Bovine Papillomavirus Type 1 Infection in an Equine Congenital Papilloma
Abstract
1. Introduction
- classic equine viral papillomatosis via EcPV-1, which usually affects the muzzle and lips of young horses (<3 years of age) and regresses spontaneously within 2–3 months;
- equine genital papillomas caused by EcPV-2, which affect older horses and do not resolve spontaneously;
- equine ear papillomas caused by EcPV3-6, which present with bilateral and symmetric white hyperkeratotic plaques confined to the pinnae, and rarely resolve spontaneously [3].
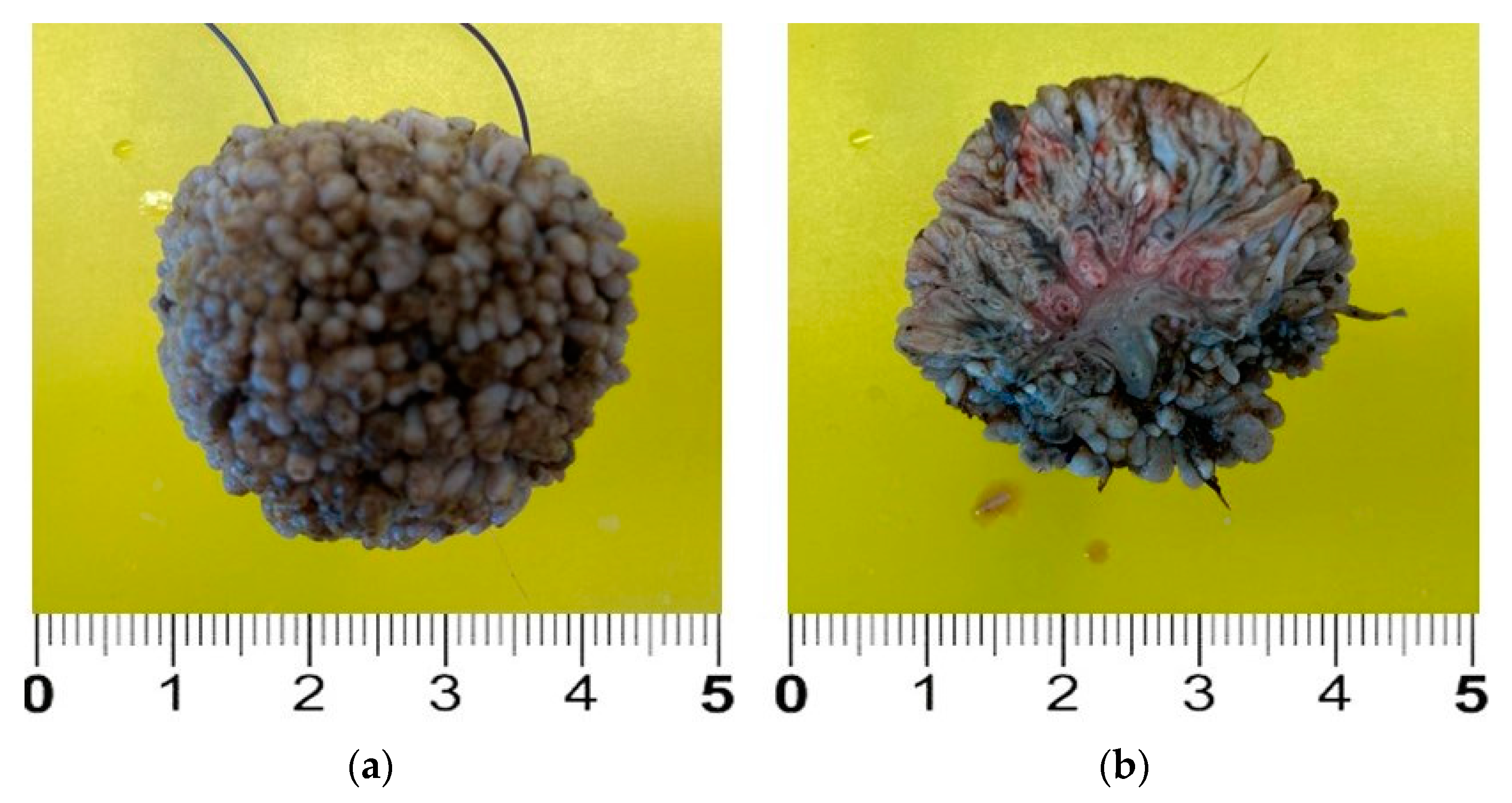
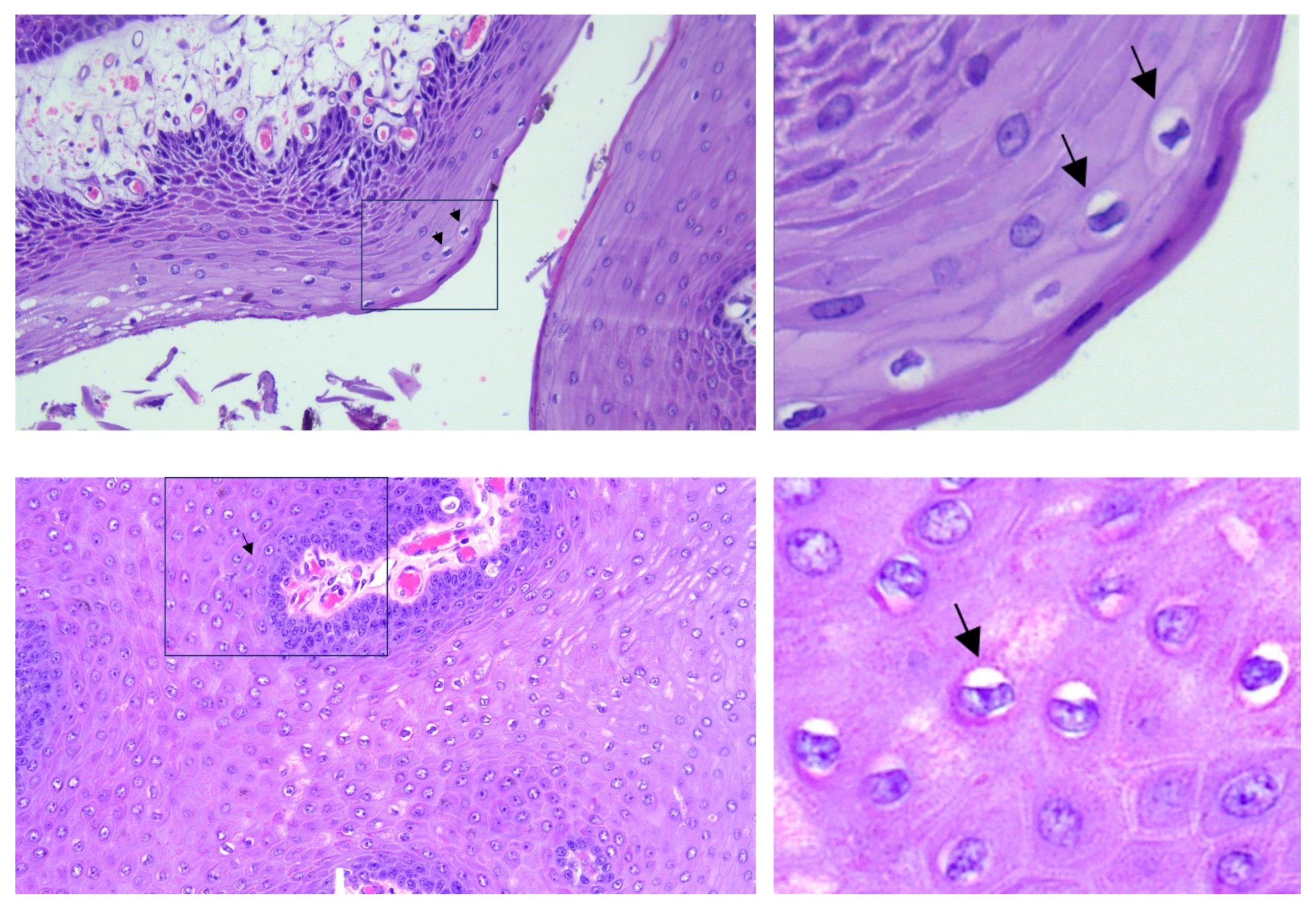
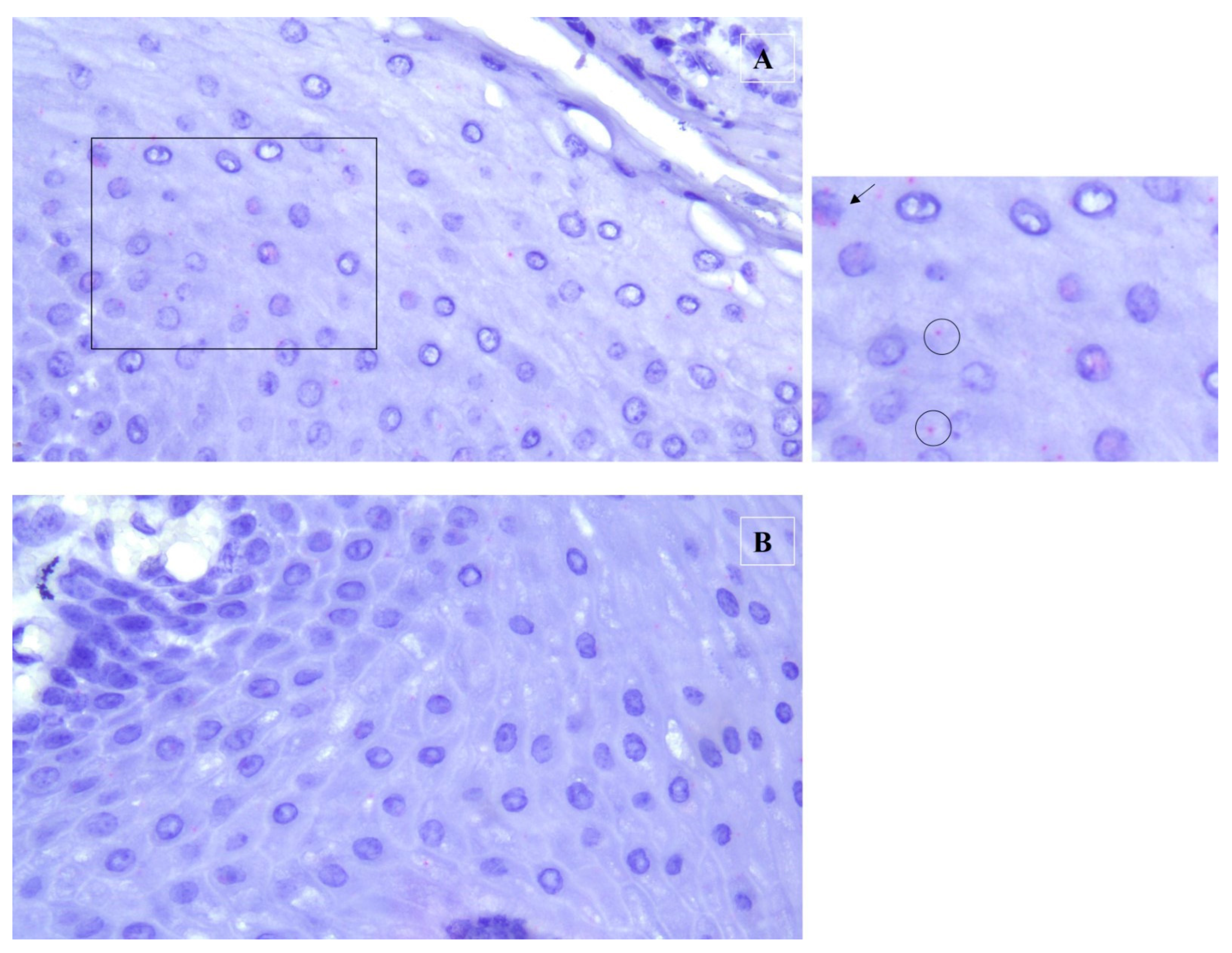
2. Case Description
3. Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kumar, V.; Abbas, A.K.; Fausto, N. Neoplasia. In Pathologic Basis of Disease, 7th ed.; Kumar, V., Abbas, A.K., Fausto, N., Eds.; Elsevier Saunders: Philadelphia, PA, USA, 2005; pp. 269–342. [Google Scholar]
- de Villiers, E.M.; Fauquet, C.; Broker, T.R.; Bernard, H.U.; zur Hausen, H. Classification of papillomaviruses. Virology 2004, 324, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Mauldin, E.A.; Peters-Kennedy, J. Integumentary System. In Pathology of Domestic Animals, 6th ed.; Maxie, M.G., Ed.; Elsevier: Alpharetta, GA, USA, 2015; pp. 509–736. [Google Scholar]
- Scott, D.W. Neoplastic diseases. In Large Animal Dermatology; Scott, D.W., Ed.; W.B. Saunders Company: Philadelphia, PA, USA, 1998; pp. 419–468. [Google Scholar]
- Goodrich, L.; Gerber, H.; Marti, E.; Antczak, D.F. Equine sarcoids. Vet. Clin. N. Am. Equine Pract. 1998, 14, 607–623. [Google Scholar] [CrossRef] [PubMed]
