Analyses of Mosquito Species Composition, Blood-Feeding Habits and Infection with Insect-Specific Flaviviruses in Two Arid, Pastoralist-Dominated Counties in Kenya
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Sampling Sites, Mosquito Surveys and Processing
2.3. Blood-meal Source Identification
2.4. Detection and Identification of Viruses
2.5. Virus Isolation and Screening of Cultures
2.6. Next-generation Sequencing (NGS)
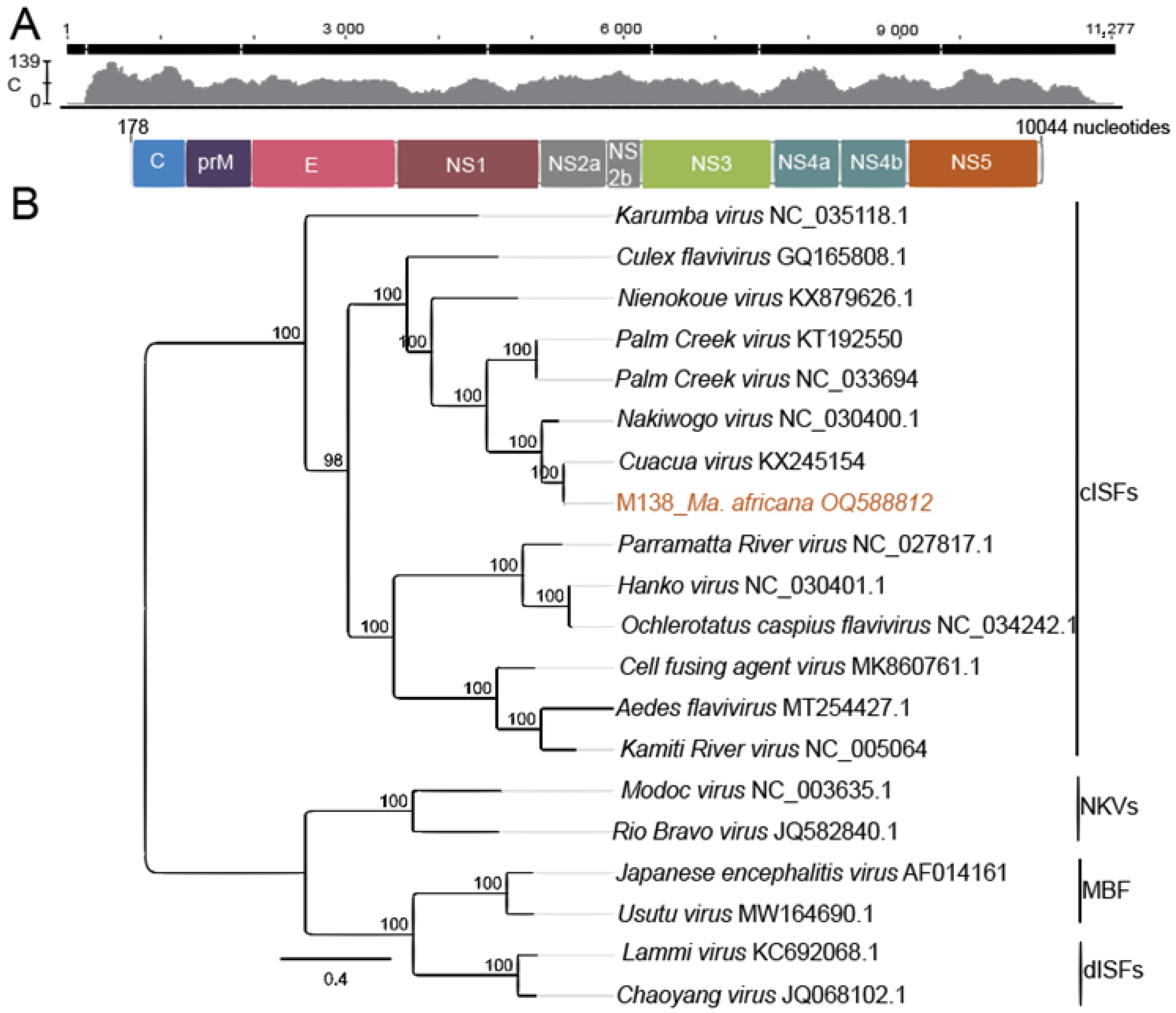
2.7. Phylogenetic Analysis and Genome Characterization
2.8. Statistical Analysis
2.9. Sequence Accession Numbers
3. Results
3.1. Mosquito Composition
3.2. Mosquito Blood-Meal Host Sources
3.3. Flavivirus Sequence Detections in Mosquitoes
3.4. Virus Isolation and Genome Organization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Simmonds, P.; Becher, P.; Bukh, J.; Gould, E.A.; Meyers, G.; Monath, T.; Muerhoff, S.; Pletnev, A.; Rico-Hesse, R.; Smith, D.B.; et al. ICTV Virus Taxonomy Profile: Flaviviridae. J. Gen. Virol. 2017, 98, 2–3. [Google Scholar] [CrossRef]
- Reiter, P.; Gitau, L.G.; Cordellier, R.; Gubler, D.J.; Cropp, C.B.; Agata, N.N.; Ouma, J.O.; Tukei, P.M.; Sanders, E.J.; Savage, H.M.; et al. First recorded outbreak of yellow fever in Kenya, 1992-1993. II. Entomologic investigations. Am. J. Trop. Med. Hyg. 1998, 59, 650–656. [Google Scholar] [CrossRef]
- Lutomiah, J.; Barrera, R.; Makio, A.; Mutisya, J.; Koka, H.; Owaka, S.; Koskei, E.; Nyunja, A.; Eyase, F.; Coldren, R.; et al. Dengue Outbreak in Mombasa City, Kenya, 2013–2014: Entomologic Investigations. PLoS Neglected Trop. Dis. 2016, 10, e0004981. [Google Scholar] [CrossRef] [PubMed]
- Yellow Fever—Kenya. Available online: https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON361 (accessed on 25 May 2022).
