Clinically Isolated β-Lactam-Resistant Gram-Negative Bacilli in a Philippine Tertiary Care Hospital Harbor Multi-Class β-Lactamase Genes
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection and Characterization of Bacterial Isolates
2.2. Detection of β-Lactamase Production
2.2.1. Disk-Diffusion-Based ESBL Test
2.2.2. Modified Carbapenem Inactivation (mCIM), and Ethylenediamine Tetraacetic Acid (EDTA)-Modified Carbapenem Inactivation Methods (eCIM)
2.3. Molecular Characterization of Study Isolates
2.4. Sequence Analysis of the 16S rRNA and β-Lactamase Genes
3. Results
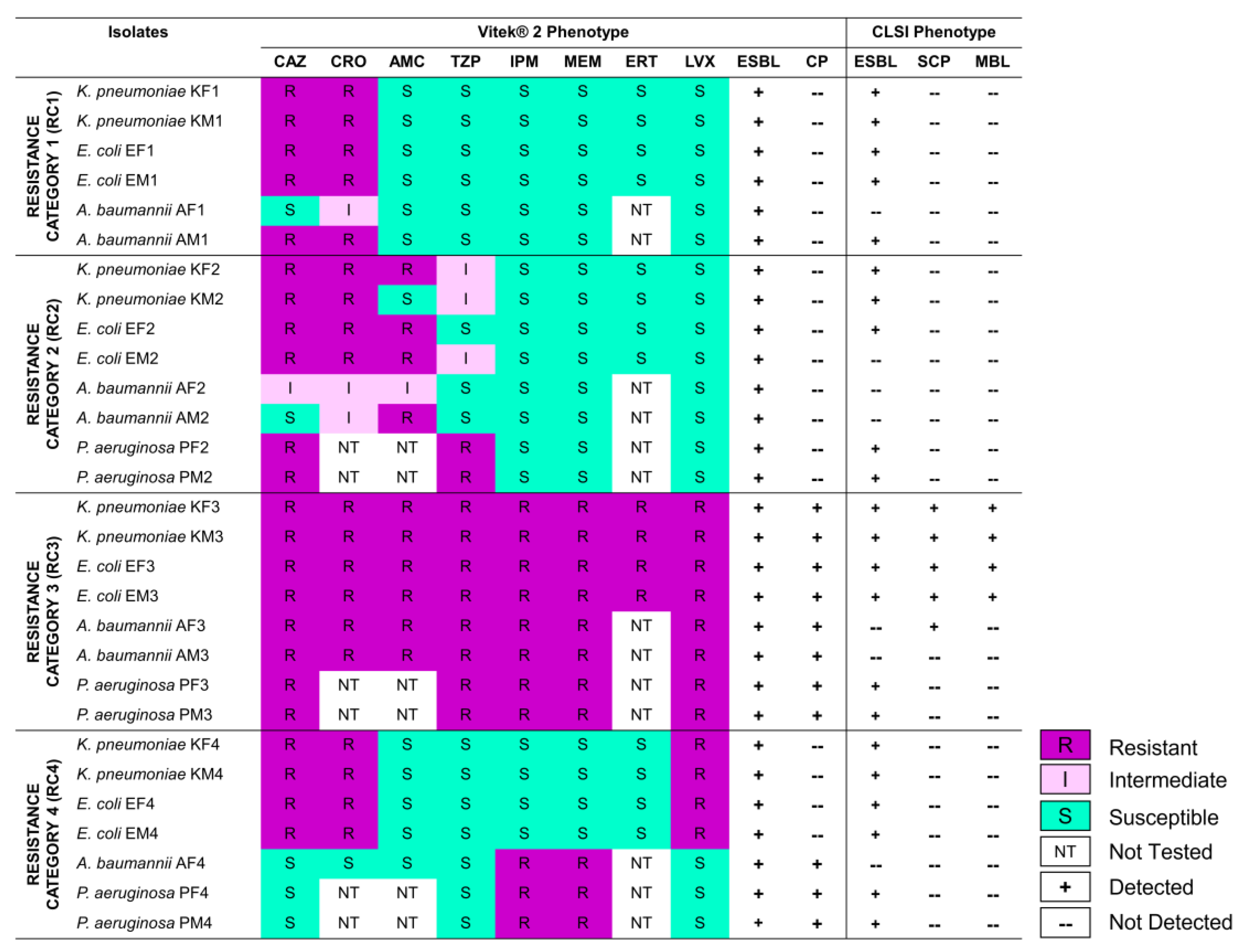