- White, K.S.; Fuji, R.N.; Valentine, B.A.; Bildfell, R.J. Equine congenital papilloma: Pathological findings and results of papillomavirus immunohistochemistry in five cases. Vet. Dermatol. 2004, 15, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Postey, R.C.; Appleyard, G.D.; Kidney, B.A. Evaluation of equine papillomas, aural plaques, and sarcoids for the presence of Equine papillomavirus DNA and Papillomavirus antigen. Can. J. Vet. Res. 2007, 71, 28–33. [Google Scholar] [PubMed]
- Christmann, U. Handling and restraint. In Equine Reproductive Procedures, 2nd ed.; Dascanio, J., McCue, P., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2021; pp. 681–686. [Google Scholar]
- Wang, F.; Flanagan, J.; Su, N.; Wang, L.C.; Bui, S.; Nielson, A.; Wu, X.; Vo, H.T.; Ma, X.J.; Luo, Y. RNAscope: A novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 2012, 14, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Campo, M.S. Papillomavirus and disease in humans and animals. Vet. Comp. Oncol. 2003, 1, 3–14. [Google Scholar] [CrossRef]
- Araldi, R.P.; Assaf, S.M.R.; Carvalho, R.F.; Carvalho, M.A.C.R.; Souza, J.M.; Magnelli, R.F.; Mόdolo, D.G.; Roperto, F.P.; de Cassia Stocco, R.; Beçak, W. Papillomaviruses: A systematic review. Genet. Mol. Biol. 2017, 40, 1–21. [Google Scholar] [CrossRef]
- Okunade, K.S. Human papillomavirus and cervical cancer. J. Obstet. Gynaecol. 2020, 40, 602–608. [Google Scholar] [CrossRef]
- Tumban, E. A Current Update on Human Papillomavirus-Associated Head and Neck Cancers. Viruses 2019, 11, 922. [Google Scholar] [CrossRef]
- Hainisch, E.K.; Jindra, C.; Kirnbauer, R.; Brandt, S. Papillomavirus-like Particles in Equine Medicine. Viruses 2023, 15, 345. [Google Scholar] [CrossRef]
- Straticò, P.; Razzuoli, E.; Hattab, J.; Guerri, G.; Celani, G.; Palozzo, A.; Bonanni, D.; Fruscione, F.; Varasano, V.; Petrizzi, L.; et al. Equine Gastric Squamous Cell Carcinoma in a Friesian Stallion. J. Equine Vet. Sci. 2022, 117, 104087. [Google Scholar] [CrossRef] [PubMed]