- World Health Organization (WHO). Dengue and Severe Dengue. Key Facts. 2019. Available online: https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue (accessed on 25 May 2022).
- Hill, S.C.; Vasconcelos, J.; Neto, Z.; Jandondo, D.; Zé-Zé, L.; Aguiar, R.S.; Xavier, J.; Thézé, J.; Mirandela, M.; Cândido, A.L.M.; et al. Emergence of the Asian lineage of Zika virus in Angola: An outbreak investigation. Lancet Infect. Dis. 2019, 19, 1138–1147. [Google Scholar] [CrossRef]
- Faye, O.; Monteiro, M.D.L.; Vrancken, B.; Prot, M.; Lequime, S.; Diarra, M.; Ndiaye, O.; Valdez, T.; Tavarez, S.; Ramos, J.; et al. Genomic Epidemiology of 2015–2016 Zika Virus Outbreak in Cape Verde. Emerg. Infect. Dis. 2020, 26, 1084. [Google Scholar] [CrossRef] [PubMed]
- Ushijima, Y.; Abe, H.; Ondo, G.N.; Bikangui, R.; Loembé, M.M.; Zadeh, V.R.; Essimengane, J.G.E.; Mbouna, A.V.N.; Bache, E.B.; Agnandji, S.T.; et al. Surveillance of the major pathogenic arboviruses of public health concern in Gabon, Central Africa: Increased risk of West Nile virus and dengue virus infections. BMC Infect. Dis. 2021, 21, 265. [Google Scholar] [CrossRef] [PubMed]
- Blitvich, B.J.; Firth, A.E. Insect-Specific Flaviviruses: A Systematic Review of Their Discovery, Host Range, Mode of Transmission, Superinfection Exclusion Potential and Genomic Organization. Viruses 2015, 7, 1927–1959. [Google Scholar] [CrossRef] [PubMed]
- Blitvich, B.J.; Firth, A.E. A Review of Flaviviruses that Have No Known Arthropod Vector. Viruses 2017, 9, 154. [Google Scholar] [CrossRef]
- Sang, R.C.; Gichogo, A.; Gachoya, J.; Dunster, M.D.; Ofula, V.; Hunt, A.R.; Crabtree, M.B.; Miller, B.R.; Dunster, L.M. Isolation of a new flavivirus related to Cell fusing agent virus (CFAV) from field-collected flood-water Aedes mosquitoes sampled from a dambo in central Kenya. Arch. Virol. 2003, 148, 1085–1093. [Google Scholar] [CrossRef]
- Cholleti, H.; Hayer, J.; Abílio, A.P.; Mulandane, F.; Verner-Carlsson, J.; Falk, K.I.; Fafetine, J.M.; Berg, M.; Blomström, A.-L. Discovery of Novel Viruses in Mosquitoes from the Zambezi Valley of Mozambique. PLoS ONE 2016, 11, e0162751. [Google Scholar] [CrossRef]
- Junglen, S.; Korries, M.; Grasse, W.; Wieseler, J.; Kopp, A.; Hermanns, K.; León-Juárez, M.; Drosten, C.; Kümmerer, B.M. Host Range Restriction of Insect-Specific Flaviviruses Occurs at Several Levels of the Viral Life Cycle. mSphere 2017, 2, e00375-16. [Google Scholar] [CrossRef] [PubMed]
- Junglen, S.; Kopp, A.; Kurth, A.; Pauli, G.; Ellerbrok, H.; Leendertz, F.H. A New Flavivirus and a New Vector: Characterization of a Novel Flavivirus Isolated from Uranotaenia Mosquitoes from a Tropical Rain Forest. J. Virol. 2009, 83, 4462–4468. [Google Scholar] [CrossRef] [PubMed]
- Romo, H.; Kenney, J.L.; Blitvich, B.J.; Brault, A.C. Restriction of Zika virus infection and transmission in Aedes aegypti mediated by an insect-specific Flavivirus. Emerg. Microbes Infect. 2018, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hall-Mendelin, S.; McLean, B.J.; Bielefeldt-Ohmann, H.; Hobson-Peters, J.; Hall, R.A.; Hurk, A.F.V.D. The insect-specific Palm Creek virus modulates West Nile virus infection in and transmission by Australian mosquitoes. Parasites Vectors 2016, 9, 414. [Google Scholar] [CrossRef]
- Lutomiah, J.J.L.; Mwandawiro, C.; Magambo, J.; Sang, R.C. Infection and Vertical Transmission of Kamiti River Virus in Laboratory Bred Aedes aegypti Mosquitoes. J. Insect Sci. 2007, 7, 55. [Google Scholar] [CrossRef]
- Ajamma, Y.U.; Onchuru, T.O.; Ouso, D.O.; Omondi, D.; Masiga, D.K.; Villinger, J. Vertical transmission of naturally occurring Bunyamwera and insect-specific flavivirus infections in mosquitoes from islands and mainland shores of Lakes Victoria and Baringo in Kenya. PLoS Neglected Trop. Dis. 2018, 12, e0006949. [Google Scholar] [CrossRef]
- Chiuya, T.; Masiga, D.K.; Falzon, L.C.; Bastos, A.D.S.; Fèvre, E.M.; Villinger, J. A survey of mosquito-borne and insect-specific viruses in hospitals and livestock markets in western Kenya. PLoS ONE 2021, 16, e0252369. [Google Scholar] [CrossRef]
