Molecular Epidemiology of Streptococcus pneumoniae Detected in Hospitalized Pediatric Acute Respiratory Infection Cases in Central Vietnam
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Patients
2.3. Specimen Collection
2.4. Culture, Identification and DNA Extraction of S. pneumoniae
2.5. Serotyping
2.6. Antimicrobial Susceptibility Testing
2.7. Penicillin and Erythromycin Resistance Genes
2.8. Multi-Locus Sequence Typing
2.9. Statistical Analysis
3. Results
3.1. Demographic and Clinical Characteristics of Study Participants
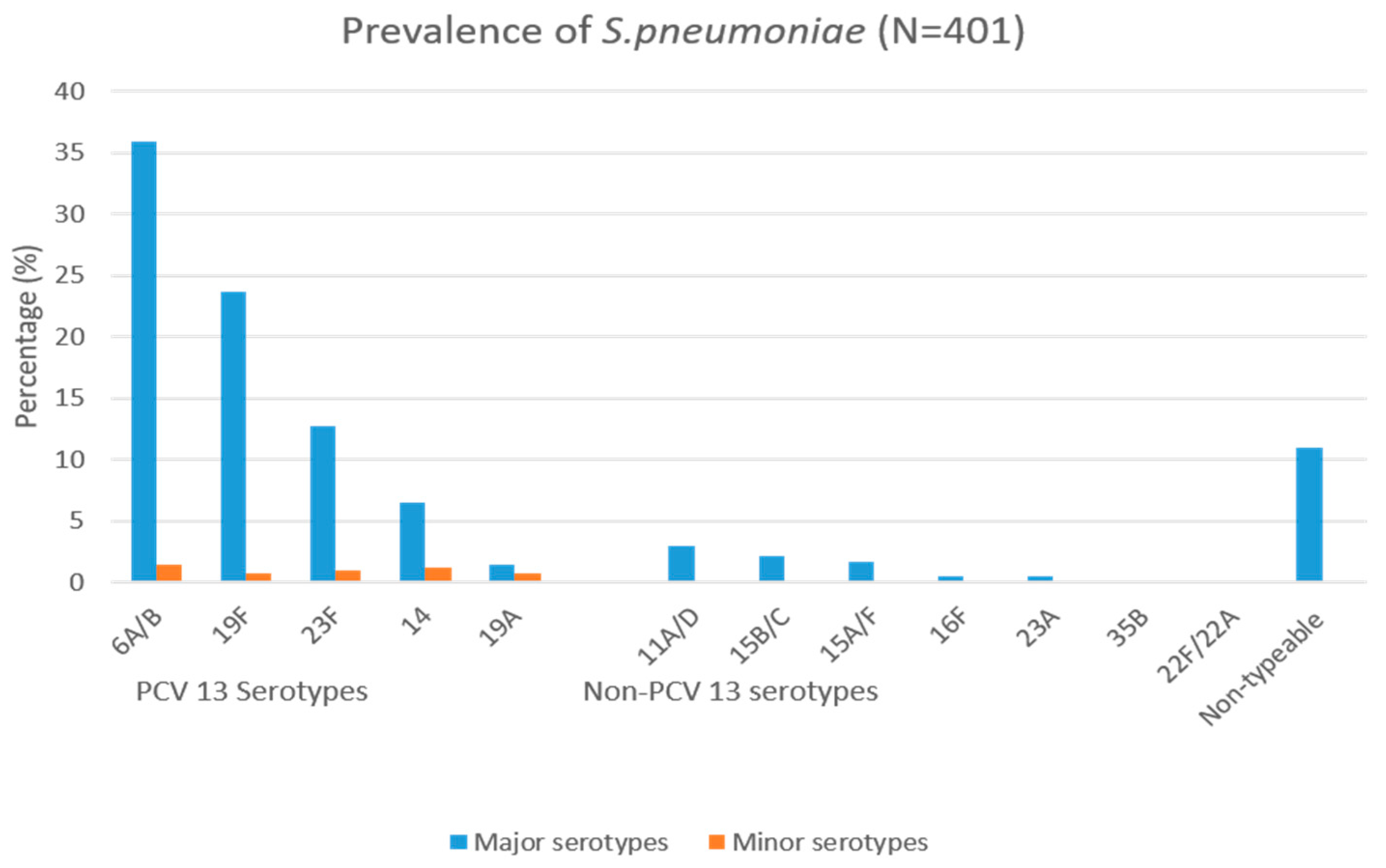
3.2. Serotyping
3.3. Antimicrobial Susceptibility Testing
3.4. Penicillin and Erythromycin Resistance Genes
3.5. Multi-Locus Sequence Typing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Safiri, S.; Mahmoodpoor, A.; Kolahi, A.A.; Nejadghaderi, S.A.; Sullman, M.J.M.; Mansournia, M.A.; Ansarin, K.; Collins, G.S.; Kaufman, J.S.; Abdollahi, M. Global burden of lower respiratory infections during the last three decades. Front. Public Health 2023, 10, 1028525. [Google Scholar] [CrossRef] [PubMed]
- Global Burden of Disease Collaborative Network Global Burden of Disease Collaborative Network. Global Burden of Disease Study 2019 (GBD 2019). Results. Institute for Health Metrics and Evaluation (IHME): Seattle, WA, USA, 2020. Available online: https://vizhub.healthdata.org/gbd-results/ (accessed on 7 June 2023).
- Bogaert, D.; De Groot, R.; Hermans, P.W.M. Streptococcus pneumoniae colonisation: The key to pneumococcal disease. Lancet Infect. Dis. 2004, 4, 144–154. [Google Scholar] [CrossRef] [PubMed]