- Linder, K.E.; Bizikova, P.; Luff, J.; Zhou, D.; Yuan, H.; Breuhaus, B.; Nelson, E.; Mackay, R. Generalized papillomatosis in three horses associated with a novel equine papillomavirus (EcPV8). Vet. Dermatol. 2018, 29, 72-e30. [Google Scholar] [CrossRef] [PubMed]
- Li, C.X.; Chang, W.S.; Mitsakos, K.; Rodger, J.; Holmes, E.C.; Hudson, B.J. Identification of a Novel Equine Papillomavirus in Semen from a Thoroughbred Stallion with a Penile Lesion. Viruses 2019, 11, 713. [Google Scholar] [CrossRef] [PubMed]
- Turco, S.; Gabbianelli, F.; Mavian, C.N.; Pietrucci, D.; De Paolis, L.; Gialletti, R.; Mechelli, L.; De Ciucis, C.G.; Cappelli, K.; Dell’Anno, F.; et al. Genetic Characterization of a Novel Equus caballus Papillomavirus Isolated from a Thoroughbred Mare. Viruses 2023, 15, 650. [Google Scholar] [CrossRef]
- Campo, M.S. Animal models of papillomavirus pathogenesis. Virus Res. 2002, 89, 249–261. [Google Scholar] [CrossRef]
- Savini, F.; Gallina, L.; Prosperi, A.; Puleio, R.; Lavazza, A.; Di Marco, P.; Tumino, S.; Moreno, A.; Lelli, D.; Guercio, A.; et al. Bovine Papillomavirus 1 Gets Out of the Flock: Detection in an Ovine Wart in Sicily. Pathogens 2020, 9, 429. [Google Scholar] [CrossRef]
- Trewby, H.; Ayele, G.; Borzacchiello, G.; Brandt, S.; Campo, M.S.; Del Fava, C.; Marais, J.; Leonardi, L.; Vanselow, B.; Biek, R.; et al. Analysis of the long control region of bovine papillomavirus type 1 associated with sarcoids in equine hosts indicates multiple cross-species transmission events and phylogeographical structure. J. Gen. Virol. 2014, 95, 2748–2756. [Google Scholar] [CrossRef][Green Version]
- Nasir, L.; Reid, S.W.J. Bovine papillomaviruses and equine sarcoids. In Papillomavirus Research: From Natural History to Vaccines and Beyond; Campo, M.S., Ed.; Caister Academic Press: Norfolk, UK, 2006; pp. 389–397. [Google Scholar]