- Kamau, W.W.; Sang, R.; Ogola, E.O.; Rotich, G.; Getugi, C.; Agha, S.B.; Menza, N.; Torto, B.; Tchouassi, D.P. Survival rate, blood feeding habits and sibling species composition of Aedes simpsoni complex: Implications for arbovirus transmission risk in East Africa. PLoS Neglected Trop. Dis. 2022, 16, e0010171. [Google Scholar] [CrossRef] [PubMed]
- Chepkorir, E.; Venter, M.; Lutomiah, J.; Mulwa, F.; Arum, S.; Tchouassi, D.; Sang, R. The occurrence, diversity and blood feeding patterns of potential vectors of dengue and yellow fever in Kacheliba, West Pokot County, Kenya. Acta Trop. 2018, 186, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Ogola, E.; Villinger, J.; Mabuka, D.; Omondi, D.; Orindi, B.; Mutunga, J.; Owino, V.; Masiga, D.K. Composition of Anopheles mosquitoes, their blood-meal hosts, and Plasmodium falciparum infection rates in three islands with disparate bed net coverage in Lake Victoria, Kenya. Malar. J. 2017, 16, 360. [Google Scholar] [CrossRef]
- Chaves, L.F.; Harrington, L.C.; Keogh, C.L.; Nguyen, A.M.; Kitron, U.D. Blood feeding patterns of mosquitoes: Random or structured? Front. Zool. 2010, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- McBride, C.S.; Baier, F.; Omondi, A.B.; Spitzer, S.A.; Lutomiah, J.; Sang, R.; Ignell, R.; Vosshall, L.B. Evolution of mosquito preference for humans linked to an odorant receptor. Nature 2014, 515, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Crawford, J.E.; Alves, J.M.; Palmer, W.J.; Day, J.P.; Sylla, M.; Ramasamy, R.; Surendran, S.N.; Black, W.C.; Pain, A.; Jiggins, F.M. Population genomics reveals that an anthropophilic population of Aedes aegypti mosquitoes in West Africa recently gave rise to American and Asian populations of this major disease vector. BMC Biol. 2017, 15, 16. [Google Scholar] [CrossRef] [PubMed]
- Omondi, D.; Masiga, D.K.; Ajamma, Y.U.; Fielding, B.C.; Njoroge, L.; Villinger, J. Unraveling Host-Vector-Arbovirus Interactions by Two-Gene High Resolution Melting Mosquito Bloodmeal Analysis in a Kenyan Wildlife-Livestock Interface. PLoS ONE 2015, 10, e0134375. [Google Scholar] [CrossRef] [PubMed]
- Musa, A.A.; Muturi, M.W.; Musyoki, A.M.; Ouso, D.O.; Oundo, J.W.; Makhulu, E.E.; Wambua, L.; Villinger, J.; Jeneby, M.M. Arboviruses and Blood Meal Sources in Zoophilic Mosquitoes at Human-Wildlife Interfaces in Kenya. Vector-Borne Zoonotic Dis. 2020, 20, 444–453. [Google Scholar] [CrossRef]
- Swei, A.; Kwan, J.Y. Tick microbiome and pathogen acquisition altered by host blood meal. ISME J. 2016, 11, 813–816. [Google Scholar] [CrossRef]
- Muturi, E.J.; Dunlap, C.; Ramirez, J.L.; Rooney, A.P.; Kim, C.-H. Host blood meal source has a strong impact on gut microbiota of Aedes aegypti. FEMS Microbiol. Ecol. 2018, 95, fiy213. [Google Scholar] [CrossRef]
- Agha, S.B.; Tchouassi, D.P.; Bastos, A.D.S.; Sang, R. Assessment of risk of dengue and yellow fever virus transmission in three major Kenyan cities based on Stegomyia indices. PLoS Neglected Trop. Dis. 2017, 11, e0005858. [Google Scholar] [CrossRef]
- Tchouassi, D.P.; Marklewitz, M.; Chepkorir, E.; Zirkel, F.; Agha, S.B.; Tigoi, C.C.; Koskei, E.; Drosten, C.; Borgemeister, C.; Torto, B.; et al. Sand Fly–Associated Phlebovirus with Evidence of Neutralizing Antibodies in Humans, Kenya. Emerg. Infect. Dis. 2019, 25, 681–690. [Google Scholar] [CrossRef]
- Marklewitz, M.; Tchouassi, D.P.; Hieke, C.; Heyde, V.; Torto, B.; Sang, R.; Junglen, S. Insights into the Evolutionary Origin of Mediterranean Sandfly Fever Viruses. mSphere 2020, 5, e00598-20. [Google Scholar] [CrossRef]
- Ogola, E.O.; Kopp, A.; Bastos, A.D.S.; Slothouwer, I.; Marklewitz, M.; Omoga, D.; Rotich, G.; Getugi, C.; Sang, R.; Torto, B.; et al. Jingmen Tick Virus in Ticks from Kenya. Viruses 2022, 14, 1041. [Google Scholar] [CrossRef] [PubMed]
- QGIS Development Team. Welcome to the QGIS Project! Available online: https://qgis.org/en/site/ (accessed on 11 June 2019).