3.1. Selected Gram-Negative Bacilli Exhibit Varying Resistance to β-Lactams, and Produce ESBLs and CPs
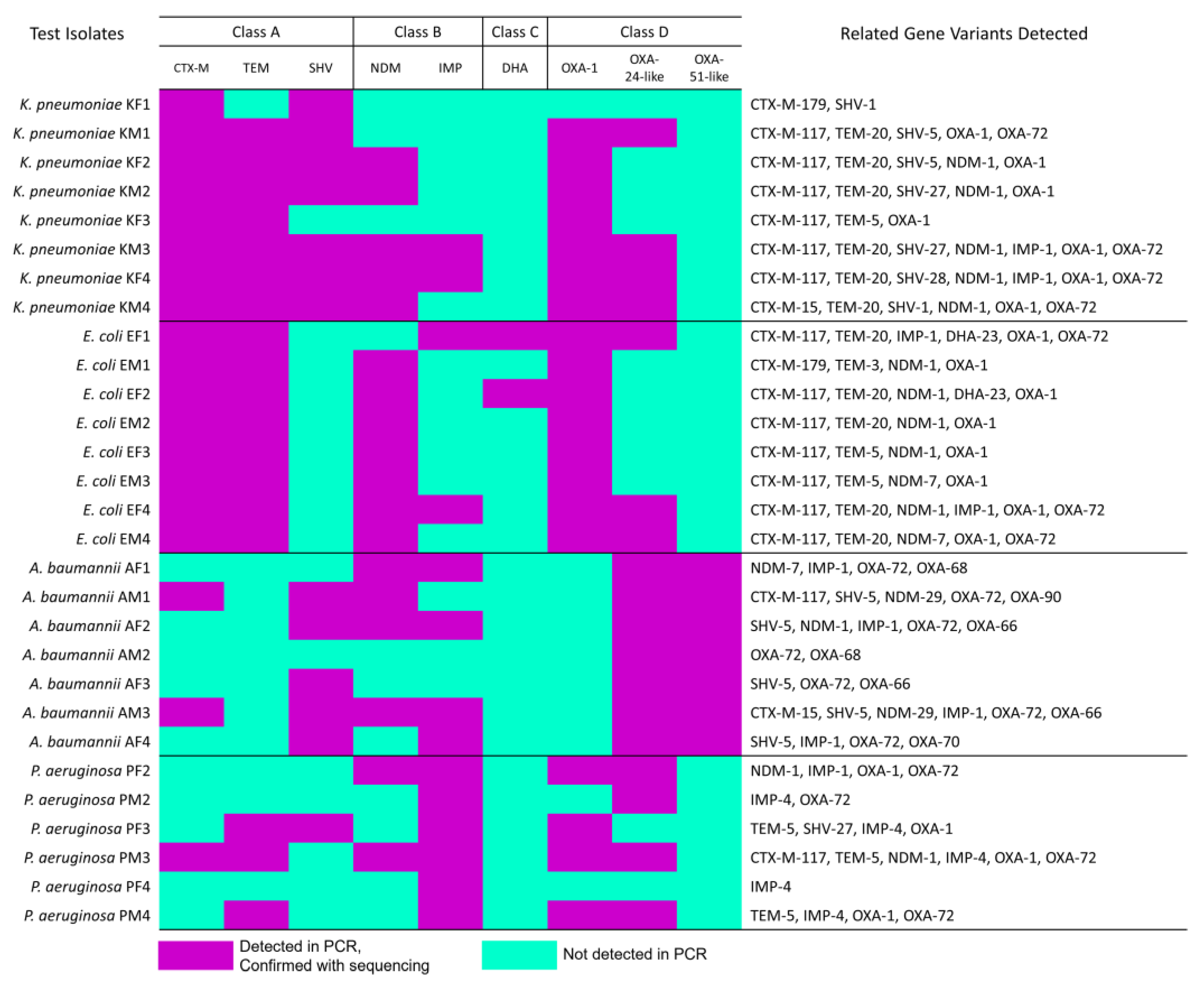
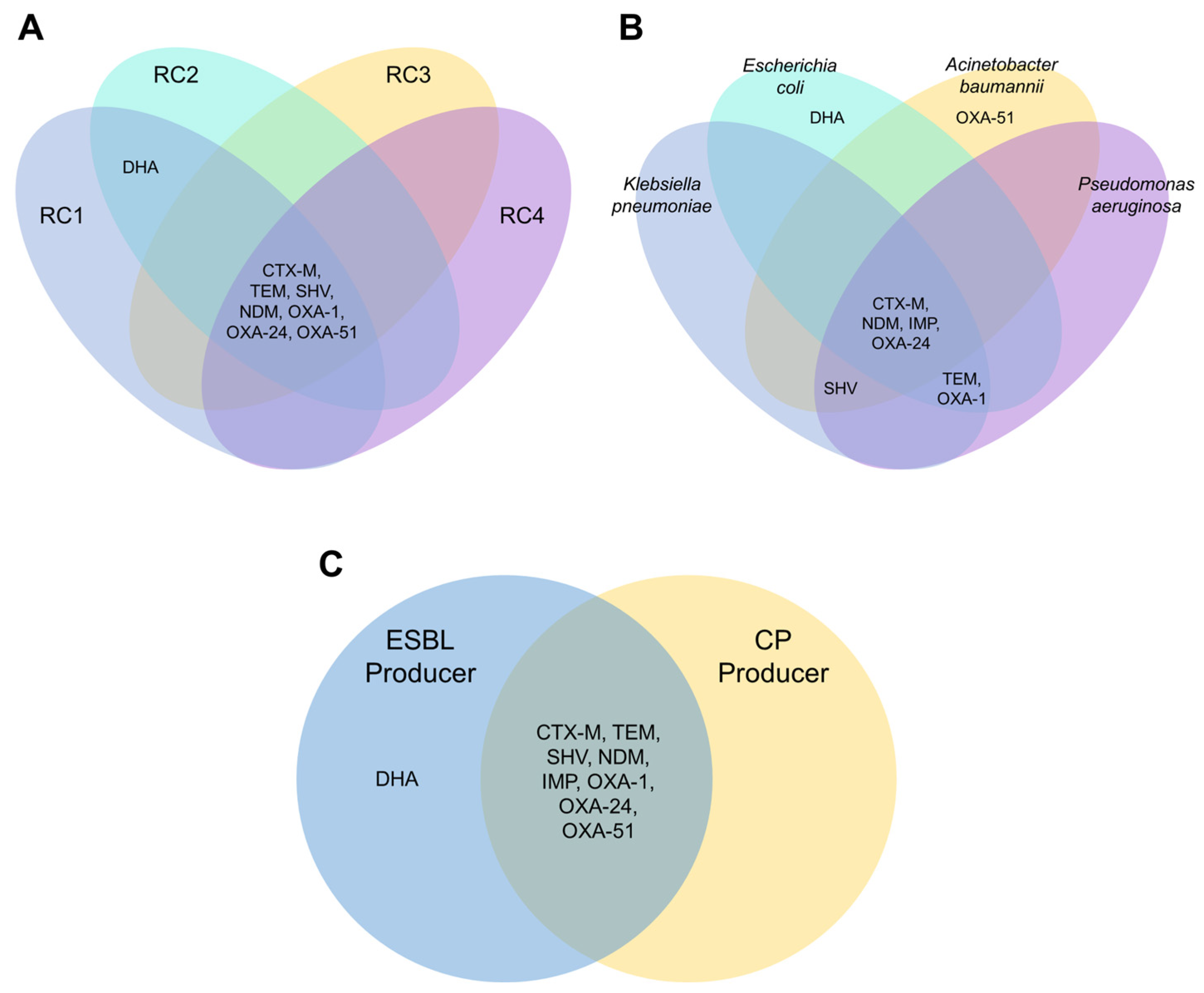
3.2. β-Lactam-Resistant Gram-Negative Bacilli Harbor Multi-Class BL Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 16 August 2022).