- Brandt, S.; Tober, R.; Corteggio, A.; Burger, S.; Sabitzer, S.; Walter, I.; Kainzbauer, C.; Steinborn, R.; Nasir, L.; Borzacchiello, G. BPV-1 infection is not confined to the dermis but also involves the epidermis of equine sarcoids. Vet. Microbiol. 2011, 150, 35–40. [Google Scholar] [CrossRef]
- Hartl, B.; Hainisch, E.K.; Shafti-Keramat, S.; Kirnbauer, R.; Corteggio, A.; Borzacchiello, G.; Tober, R.; Kainzbauer, C.; Pratscher, B.; Brandt, S. Inoculation of young horses with bovine papillomavirus type 1 virions leads to early infection of PBMCs prior to pseudo-sarcoid formation. J. Gen. Virol. 2011, 92, 2437–2445. [Google Scholar] [CrossRef]
- Savini, F.; Gallina, L.; Mazza, F.; Mariella, J.; Castagnetti, C.; Scagliarini, A. Molecular Detection of Bovine Papillomavirus DNA in the Placenta and Blood of Healthy Mares and Respective Foals. Vet. Sci. 2019, 6, 14. [Google Scholar] [CrossRef]
- Brandt, S.; Schoster, A.; Tober, R.; Kainzbauer, C.; Burgstaller, J.P.; Haralambus, R.; Steinborn, R.; Hinterhofer, C.; Stanek, C. Consistent detection of bovine papillomavirus in lesions, intact skin and peripheral blood mononuclear cells of horses affected by hoof canker. Equine Vet. J. 2011, 43, 202–209. [Google Scholar] [CrossRef]
- Silva, M.A.; Silva, K.M.; Jesus, A.L.; Barros, L.O.; Corteggio, A.; Altamura, G.; Borzacchiello, G.; Freitas, A.C. The presence and gene expression of bovine papillomavirus in the peripheral blood and semen of healthy horses. Transbound. Emerg. Dis. 2014, 61, 329–333. [Google Scholar] [CrossRef]
- Armbruster-Moraes, E.; Ioshimoto, L.M.; Leão, E.; Zugaib, M. Presence of human papillomavirus DNA in amniotic fluids of pregnant women with cervical lesions. Gynecol. Oncol. 1994, 54, 152–158. [Google Scholar] [CrossRef]
- Rombaldi, R.L.; Serafini, E.P.; Mandelli, J.; Zimmermann, E.; Losquiavo, K.P. Perinatal transmission of human papilomavirus DNA. Virol. J. 2009, 6, 83. [Google Scholar] [CrossRef]