- Guggemos, H.D.; Fendt, M.; Hieke, C.; Heyde, V.; Mfune, J.K.E.; Borgemeister, C.; Junglen, S. Simultaneous circulation of two West Nile virus lineage 2 clades and Bagaza virus in the Zambezi region, Namibia. PLoS Neglected Trop. Dis. 2021, 15, e0009311. [Google Scholar] [CrossRef] [PubMed]
- Gillies, M.T.; De Meillon, B. The Anophelinae of Africa South of the Sahara (Ethiopian Zoogeographical Region); South African Institute for Medical Research: Johannesburg, South Africa, 1968; Volume 54. [Google Scholar]
- Gillies, M.; Coetzee, M. A Supplement to the Anophelinae of Africa South of the Sahara; South African Institute for Medical Research: Johannesburg, South Africa, 1987; Volume 55. [Google Scholar]
- Reeves, L.E.; Gillett-Kaufman, J.L.; Kawahara, A.Y.; Kaufman, P.E. Barcoding blood meals: New vertebrate-specific primer sets for assigning taxonomic identities to host DNA from mosquito blood meals. PLoS Neglected Trop. Dis. 2018, 12, e0006767. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. BOLD: The Barcode of Life Data System (http://www.barcodinglife.org). Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 2014, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Crochu, S.; Cook, S.; Attoui, H.; Charrel, R.N.; De Chesse, R.; Belhouchet, M.; Lemasson, J.-J.; de Micco, P.; de Lamballerie, X. Sequences of flavivirus-related RNA viruses persist in DNA form integrated in the genome of Aedes spp. mosquitoes. J. Gen. Virol. 2004, 85, 1971–1980. [Google Scholar] [CrossRef]
- Moureau, G.; Temmam, S.; Gonzalez, J.; Charrel, R.N.; Grard, G.; de Lamballerie, X. A Real-Time RT-PCR Method for the Universal Detection and Identification of Flaviviruses. Vector-Borne Zoonotic Dis. 2007, 7, 467–478. [Google Scholar] [CrossRef]
- Armstrong, P.M.; Andreadis, T.G.; Finan, S.L.; Shepard, J.J.; Thomas, M.C. Detection of Infectious Virus from Field-collected Mosquitoes by Vero Cell Culture Assay. J. Vis. Exp. 2011, 52, e2889. [Google Scholar] [CrossRef]
- Marklewitz, M.; Dutari, L.C.; Paraskevopoulou, S.; Page, R.A.; Loaiza, J.; Junglen, S. Diverse novel phleboviruses in sandflies from the Panama Canal area, Central Panama. J. Gen. Virol. 2019, 100, 938–949. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Kazutaka, K.; Misakwa, K.; Kei-ichi, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Dunn, P.K.; Smyth, G.K. Chapter 9: Models for proportions: Binomial GLMs. In Generalized Linear Models with Examples in R; Springer Nature: Berlin/Heidelberg, Germany, 2018; pp. 333–369. [Google Scholar] [CrossRef]
- Ségard, A.; Gardès, L.; Jacquier, E.; Grillet, C.; Mathieu, B.; Rakotoarivony, I.; Setier-Rio, M.-L.; Chavernac, D.; Cêtre-Sossah, C.; Balenghien, T.; et al. Schmallenberg virus in Culicoides latreille (Diptera: Ceratopogonidae) populations in France during 2011–2012 outbreak. Transbound. Emerg. Dis. 2017, 65, e94–e103. [Google Scholar] [CrossRef] [PubMed]
- Dormann, C.F.; Gruber, B.; Fründ, J. Introducing the bipartite package: Analysing ecological networks. Interaction 2008, 8, 8–11. [Google Scholar]
- Ebhodaghe, F.I.; Okal, M.N.; Kalayou, S.; Bastos, A.D.S.; Masiga, D.K. Tsetse Bloodmeal Analyses Incriminate the Common Warthog Phacochoerus africanus as an Important Cryptic Host of Animal Trypanosomes in Smallholder Cattle Farming Communities in Shimba Hills, Kenya. Pathogens 2021, 10, 1501. [Google Scholar] [CrossRef]