- Carascal, M.B.; Dela Cruz-Papa, D.M.; Remenyi, R.; Cruz, M.C.B.; Destura, R.V. Phage Revolution against Multidrug-Resistant Clinical Pathogens in Southeast Asia. Front. Microbiol. 2022, 13, 820572. [Google Scholar] [CrossRef] [PubMed]
- Antimicrobial Resistance Surveillance Program Annual Report. 2022. Available online: https://arsp.com.ph/download/1864/?tmstv=1690439576 (accessed on 26 July 2023).
- World Health Organization. New Report Calls for Urgent Action to Avert Antimicrobial Resistance Crisis. Available online: https://www.who.int/news/item/29-04-2019-new-report-calls-for-urgent-action-to-avert-antimicrobial-resistance-crisis (accessed on 1 September 2022).
- World Health Organization. Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report: 2021. Available online: https://www.who.int/publications/i/item/9789240027336 (accessed on 29 August 2022).
- Jean, S.S.; Coombs, G.; Ling, T.; Balaji, V.; Rodrigues, C.; Mikamo, H.; Kim, M.J.; Rajasekaram, D.G.; Mendoza, M.; Tan, T.Y.; et al. Epidemiology and antimicrobial susceptibility profiles of pathogens causing urinary tract infections in the Asia-Pacific region: Results from the Study for Monitoring Antimicrobial Resistance Trends (SMART), 2010–2013. Int. J. Antimicrob. Agents 2016, 47, 328–334. [Google Scholar] [CrossRef]
- Cruz, M.; Hedreyda, C.T. Detection of plasmid-borne b-lactamase genes in extended-spectrum b-lactamase (ESBL) and non-ESBL-producing Escherichia coli clinical isolates. Phil. J. Sci. 2017, 146, 167–175. [Google Scholar]
- De Angelis, G.; Del Giacomo, P.; Posteraro, B.; Sanguinetti, M.; Tumbarello, M. Molecular Mechanisms, Epidemiology, and Clinical Importance of β-Lactam Resistance in Enterobacteriaceae. Int. J. Mol. Sci. 2020, 21, 5090. [Google Scholar] [CrossRef] [PubMed]
- Naas, T.; Oueslati, S.; Bonnin, R.A.; Dabos, M.L.; Zavala, A.; Dortet, L.; Retailleau, P.; Iorga, B.I. Beta-lactamase database (BLDB)—Structure and function. J. Enzyme Inhib. Med. Chem. 2017, 32, 917–919. [Google Scholar] [CrossRef] [PubMed]
- Ambler, R.P. The structure of beta-lactamases. Philos. Trans. R Soc. Lond. B Biol. Sci. 1980, 289, 321–331. [Google Scholar] [PubMed]
- Tamma, P.D.; Aitken, S.L.; Bonomo, R.A.; Mathers, A.J.; van Duin, D.; Clancy, C.J. Infectious Diseases Society of America Guidance on the Treatment of AmpC β-Lactamase-Producing Enterobacterales, Carbapenem-Resistant Acinetobacter baumannii, and Stenotrophomonas maltophilia Infections. Clin. Infect. Dis. 2022, 74, 2089–2114. [Google Scholar] [CrossRef]
- Szczypta, A.; Talaga-Ćwiertnia, K.; Kielar, M.; Krzyściak, P.; Gajewska, A.; Szura, M.; Bulanda, M.; Chmielarczyk, A. Investigation of Acinetobacter baumannii Activity in Vascular Surgery Units through Epidemiological Management Based on the Analysis of Antimicrobial Resistance, Biofilm Formation and Genotyping. Int. J. Environ. Res. Public Health 2021, 18, 1563. [Google Scholar] [CrossRef]