- Weiser, J.N.; Ferreira, D.M.; Paton, J.C. Streptococcus pneumoniae: Transmission, colonization and invasion. Nat. Rev. Microbiol. 2018, 16, 355–367. [Google Scholar] [CrossRef]
- O’Brien, K.L.; Wolfson, L.J.; Watt, J.P.; Henkle, E.; Deloria-Knoll, M.; McCall, N.; Lee, E.; Mulholland, K.; Levine, O.S.; Cherian, T. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: Global estimates. Lancet 2009, 374, 893–902. [Google Scholar] [CrossRef]
- Johnson, H.L.; Deloria-Knoll, M.; Levine, O.S.; Stoszek, S.K.; Hance, L.F.; Reithinger, R.; Muenz, L.R.; O’Brien, K.L. Systematic evaluation of serotypes causing invasive pneumococcal disease among children under five: The pneumococcal global serotype project. PLoS Med. 2010, 7, e1000348. [Google Scholar] [CrossRef] [PubMed]
- Song, J.Y.; Nahm, M.H.; Moseley, M.A. Clinical implications of pneumococcal serotypes: Invasive disease potential, clinical presentations, and antibiotic resistance. J. Korean Med. Sci. 2013, 28, 4–15. [Google Scholar] [CrossRef]
- Ganaie, F.; Saad, J.S.; McGee, L.; van Tonder, A.J.; Bentley, S.D.; Lo, S.W.; Gladstone, R.A.; Turner, P.; Keenan, J.D.; Breiman, R.F.; et al. A new pneumococcal capsule type, 10D, is the 100th serotype and has a large cps fragment from an oral streptococcus. MBio 2020, 11, e00937-20. [Google Scholar] [CrossRef]
- World Health Organization (WHO) Pneumococcal conjugate vaccines in infants and children under 5 years of age: WHO position paper—February 2019. Wkly. Epidemiol. Rec. 2019, 94, 85–104.