- Bolling, B.G.; Tesh, R.B.; Thangamani, S.; Guzman, H.; Popov, V.L.; Wood, T.G.; Vasilakis, N.; Widen, S.G. Insect-Specific Viruses Detected in Laboratory Mosquito Colonies and Their Potential Implications for Experiments Evaluating Arbovirus Vector Competence. Am. J. Trop. Med. Hyg. 2015, 92, 422–428. [Google Scholar] [CrossRef]
- Guindo-Coulibaly, N.; Adja, A.; Coulibaly, J.; Kpan, M.; Adou, K.; Zoh, D. Expansion of Aedes africanus (Diptera: Culicidae), a sylvatic vector of arboviruses, into an urban environment of Abidjan, Côte d’Ivoire. J. Vector Ecol. 2019, 44, 248–255. [Google Scholar] [CrossRef]
- Santos, J.M.; Capinha, C.; Rocha, J.; Sousa, C.A. The current and future distribution of the yellow fever mosquito (Aedes aegypti) on Madeira Island. PLoS Neglected Trop. Dis. 2022, 16, e0010715. [Google Scholar] [CrossRef]
- Bett, B.; Said, M.Y.; Sang, R.; Bukachi, S.; Wanyoike, S.; Kifugo, S.C.; Otieno, F.; Ontiri, E.; Njeru, I.; Lindahl, J.; et al. Effects of flood irrigation on the risk of selected zoonotic pathogens in an arid and semi-arid area in the eastern Kenya. PLoS ONE 2017, 12, e0172626. [Google Scholar] [CrossRef]
- Li, S.L.; Acosta, A.L.; Hill, S.C.; Brady, O.J.; de Almeida, M.A.B.; Cardoso, J.d.C.; Hamlet, A.; Mucci, L.F.; de Deus, J.T.; Iani, F.C.M.; et al. Mapping environmental suitability of Haemagogus and Sabethes spp. mosquitoes to understand sylvatic transmission risk of yellow fever virus in Brazil. PLoS Neglected Trop. Dis. 2022, 16, e0010019. [Google Scholar] [CrossRef]
- Tchouassi, D.P.; Torto, B.; Sang, R.; Riginos, C.; Ezenwa, V.O. Large herbivore loss has complex effects on mosquito ecology and vector-borne disease risk. Transbound. Emerg. Dis. 2020, 68, 2503–2513. [Google Scholar] [CrossRef] [PubMed]
- García-Carrasco, J.-M.; Muñoz, A.-R.; Olivero, J.; Segura, M.; Real, R. Mapping the Risk for West Nile Virus Transmission, Africa. Emerg. Infect. Dis. 2022, 28, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Boyer, S.; Durand, B.; Yean, S.; Brengues, C.; Maquart, P.-O.; Fontenille, D.; Chevalier, V. Host-Feeding Preference and Diel Activity of Mosquito Vectors of the Japanese Encephalitis Virus in Rural Cambodia. Pathogens 2021, 10, 376. [Google Scholar] [CrossRef]
- Villinger, J.; Mbaya, M.K.; Ouso, D.; Kipanga, P.N.; Lutomiah, J.; Masiga, D.K. Arbovirus and insect-specific virus discovery in Kenya by novel six genera multiplex high-resolution melting analysis. Mol. Ecol. Resour. 2016, 17, 466–480. [Google Scholar] [CrossRef] [PubMed]
- Iwashita, H.; Higa, Y.; Futami, K.; Lutiali, P.A.; Njenga, S.M.; Nabeshima, T.; Minakawa, N. Mosquito arbovirus survey in selected areas of Kenya: Detection of insect-specific virus. Trop. Med. Health 2018, 46, 19. [Google Scholar] [CrossRef] [PubMed]
- Fauver, J.R.; Grubaugh, N.D.; Krajacich, B.J.; Weger-Lucarelli, J.; Lakin, S.M.; Fakoli, L.S., III; Bolay, F.K.; Diclaro, J.W., II; Dabiré, K.R.; Foy, B.D.; et al. West African Anopheles gambiae mosquitoes harbor a taxonomically diverse virome including new insect-specific flaviviruses, mononegaviruses, and totiviruses. Virology 2016, 498, 288–299. [Google Scholar] [CrossRef]
- Guégan, M.; Zouache, K.; Démichel, C.; Minard, G.; Van, V.T.; Potier, P.; Mavingui, P.; Moro, C.V. The mosquito holobiont: Fresh insight into mosquito-microbiota interactions. Microbiome 2018, 6, 49. [Google Scholar] [CrossRef]