- Castanheira, M.; Farrell, S.E.; Krause, K.M.; Jones, R.N.; Sader, H.S. Contemporary diversity of β-lactamases among Enterobacteriaceae in the nine U.S. census regions and ceftazidime-avibactam activity tested against isolates producing the most prevalent β-lactamase groups. Antimicrob. Agents Chemother. 2014, 58, 833–838. [Google Scholar] [CrossRef]
- Coşkun, S.; Altanlar, N. Detection of plasmid-mediated AmpC beta-lactamase in clinical isolates of cefoxitin-resistant Escherichia coli and Klebsiella pneumoniae. Mikrobiyol. Bul. 2012, 46, 375–385. [Google Scholar]
- Ruh, E.; Zakka, J.; Hoti, K.; Fekrat, A.; Guler, E.; Gazi, U.; Erdogmus, Z.; Suer, K. Extended-spectrum β-lactamase, plasmid-mediated AmpC β-lactamase, fluoroquinolone resistance, and decreased susceptibility to carbapenems in Enterobacteriaceae: Fecal carriage rates and associated risk factors in the community of Northern Cyprus. Antimicrob. Resist. Infect. Control. 2019, 8, 98. [Google Scholar] [CrossRef]
- Poulou, A.; Grivakou, E.; Vrioni, G.; Koumaki, V.; Pittaras, T.; Pournaras, S.; Tsakris, A. Modified CLSI extended-spectrum β-lactamase (ESBL) confirmatory test for phenotypic detection of ESBLs among Enterobacteriaceae producing various β-lactamases. J. Clin. Microbiol. 2014, 52, 1483–1489. [Google Scholar] [CrossRef] [PubMed]
- Tenover, F.C.; Dela Cruz, C.M.; Dewell, S.; Le, V.M.; Tickler, I.A. Does the presence of multiple β-lactamases in Gram-negative bacilli impact the results of antimicrobial susceptibility tests and extended-spectrum β-lactamase and carbapenemase confirmation methods? J. Glob. Antimicrob. Resist. 2020, 23, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, E.C.; Rodriguez, R.D. First report on the occurrence of SHV-12 extended-spectrum beta-lactamase-producing Enterobacteriaceae in the Philippines. J. Microbiol. Immunol. Infect. 2009, 42, 74–85. [Google Scholar]
- Velasco, J.M.; Valderama, M.T.; Peacock, T.; Warawadee, N.; Nogrado, K.; Navarro, F.C.; Chua, D., Jr.; Apichai, S.; Sirigade, R.; Macareo, L.R.; et al. Carbapenemase-Producing Enterobacteriaceae and Nonfermentative Bacteria, the Philippines, 2013–2016. Emerg. Infect. Dis. 2017, 23, 1597–1598. [Google Scholar] [CrossRef] [PubMed]
- Cornista, J.C.; Cuna, A.M.D.; Sanchez, H.J.A.; Balolong, M.P. Detection of Extended-spectrum β-Lactamase-producing Klebsiella pneumoniae Isolated from Four Provincial Hospitals in Luzon and Genotyping of β-Lactamase (blaCTX-M, blaTEM, blaSHV, and blaOXA-1) Genes. Phil. J. Sci. 2019, 148, 277–287. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 32nd ed.; CLSI supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2022. [Google Scholar]
- Park, S.H. Third-generation cephalosporin resistance in gram-negative bacteria in the community: A growing public health concern. Korean J. Intern. Med. 2014, 29, 27–30. [Google Scholar] [CrossRef]
