fever virus to domestic pigs: Current knowledge and future research directions. Vet. Rec. 2016, 178, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Cadenas-Fernández, E.; Ito, S.; Aguilar-Vega, C.; Sánchez-Vizcaíno, J.M.; Bosch, J. The Role of the Wild Boar Spreading African Swine Fever Virus in Asia: Another Underestimated Problem. Front. Vet. Sci. 2022, 9, 844209. [Google Scholar] [CrossRef] [PubMed]
- Pautienius, A.; Grigas, J.; Pileviciene, S.; Zagrabskaite, R.; Buitkuviene, J.; Pridotkas, G.; Stankevicius, R.; Streimikyte, Z.; Salomskas, A.; Zienius, D.; et al. Prevalence and spatiotemporal distribution of African swine fever in Lithuania, 2014–2017. Virol. J. 2018, 15, 177. [Google Scholar] [CrossRef] [PubMed]
- Cwynar, P.; Stojkov, J.; Wlazlak, K. African Swine Fever Status in Europe. Viruses 2019, 11, 310. [Google Scholar] [CrossRef]
- Guberti, V.; Khomenko, S.; Masiulis, M.; Kerba, S. African Swine Fever in Wild Boar—Ecology and Biosecurity, 2nd ed.; FAO Animal Production and Health Manual No. 28; FAO; World Organisation for Animal Health and European Commission: Rome, Italy, 2022. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© FAO, 2023. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY NC SA) license (https://creativecommons.org/licenses/by-nc-sa/3.0/igo/).