- Bolling, B.G.; Olea-Popelka, F.J.; Eisen, L.; Moore, C.G.; Blair, C.D. Transmission dynamics of an insect-specific flavivirus in a naturally infected Culex pipiens laboratory colony and effects of co-infection on vector competence for West Nile virus. Virology 2012, 427, 90–97. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhang, C.; Zhang, L.; Yang, Y.; Yu, X.; Wang, J.; Liu, Q.; Wang, P.; Cheng, G. A human-blood-derived microRNA facilitates flavivirus infection in fed mosquitoes. Cell Rep. 2021, 37, 110091. [Google Scholar] [CrossRef]
- Sant’Anna, M.R.; Nascimento, A.; Alexander, B.; Dilger, E.; Cavalcante, R.R.; Diaz-Albiter, H.M.; Bates, P.A.; Dillon, R.J. Chicken blood provides a suitable meal for the sand fly Lutzomyia longipalpis and does not inhibit leishmania development in the gut. Parasites Vectors 2010, 3, 3. [Google Scholar] [CrossRef] [PubMed]
- Souza, A.V.G.; Petretski, J.H.; Demasi, M.; Bechara, E.; Oliveira, P.L. Urate Protects a Blood-Sucking Insect Against Hemin-Induced Oxidative Stress. Free Radic. Biol. Med. 1997, 22, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gilbreath, T.M.; Kukutla, P.; Yan, G.; Xu, J. Dynamic gut microbiome across life history of the malaria mosquito Anopheles gambiae in Kenya. PLoS ONE 2011, 6, e24767. [Google Scholar] [CrossRef] [PubMed]
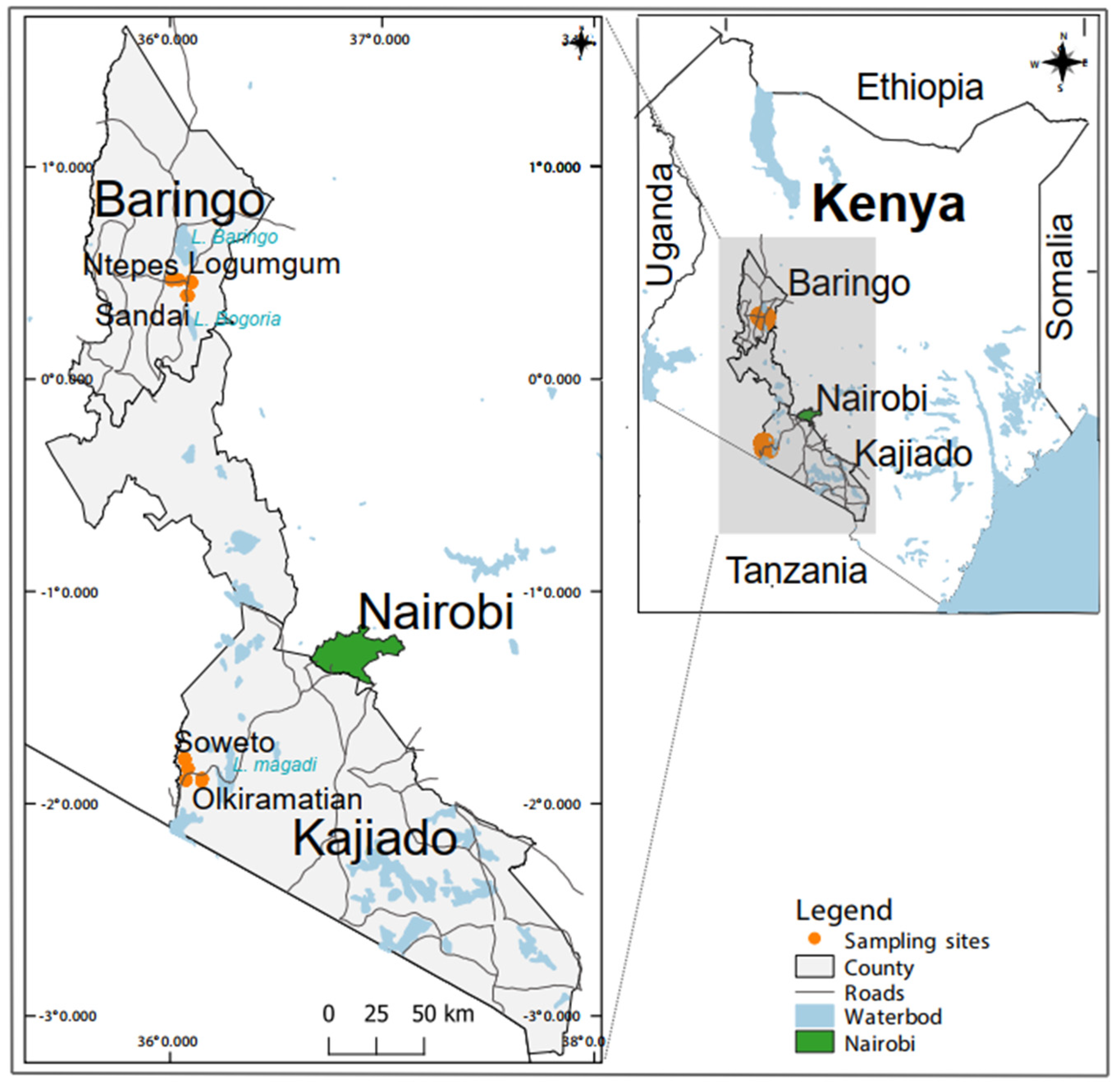


| Baringo County | Kajiado County | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ntepes | Sandai | Logumgum | Soweto | Olkiramatian | n (%) | |||||||||||
| Sampling method | BG | CDC-LT | CDC-Gravid | BG | CDC-LT | CDC-Gravid | BG | CDC-LT | CDC-Gravid | BG | CDC-LT | CDC-Gravid | BG | CDC-LT | CDC-Gravid | |
| 370 | 313 | 35 | 479 | 257 | 19 | 395 | 454 | 7 | 356 | 2303 | 0 | 638 | 1036 | 3 | 6665 | |
| Sex | ||||||||||||||||
| Female | 364 | 310 | 31 | 476 | 254 | 18 | 394 | 446 | 4 | 316 | 1065 | 0 | 620 | 935 | 3 | 5236 (78.6) |
| Male | 6 | 3 | 4 | 3 | 3 | 1 | 1 | 8 | 3 | 40 | 1238 | 0 | 18 | 101 | 0 | 1429 (21.4) |
| Mosquito species | ||||||||||||||||
| Uranotaenia nigromaculata | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 57 | 1387 | 0 | 0 | 62 | 0 | 1506 (22.6) |
| Mansonia africana | 246 | 129 | 0 | 98 | 24 | 0 | 247 | 346 | 5 | 1 | 0 | 0 | 177 | 151 | 0 | 1424 (21.4) |
| Culex univittatus | 0 | 16 | 0 | 5 | 2 | 0 | 0 | 0 | 0 | 97 | 255 | 0 | 260 | 313 | 0 | 948 (14.2) |
| Mansonia uniformis | 50 | 14 | 0 | 325 | 202 | 15 | 122 | 69 | 0 | 0 | 2 | 0 | 17 | 19 | 0 | 835 (12.5) |
| Anopheles funestus s.l. | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 40 | 157 | 0 | 37 | 89 | 0 | 325 (4.9) |
| Culex vansomereni | 3 | 3 | 0 | 0 | 3 | 0 | 0 | 6 | 0 | 19 | 22 | 0 | 53 | 156 | 0 | 265 (4.0) |
| Culex pipiens | 7 | 64 | 5 | 15 | 8 | 1 | 4 | 1 | 0 | 16 | 35 | 0 | 46 | 58 | 0 | 260 (3.9) |
| Culex annulioris | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 86 | 152 | 0 | 0 | 19 | 0 | 258 (3.9) |
| Culex cinereus | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 102 | 0 | 0 | 4 | 0 | 107 (1.6) |
| Aedes aegypti | 29 | 0 | 23 | 20 | 1 | 2 | 0 | 1 | 0 | 3 | 0 | 0 | 17 | 0 | 0 | 96 (1.4) |
| Aedes mcintoshi | 4 | 5 | 0 | 1 | 0 | 0 | 4 | 0 | 0 | 4 | 24 | 0 | 7 | 43 | 0 | 92 (1.4) |
| Anopheles coustani | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 31 | 0 | 5 | 25 | 0 | 68 (1.0) |
| Anopheles gambiae s.l. | 4 | 24 | 4 | 0 | 1 | 0 | 1 | 2 | 1 | 4 | 4 | 0 | 6 | 15 | 0 | 66 (1.0) |
| Ficalbia mediolineata | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 35 | 0 | 0 | 12 | 0 | 58 (0.9) |
| Ficalbia splendens | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 28 | 0 | 0 | 27 | 0 | 57 (0.9) |
| Mansonia spp. | 3 | 12 | 0 | 0 | 0 | 0 | 11 | 5 | 0 | 0 | 0 | 0 | 3 | 2 | 0 | 36 (0.5) |