- Jones, B.M.; Wagner, J.L.; Chastain, D.B.; Bookstaver, P.B.; Stover, K.; Lin, J.; Matson, H.; White, N.; Motesh, M.; Bland, C.M. Real-world, multicentre evaluation of the incidence and risk factors for non-susceptible Stenotrophomonas maltophilia isolates. J. Glob. Antimicrob. Resist. 2022, 28, 282–287. [Google Scholar] [CrossRef]
- Bassetti, M.; Giacobbe, D.R.; Castaldo, N.; Russo, A.; Vena, A. Role of new antibiotics in extended-spectrum β-lactamase-, AmpC-infections. Curr. Opin. Infect. Dis. 2021, 34, 748–755. [Google Scholar] [CrossRef]
- Li, S.; Jia, X.; Li, C.; Zou, H.; Liu, H.; Guo, Y.; Zhang, L. Carbapenem-resistant and cephalosporin-susceptible Pseudomonas aeruginosa: A notable phenotype in patients with bacteremia. Infect. Drug. Resist. 2018, 11, 1225–1235. [Google Scholar] [CrossRef]
- Dallenne, C.; Da Costa, A.; Decré, D.; Favier, C.; Arlet, G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Walsh, T.R.; Cuvillier, V.; Nordmann, P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 2011, 70, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Woodford, N.; Ellington, M.J.; Coelho, J.M.; Turton, J.F.; Ward, M.E.; Brown, S.; Amyes, S.G.; Livermore, D.M. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents. 2006, 27, 351–353. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, R.; Karaoz, U.; Volegova, M.; MacKichan, J.; Kato-Maeda, M.; Miller, S.; Nadarajan, R.; Brodie, E.L.; Lynch, S.V. Use of 16S rRNA Gene for Identification of a Broad Range of Clinically Relevant Bacterial Pathogens. PLoS ONE 2015, 10, e0117617. [Google Scholar] [CrossRef]
- Weill, F.X.; Demartin, M.; Tandé, D.; Espié, E.; Rakotoarivony, I.; Grimont, P.A. SHV-12-like extend-ed-spectrum-beta-lactamase-producing strains of Salmonella enterica serotypes Babelsberg and Enteritidis isolated in France among infants adopted from Mali. J. Clin. Microbiol. 2004, 42, 2432–2437. [Google Scholar] [CrossRef]
- Pérez-Pérez, F.J.; Hanson, N.D. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 2002, 40, 2153–2162. [Google Scholar] [CrossRef]
- Juan, C.; Beceiro, A.; Gutiérrez, O.; Albertí, S.; Garau, M.; Pérez, J.L.; Bou, G.; Oliver, A. Characterization of the new metallo-beta-lactamase VIM-13 and its integron-borne gene from a Pseudomonas aeruginosa clinical isolate in Spain. J. Antimicrob. Chemother. 2008, 52, 3589–3596. [Google Scholar] [CrossRef]
- Mohammed, Y.; Zailani, S.B.; Onipede, A.O. Characterization of KPC, NDM and VIM type carbapenem resistance Enterobacteriaceae from north eastern Nigeria. J. Biosci. Med. 2015, 3, 100–107. [Google Scholar] [CrossRef]
- Mlynarcik, P.; Chalachanova, A.; Vagnerovă, I.; Holy, O.; Zatloukalova, S.; Kolar, M. PCR Detection of Oxacillinases in Bacteria. Microb. Drug Resist. 2020, 26, 1023–1037. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, P.; Song, P.; Li, X. Carbapenemase OXA-423: A Novel OXA-23 Variant in Acinetobacter baumannii. Infect. Drug. Resist. 2020, 13, 4069–4075. [Google Scholar] [CrossRef]