- Freitas, A.C.; Mariz, F.C.; Silva, M.A.; Jesus, A.L. Human papillomavirus vertical transmission: Review of current data. Clin. Infect. Dis. 2013, 56, 1451–1456. [Google Scholar] [CrossRef] [PubMed]
- Roperto, S.; Borzacchiello, G.; Esposito, I.; Riccardi, M.; Urraro, C.; Lucà, R.; Corteggio, A.; Tatè, R.; Cermola, M.; Paciello, O.; et al. Productive infection of bovine papillomavirus type 2 in the placenta of pregnant cows affected with urinary bladder tumors. PLoS ONE 2012, 7, 33569. [Google Scholar] [CrossRef] [PubMed]
- Roperto, S.; Russo, V.; De Falco, F.; Taulescu, M.; Roperto, F. Congenital papillomavirus infection in cattle: Evidence for transplacental transmission. Vet. Microbiol. 2019, 230, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Roperto, S.; Russo, V.; Corrado, F.; De Falco, F.; Munday, J.S.; Roperto, F. Oral fibropapillomatosis and epidermal hyperplasia of the lip in newborn lambs associated with bovine Deltapapillomavirus. Sci. Rep. 2018, 8, 13310. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.A.R.; Batista, M.V.A.; Pontes, N.E.; Santos, E.U.D.; Coutinho, L.C.A.; Castro, R.S.; Balbino, V.Q.; Freitas, A.C. Comparison of two PCR strategies for the detection, of bovine papillomavirus. J. Virol. Methods 2013, 192, 55–58. [Google Scholar] [CrossRef]
- Foresta, C.; Patassini, C.; Bertoldo, A.; Menegazzo, M.; Francavilla, F.; Barzon, L.; Ferlin, A. Mechanism of human papillomavirus binding to human spermatozoa and fertilizing ability of infected spermatozoa. PLoS ONE 2011, 6, e15036. [Google Scholar] [CrossRef]
- Bogaert, L.; Martens, A.; Van Poucke, M.; Ducatelle, R.; De Cock, H.; Dewulf, J.; De Baere, C.; Peelman, L.; Gasthuys, F. High prevalence of bovine papillomaviral DNA in the normal skin of equine sarcoid-affected and healthy horses. Vet. Microbiol. 2008, 129, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Pratscher, B.; Hainisch, K.; Sykora, S.; Brandt, S.; Jindra, C. No evidence of bovine papillomavirus type 1 or 2 infection in healthy equids. Equine Vet. J. 2019, 51, 612–616. [Google Scholar] [CrossRef] [PubMed]