| Aedes tricholabis | 13 | 8 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 23 (0.3) |
| Aedes vittatus | 1 | 10 | 0 | 0 | 0 | 0 | 4 | 5 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 21 (0.3) |
| Anopheles spp. | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 5 | 5 | 0 | 20 (0.3) |
| Culex ethiopicus | 0 | 0 | 0 | 2 | 7 | 0 | 0 | 1 | 0 | 4 | 5 | 0 | 0 | 1 | 0 | 20 (0.3) |
| Orthopodomyia spp. | 1 | 14 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 18 (0.3) |
| Uranotaenia philaroxia | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 16 | 0 | 0 | 0 | 0 | 17 (0.3) |
| Uranotaenia spp. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15 | 0 | 0 | 2 | 0 | 17 (0.3) |
| Culex spp. | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 2 | 0 | 1 | 7 | 3 | 17 (0.3) |
| Culex tigriepes | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 9 | 0 | 1 | 4 | 0 | 16 (0.2) |
| Aedes simpsoni s.l. | 5 | 0 | 0 | 7 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 15 (0.2) |
| Anopheles pharoensis | 0 | 5 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 4 | 0 | 13 (0.2) |
| Culex poicilipes | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 1 | 5 | 0 | 12 (0.2) |
| Aedes spp. | 1 | 3 | 1 | 2 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 10 (0.2) |
| Anopheles maculipalpis | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 8 (0.1) |
| Culex bitaeniorhynhus | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 (0.1) |
| Ficalbia spp. | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 7 (0.1) |
| Aedes ethiopicus | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 (0.1) |
| Aedes metallicus | 3 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 (0.1) |
| Eretmapodite spp. | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 (0.0) |
| Ficalbia uniformis | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 3 (0.0) |
| Uranotaenia pallidoephala | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 3 (0.0) |
| Anophelessquamosus | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (0.0) |
| Coquillettidia aurites | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (0.0) |
| Coquillettidia fuscopenatta | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 (0.0) |
| Culex zombaensis | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 (0.0) |
| Eretmapodite chrystogastar | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 (0.0) |
| n | 370 | 313 | 35 | 479 | 257 | 19 | 395 | 454 | 7 | 356 | 2303 | 0 | 638 | 1036 | 3 | 6665 |
| Blood-fed | 12 | 34 | 0 | 1 | 0 | 0 | 3 | 6 | 1 | 3 | 7 | 0 | 5 | 8 | 0 | 80 (1.2) |
| County | Study Area | Species | n | Human | Cattle | Goat | Sheep | Amphibian | Bird | Antelope | Leopard | Buffalo | Hippopotamus | Bat | UN |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Baringo | Ntepes | Culex pipiens | 20 | 6 | 9 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 |
| Mansonia africana | 17 | 3 | 4 | 5 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | ||
| Anopheles gambiae s.l. | 5 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Aedes mcintoshi | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Aedes tricholabis | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Mansonia uniformis | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Logumgum | Mansonia africana | 8 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | |
| Ae. mcintoshi | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | ||