- Salloum, N.A.; Kissoyan, K.A.B.; Fadlallah, S.; Cheaito, K.; Araj, G.F.; Wakim, R.; Kanj, S.; Kanafani, Z.; Dbaibo, G.; Matar, G.M. Assessment of combination therapy in BALB/c mice injected with carbapenem-resistant Enterobacteriaceae strains. Front. Microbiol. 2015, 6, 999. [Google Scholar] [CrossRef]
- Hu, W.S.; Yao, S.M.; Fung, C.P.; Hsieh, Y.P.; Liu, C.P.; Lin, J.F. An OXA-66/OXA-51-like carbapenemase and possibly an efflux pump are associated with resistance to imipenem in Acinetobacter baumannii. J. Antimicrob. Chemother. 2007, 51, 3844–3852. [Google Scholar] [CrossRef]
- Sohrabi, N.; Farajnia, S.; Akhi, M.T.; Nahaei, M.R.; Naghili, B.; Peymani, A.; Amiri, Z.; Rezaee, M.A.; Saeedi, N.; Nikoo, H.R.; et al. Prevalence of OXA-Type β-Lactamases among Acinetobacter baumannii Isolates from Northwest of Iran. Microb. Drug Resist. 2012, 18, 385–389. [Google Scholar] [CrossRef]
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Bush, K.; Bradford, P.A. Epidemiology of β-Lactamase-Producing Pathogens. Clin. Microbiol. Rev. 2020, 33, e00047-19. [Google Scholar] [CrossRef] [PubMed]
- Schwaber, M.J.; Navon-Venezia, S.; Chmelnitsky, I.; Leavitt, A.; Schwartz, D.; Carmeli, Y. Utility of the VITEK 2 Advanced Expert System for identification of extended-spectrum beta-lactamase production in Enterobacter spp. J. Clin. Microbiol. 2006, 44, 241–243. [Google Scholar] [CrossRef] [PubMed]
- Rice, K.S.; Luethy, P.M. Overprediction of Carbapenemase-Containing Isolates by an Automated AST Instrument’s Computer Algorithm. J. Appl. Lab. Med. 2022, 7, 1283–1289. [Google Scholar] [CrossRef]
- Lee, C.R.; Lee, J.H.; Park, M.; Park, K.S.; Bae, I.K.; Kim, Y.B.; Cha, C.J.; Jeong, B.C.; Lee, S.H. Biology of Acinetobacter baumannii: Pathogenesis, Antibiotic Resistance Mechanisms, and Prospective Treatment Options. Front. Cell. Infect. Microbiol. 2017, 7, 55. [Google Scholar] [CrossRef] [PubMed]
- Stasiak, M.; Maćkiw, E.; Kowalska, J.; Kucharek, K.; Postupolski, J. Silent Genes: Antimicrobial Resistance and Antibiotic Production. Pol. J. Microbiol. 2021, 70, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Picão, R.C.; Carrara-Marroni, F.E.; Gales, A.C.; Venâncio, E.J.; Xavier, D.E.; Tognim, M.C.; Pelayo, J.S. Metallo-β-lactamase-production in meropenem-susceptible Pseudomonas aeruginosa isolates: Risk for silent spread. Mem. Inst. Oswaldo Cruz. 2012, 107, 747–751. [Google Scholar] [CrossRef]
- Kaye, K.S.; Gold, H.S.; Schwaber, M.J.; Venkataraman, L.; Qi, Y.; De Girolami, P.C.; Samore, M.H.; Anderson, G.; Rasheed, J.K.; Tenover, F.C. Variety of beta-lactamases produced by amoxicillin-clavulanate-resistant Escherichia coli isolated in the northeastern United States. Antimicrob. Agents Chemother. 2004, 48, 1520–1525. [Google Scholar] [CrossRef]
- Birgy, A.; Madhi, F.; Jung, C.; Levy, C.; Cointe, A.; Bidet, P.; Hobson, C.A.; Bechet, S.; Sobral, E.; Vuthien, H.; et al. Clavulanate combinations with mecillinam, cefixime or cefpodoxime against ESBL-producing Enterobacterales frequently associated with blaOXA-1 in a paediatric population with febrile urinary tract infections. J. Antimicrob. Chemother. 2021, 76, 2839–2846. [Google Scholar] [CrossRef] [PubMed]