- Huang, S.S.; Platt, R.; Rifas-Shiman, S.L.; Pelton, S.I.; Goldmann, D.; Finkelstein, J.A. Post-PCV7 changes in colonizing pneumococcal serotypes in 16 Massachusetts communities, 2001 and 2004. Pediatrics 2005, 116, e408–e413. [Google Scholar] [CrossRef]
- Kyaw, M.H.; Lynfield, R.; Schaffner, W.; Craig, A.S.; Hadler, J.; Reingold, A.; Thomas, A.R.; Harrison, L.H.; Bennett, N.M.; Farley, M.M.; et al. Effect of Introduction of the Pneumococcal Conjugate Vaccine on Drug-Resistant Streptococcus pneumoniae. N. Engl. J. Med. 2006, 354, 1455–1463. [Google Scholar] [CrossRef]
- Ladhani, S.N.; Slack, M.P.E.; Andrews, N.J.; Waight, P.A.; Borrow, R.; Miller, E. Invasive pneumococcal disease after routine pneumococcal conjugate vaccination in children, England and Wales. Emerg. Infect. Dis. 2013, 19, 61–68. [Google Scholar] [CrossRef]
- Brandileone, M.C.C.; Almeida, S.C.G.; Bokermann, S.; Minamisava, R.; Berezin, E.N.; Harrison, L.H.; Andrade, A.L. Dynamics of antimicrobial resistance of Streptococcus pneumoniae following PCV10 introduction in Brazil: Nationwide surveillance from 2007 to 2019. Vaccine 2021, 39, 3207–3215. [Google Scholar] [CrossRef] [PubMed]
- Temple, B.; Nation, M.L.; Dai, V.T.T.; Beissbarth, J.; Bright, K.; Dunne, E.M.; Hinds, J.; Hoan, P.T.; Lai, J.; Nguyen, C.D.; et al. Effect of a 2+1 schedule of ten-valent versus 13-valent pneumococcal conjugate vaccine on pneumococcal carriage: Results from a randomised controlled trial in Vietnam. Vaccine 2021, 39, 2303–2310. [Google Scholar] [CrossRef] [PubMed]
- Løchen, A.; Croucher, N.J.; Anderson, R.M. Divergent serotype replacement trends and increasing diversity in pneumococcal disease in high income settings reduce the benefit of expanding vaccine valency. Sci. Rep. 2020, 10, 18977. [Google Scholar] [CrossRef] [PubMed]
- Nagaraj, S.; Kalal, B.S.; Manoharan, A.; Shet, A. Streptococcus pneumoniae serotype prevalence and antibiotic resistance among young children with invasive pneumococcal disease: Experience from a tertiary care center in South India. Germs 2017, 7, 78–85. [Google Scholar] [CrossRef]
- Shak, J.R.; Vidal, J.E.; Klugman, K.P. Influence of bacterial interactions on pneumococcal colonization of the nasopharynx. Trends Microbiol. 2013, 21, 129–135. [Google Scholar] [CrossRef]
- Hakenbeck, R.; Balmelle, N.; Weber, B.; Gardès, C.; Keck, W.; De Saizieu, A. Mosaic genes and mosaic chromosomes: Intra- and interspecies genomic variation of Streptococcus pneumoniae. Infect. Immun. 2001, 69, 2477–2486. [Google Scholar] [CrossRef]
- Noguchi, N.; Tano, J.; Nasu, Y.; Koyama, M.; Narui, K.; Kamishima, H.; Saito, T.; Tsuyuki, K.; Sasatsu, M. Antimicrobial susceptibilities and distribution of resistance genes for β-lactams and macrolides in Streptococcus pneumoniae isolated between 2002 and 2004 in Tokyo. Int. J. Antimicrob. Agents 2007, 29, 26–33. [Google Scholar] [CrossRef]
- Dadgostar, P. Antimicrobial resistance: Implications and costs. Infect. Drug Resist. 2019, 12, 3903–3910. [Google Scholar] [CrossRef]
- Nguyen, H.A.T.; Fujii, H.; Vu, H.T.T.; Parry, C.M.; Dang, A.D.; Ariyoshi, K.; Yoshida, L.M. An alarmingly high nasal carriage rate of Streptococcus pneumoniae serotype 19F non-susceptible to multiple beta-lactam antimicrobials among Vietnamese children. BMC Infect. Dis. 2019, 19, 241. [Google Scholar] [CrossRef]