| Anopheles gambiae s.l. | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Sandai | Mansonia africana | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| n (%) | 57 (71.3) | 14 (17.5) | 20 (25.0) | 8 (10.0) | 3 (3.8) | 0 | 0 | 0 | 0 | 0 | 2 (2.5) | 0 | 10 (12.5) | ||
| Kajiado | Olkiramatian | Culexunivittatus | 6 | 1 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 |
| Culex sp. | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | ||
| Aedes mcintoshi | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | ||
| Culexpipiens | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Mansonia africana | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | ||
| Soweto | Culex univittatus | 5 | 2 * | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 1 * | 0 | |
| Uranotaenia nigromaculata | 5 | 3 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Aedes aegypti | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| n (%) | 23 (28.8) | 8 (10.0) * | 3 (3.8) | 0 | 0 | 2 (2.5) | 3 (3.8) | 2 (2.5) | 1 (1.3) | 2 (2.5) | 0 | 1 (1.3) | 2 (2.5) | ||
| n (%) | 80 | 22 (27.2) | 23 (28.4) | 8 (9.9) | 3 (3.8) | 2 (2.5) | 3 (3.8) | 2 (2.5) | 1 (1.3) | 2 (2.5) | 2 (2.5) | 1 (1.3) | 12 (14.8) |
| Sample Code | Mosquito Species | No of Mosquitoes | Blood-Meal Source/ Abdomen Status | Closely Related Flavivirus Species | % ID (nt) | Sequence Length (bp) | Genbank Acession No. |
|---|---|---|---|---|---|---|---|
| M138 | Mansonia africana | 1 | Human | CuaCua virus | 94 | 218 | OQ588809 |
| M287 | Mansonia africana | 1 | Unknown | CuaCua virus | 94 | 201 | OQ588808 |
| M422 | Mansonia africana | 1 | Unknown | CuaCua virus | 94 | 201 | OQ588810 |
| M211B | Anopheles gambiae s.l. | 1 | Human | Aedes flavivirus | 72 | 211 | OQ588805 |
| M317B | Mansonia africana | 1 | Human | Aedes flavivirus | 72 | 212 | OQ588802 |
| KM151 | Culex univittatus | 1 | Human | Aedes flavivirus | 72 | 212 | OQ588803 |
| KM174 | Aedes aegypti | 1 | Human | Aedes flavivirus | 72 | 219 | OQ588804 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ogola, E.O.; Bastos, A.D.S.; Rotich, G.; Kopp, A.; Slothouwer, I.; Omoga, D.C.A.; Sang, R.; Torto, B.; Junglen, S.; Tchouassi, D.P. Analyses of Mosquito Species Composition, Blood-Feeding Habits and Infection with Insect-Specific Flaviviruses in Two Arid, Pastoralist-Dominated Counties in Kenya. Pathogens 2023, 12, 967. https://doi.org/10.3390/pathogens12070967
Ogola EO, Bastos ADS, Rotich G, Kopp A, Slothouwer I, Omoga DCA, Sang R, Torto B, Junglen S, Tchouassi DP. Analyses of Mosquito Species Composition, Blood-Feeding Habits and Infection with Insect-Specific Flaviviruses in Two Arid, Pastoralist-Dominated Counties in Kenya. Pathogens. 2023; 12(7):967. https://doi.org/10.3390/pathogens12070967
Chicago/Turabian StyleOgola, Edwin O., Armanda D. S. Bastos, Gilbert Rotich, Anne Kopp, Inga Slothouwer, Dorcus C. A. Omoga, Rosemary Sang, Baldwyn Torto, Sandra Junglen, and David P. Tchouassi. 2023. "Analyses of Mosquito Species Composition, Blood-Feeding Habits and Infection with Insect-Specific Flaviviruses in Two Arid, Pastoralist-Dominated Counties in Kenya" Pathogens 12, no. 7: 967. https://doi.org/10.3390/pathogens12070967
APA StyleOgola, E. O., Bastos, A. D. S., Rotich, G., Kopp, A., Slothouwer, I., Omoga, D. C. A., Sang, R., Torto, B., Junglen, S., & Tchouassi, D. P. (2023). Analyses of Mosquito Species Composition, Blood-Feeding Habits and Infection with Insect-Specific Flaviviruses in Two Arid, Pastoralist-Dominated Counties in Kenya. Pathogens, 12(7), 967. https://doi.org/10.3390/pathogens12070967