- Argimón, S.; Masim, M.A.L.; Gayeta, J.M.; Lagrada, M.L.; Macaranas, P.K.V.; Cohen, V.; Limas, M.T.; Espiritu, H.O.; Palarca, J.C.; Chilam, J.; et al. Integrating whole-genome sequencing within the National Antimicrobial Resistance Surveillance Program in the Philippines. Nat. Commun. 2020, 11, 2719. [Google Scholar] [CrossRef]
- Carlos, C.C.; Masim, M.A.L.; Lagrada, M.L.; Gayeta, J.M.; Macaranas, P.K.V.; Sia, S.B.; Facun, M.A.M.; Palarca, J.F.C.; Olorosa, A.M.; Cueno, G.A.C.; et al. Genome Sequencing Identifies Previously Unrecognized Klebsiella pneumoniae Outbreaks in Neonatal Intensive Care Units in the Philippines. Clin. Infect. Dis. 2021, 73 (Suppl. S4), S316–S324. [Google Scholar] [CrossRef] [PubMed]
- Golden, A.R.; Baxter, M.R.; Karlowsky, J.A.; Mataseje, L.; Mulvey, M.R.; Walkty, A.; Bay, D.; Schweizer, F.; Lagace-Wiens, P.R.S.; Adam, H.J.; et al. Activity of cefepime/taniborbactam and comparators against whole genome sequenced ertapenem-non-susceptible Enterobacterales clinical isolates: CANWARD 2007–19. JAC Antimicrob. Resist. 2022, 4, dlab197. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.R.; Wang, W.P.; Huang, M.; Shi, L.N.; Wang, Y.; Shao, H.F. Mechanisms of carbapenem resistance in cephalosporin-susceptible Pseudomonas aeruginosa in China. Diagn. Microbiol. Infect. Dis. 2014, 78, 268–270. [Google Scholar] [CrossRef]
- Campana, E.H.; Xavier, D.E.; Petrolini, F.V.; Cordeiro-Moura, J.R.; Araujo, M.R.; Gales, A.C. Carbapenem-resistant and cephalosporin-susceptible: A worrisome phenotype among Pseudomonas aeruginosa clinical isolates in Brazil. Braz. J. Infect. Dis. 2017, 21, 57–62. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abordo, A.M.S.; Carascal, M.B.; Remenyi, R.; Dalisay, D.S.; Saludes, J.P. Clinically Isolated β-Lactam-Resistant Gram-Negative Bacilli in a Philippine Tertiary Care Hospital Harbor Multi-Class β-Lactamase Genes. Pathogens 2023, 12, 1019. https://doi.org/10.3390/pathogens12081019
Abordo AMS, Carascal MB, Remenyi R, Dalisay DS, Saludes JP. Clinically Isolated β-Lactam-Resistant Gram-Negative Bacilli in a Philippine Tertiary Care Hospital Harbor Multi-Class β-Lactamase Genes. Pathogens. 2023; 12(8):1019. https://doi.org/10.3390/pathogens12081019
Chicago/Turabian StyleAbordo, Alecks Megxel S., Mark B. Carascal, Roland Remenyi, Doralyn S. Dalisay, and Jonel P. Saludes. 2023. "Clinically Isolated β-Lactam-Resistant Gram-Negative Bacilli in a Philippine Tertiary Care Hospital Harbor Multi-Class β-Lactamase Genes" Pathogens 12, no. 8: 1019. https://doi.org/10.3390/pathogens12081019
APA StyleAbordo, A. M. S., Carascal, M. B., Remenyi, R., Dalisay, D. S., & Saludes, J. P. (2023). Clinically Isolated β-Lactam-Resistant Gram-Negative Bacilli in a Philippine Tertiary Care Hospital Harbor Multi-Class β-Lactamase Genes. Pathogens, 12(8), 1019. https://doi.org/10.3390/pathogens12081019