- Yoshida, L.M.; Suzuki, M.; Yamamoto, T.; Nguyen, H.A.; Nguyen, C.D.; Nguyen, A.T.; Oishi, K.; Vu, T.D.; Le, T.H.; Le, M.Q.; et al. Viral Pathogens Associated with Acute Respiratory Infections in Central Vietnamese Children. Pediatr. Infect. Dis. J. 2010, 29, 75–77. [Google Scholar] [CrossRef] [PubMed]
- Pimenta, F.C.; Roundtree, A.; Soysal, A.; Bakir, M.; Du Plessis, M.; Wolter, N.; Von Gottberg, A.; McGee, L.; Da Gloria Carvalho, M.; Beall, B. Sequential Triplex Real-Time PCR Assay for Detecting 21 Pneumococcal Capsular Serotypes That Account for a High Global Disease Burden. J. Clin. Microbiol. 2013, 51, 647–652. [Google Scholar] [CrossRef] [PubMed]
- Cools, F.; Delputte, P.; Cos, P. The search for novel treatment strategies for Streptococcus pneumoniae infections. FEMS Microbiol. Rev. 2021, 45, fuaa072. [Google Scholar] [CrossRef] [PubMed]
- CLSI. M100S Performance Standards for Antimicrobial (CLSI), 26th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015; ISBN 1562389238. [Google Scholar]
- EUCAST Clinical Breakpoint Table v. 5.0, Valid from 1 January 2015. pp. 1–77. Available online: http://www.eucast.org/clinical_breakpoints/ (accessed on 2 November 2022).
- Magiorakos, A.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standarddefinitions for acquired resistance. Clin. Microbiol. Infect. 2011, 28, 250–290. [Google Scholar] [CrossRef]
- Nagai, K.; Shibasaki, Y.; Hasegawa, K.; Davies, T.A.; Jacobs, M.R.; Ubukata, K.; Appelbaum, P.C. Evaluation of PCR primers to screen for Streptococcus pneumoniae isolates and β-lactam resistance, and to detect common macrolide resistance determinants. J. Antimicrob. Chemother. 2001, 48, 915–918. [Google Scholar] [CrossRef]
- Ubukata, K.; Chiba, N.; Hasegawa, K.; Kobayashi, R.; Iwata, S.; Sunakawa, K. Antibiotic Susceptibility in Relation to Penicillin-Binding Protein Genes and Serotype Distribution of Streptococcus pneumoniae Strains Responsible for Meningitis in Japan, 1999 to 2002. Antimicrob. Agents Chemother. 2004, 48, 1488–1494. [Google Scholar] [CrossRef]
- Ubukata, K.; Takata, M.; Morozumi, M.; Chiba, N.; Wajima, T.; Hanada, S.; Shouji, M.; Sakuma, M.; Iwata, S. Effects of pneumococcal conjugate vaccine on genotypic penicillin resistance and serotype changes, Japan, 2010–2017. Emerg. Infect. Dis. 2018, 24, 2010–2020. [Google Scholar] [CrossRef]
- Ibarz Pavón, A.B.; Maiden, M.C.J. Multilocus sequence typing. Methods Mol. Biol. 2009, 551, 129–140. [Google Scholar] [CrossRef]
- Enright, M.; Spratt, B. A multilocus sequence typing scheme for Streptococcus pneumoniae: Identification of clones associated with serious invasive disease. Microbiology 1998, 144, 3049–3060. [Google Scholar] [CrossRef]
- WHO; UNICEF. Revised WHO Classification and Treatment of childhood Pneumonia at Health Facilities: Qiuck Reference Guide. Evidence for Technical Update of Pocket Book Recommendations. Geneva, WHO. 2012. Available online: https://apps.who.int/iris/bitstream/handle/10665/137332/WHO_FWC_MCA_14.9_eng.pdf (accessed on 15 July 2022).