| Target Gene | Primer Sequences | Amplicon Length | Accession Number |
|---|---|---|---|
| EcPV2-L1 | F-5′-TTGTCCAGGAGAGGGGTTAG-3′ | 80 | NC_012123.1 |
| R-5′-TGCCTTCCTTTTCTTGGTGG-3′ | |||
| p-EcPV2-L1 | FAM-CGTCCAGCACCTTCGACCACCA-TAMRA | ||
| EcPV9-L1 | F-5′-TTC ATC CCA GCT TGA GAC CA-3′ | 116 | MN117918.1 |
| R-5′-GCA GAT CAA TGG TCC AGA AGG-3′ | |||
| p-EcPV9-L1 | p-FAM-ATT GCC TCC TCA GCC ACC CG-TAMRA | ||
| EcPV10-L1 | F-5′-GTG TCA CAG GTA ACC CCC TG-3′ | 174 | OP870083 |
| R-5′-AAG CGT GTC TTC CTC CAG TG-3′ | |||
| p-EcPV10-L1 | p-FAM-TGC TGG TGG GTT GCA AGC CC-TAMRA | ||
| BPV1-L1 | F-5′-CAG GAC TGT TCA CAA CCC AA-3′ | 96 | NC_001522.1 |
| R-5′-CCC AGT TAC AGT ACC TCC AA-3′ | |||
| p-BPV1-L1 | p-FAM-TGC AGG TGT CCA GAG GGC AG-TAMRA | ||
| BPV2-L1 | F-5′-ACA GCC CGT CCA TGT GTT A-3′ | 115 | MF045490 |
| R-5′-TCA GCA GCA CCA AAC CCT AT-3′ | |||
| p-BPV2-L1 | p-FAM-AGA AAA TGG TGC GTG TCC TCC T-TAMRA | ||
| BPV1-E5 | F-5′-TGCTTCAATGCAACTGCTGCT-3′ | 77 | MZ310894 |
| R-5′-AGGAGCACTCAAAATGATCCCAG-3′ | |||
| p-BPV1-E5 | p-FAM-ACTCTTGTTTTTTCTTGTA-TAMRA | ||
| BPV1-E6 | F-5′-TGCTACTGTGGGGGCAAACT-3′ | 110 | MZ310894 |
| R-5′-CAGTCGTAGCAGCGTCCTCT-3′ | |||
| p-BPV1-E6 | p-FAM-AGCCTTTCTGCAAAACCAGAGCT-TAMRA | ||
| BPV1-E7 | F-5′-GCTGTGGAAACTGCGGAAAA-3′ | 122 | MZ310894 |
| R-5′-GCGAGATTCACAACGTGGAC-3′ | |||
| p-BPV1-E7 | p-FAM-GCTGACTTTTGCAGTGAAGACCAGC-TAMRA | ||
| BPV13-L1 | F-5′-GCA CCC CAC TTT TAA TGC CT-3′ | 87 | NC_030795 |
| R-5′-TCC TGT TTG CTT CCT GTC ATC-3′ | |||
| p-BPV13-L1 | p-FAM-AGG AAA GTG ACC AGC CAA ACA ACA-TAMRA | ||
| B2M (Beta2-microglobulin) | F-5′-GGCTACTCTCCCTGACTGG-3′ | 135 | NM_001082502.3 |
| R-5′-TCAATCTCAGGCGGATGGAA-3′ | |||
| p-B2M | p-FAM-ACTCACGTCACCCAGCAGAGA-TAMRA |
| Sample | Real-Time PCR | |||||
|---|---|---|---|---|---|---|
| B2M | EcPV2-L1 | EcPV9-L1 | EcPV10-L1 | BPV2-L1 | BPV13-L1 | |
| 29.4 ± 0.1 | >38 | >38 | >38 | >38 | >38 | |
| B2M | BPV1-E5 | BPV1-E6 | BPV1-E7 | BPV1-L1 | ||
| 29.1 ± 0.1 | 21.5 ± 0.2 | 21.7 ± 0.4 | 21.5 ± 0.2 | 22.5 ± 0.5 | ||
| Sample | Real-Time PCR | ||||
|---|---|---|---|---|---|
| B2M | BPV1-E5 | BPV1-E6 | BPV1-E7 | BPV1-L1 | |
| 31.4 ± 0.2 | 30.1 ± 0.2 | 34.2 ± 0.2 | 21.5 ± 0.2 | 37.1 ± 0.3 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maggi, R.; De Paolis, L.; De Santis, D.; Vellone, V.G.; De Ciucis, C.G.; Fruscione, F.; Mazzocco, K.; Ghelardi, A.; Marruchella, G.; Razzuoli, E. Bovine Papillomavirus Type 1 Infection in an Equine Congenital Papilloma. Pathogens 2023, 12, 1059. https://doi.org/10.3390/pathogens12081059
Maggi R, De Paolis L, De Santis D, Vellone VG, De Ciucis CG, Fruscione F, Mazzocco K, Ghelardi A, Marruchella G, Razzuoli E. Bovine Papillomavirus Type 1 Infection in an Equine Congenital Papilloma. Pathogens. 2023; 12(8):1059. https://doi.org/10.3390/pathogens12081059
Chicago/Turabian StyleMaggi, Raffaella, Livia De Paolis, Daria De Santis, Valerio Gaetano Vellone, Chiara Grazia De Ciucis, Floriana Fruscione, Katia Mazzocco, Alessandro Ghelardi, Giuseppe Marruchella, and Elisabetta Razzuoli. 2023. "Bovine Papillomavirus Type 1 Infection in an Equine Congenital Papilloma" Pathogens 12, no. 8: 1059. https://doi.org/10.3390/pathogens12081059
APA StyleMaggi, R., De Paolis, L., De Santis, D., Vellone, V. G., De Ciucis, C. G., Fruscione, F., Mazzocco, K., Ghelardi, A., Marruchella, G., & Razzuoli, E. (2023). Bovine Papillomavirus Type 1 Infection in an Equine Congenital Papilloma. Pathogens, 12(8), 1059. https://doi.org/10.3390/pathogens12081059