- Vu, H.T.T.; Yoshida, L.M.; Suzuki, M.; Nguyen, H.A.T.; Nguyen, C.D.L.; Nguyen, A.T.T.; Oishi, K.; Yamamoto, T.; Watanabe, K.; Vu, T.D.; et al. Association between Nasopharyngeal Load of Streptococcus pneumoniae, Viral Coinfection, and Radiologically Confirmed Pneumonia in Vietnamese Children. Pediatr. Infect. Dis. J. 2011, 30, 11–18. [Google Scholar] [CrossRef]
- Pernica, J.M.; Inch, K.; Alfaraidi, H.; Van Meer, A.; Carciumaru, R.; Luinstra, K.; Smieja, M. Assessment of nasopharyngeal Streptococcus pneumoniae colonization does not permit discrimination between Canadian children with viral and bacterial respiratory infection: A matched-cohort cross-sectional study. BMC Infect. Dis. 2021, 21, 509. [Google Scholar] [CrossRef]
- Tin Tin Htar, M.; Christopoulou, D.; Schmitt, H.J. Pneumococcal serotype evolution in Western Europe. BMC Infect. Dis. 2015, 15, 419. [Google Scholar] [CrossRef] [PubMed]
- Balsells, E.; Guillot, L.; Nair, H.; Kyaw, M.H. Serotype distribution of Streptococcus pneumoniae causing invasive disease in children in the post-PCV era: A systematic review and meta-analysis. PLoS ONE 2017, e0177113. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.; Shi, W.; Yu, D.; Yao, K. hu Epidemiology of non-vaccine serotypes of Streptococcus pneumoniae before and after universal administration of pneumococcal conjugate vaccines. Hum. Vaccines Immunother. 2021, 17, 5628–5637. [Google Scholar] [CrossRef]
- Mitchell, M.E.V.; Alders, R.; Unger, F.; Nguyen-Viet, H.; Le, T.T.H.; Toribio, J.A. The challenges of investigating antimicrobial resistance in Vietnam—What benefits does a One Health approach offer the animal and human health sectors? BMC Public Health 2020, 20, 213. [Google Scholar] [CrossRef] [PubMed]
- Hoa, N.Q.; Trung, N.V.; Larsson, M.; Eriksson, B.; Phuc, H.D.; Chuc, N.T.K.; Lundborg, C.S. Decreased Streptococcus pneumoniae susceptibility to oral antibiotics among children in rural Vietnam: A community study. BMC Infect. Dis. 2010, 10, 85. [Google Scholar] [CrossRef]
- Andrejko, K.; Ratnasiri, B.; Hausdorff, W.P.; Laxminarayan, R.; Lewnard, J.A. Antimicrobial resistance in paediatric Streptococcus pneumoniae isolates amid global implementation of pneumococcal conjugate vaccines: A systematic review and meta-regression analysis. Lancet Microbe 2021, 2, e450–e460. [Google Scholar] [CrossRef]
- Vu, T.V.D.; Choisy, M.; Do, T.T.N.; Nguyen, V.M.H.; Campbell, J.I.; Le, T.H.; Nguyen, V.T.; Wertheim, H.F.L.; Pham, N.T.; Nguyen, V.K.; et al. Antimicrobial susceptibility testing results from 13 hospitals in Viet Nam: VINARES 2016–2017. Antimicrob. Resist. Infect. Control 2021, 10, 78. [Google Scholar] [CrossRef]
- Fani, F.; Leprohon, P.; Légaré, D.; Ouellette, M. Whole genome sequencing of penicillin-resistant Streptococcus pneumoniae reveals mutations in penicillin-binding proteins and in a putative iron permease. Genome Biol. 2011, 12, R115. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, J.; Zhang, Z.; Cui, B.; Wang, Y.; Zhang, Y.; Xu, H.; Cheng, G.; Liu, Y.; Qin, X. Characterization of Streptococcus pneumoniae Macrolide Resistance and Its Mechanism in Northeast China over a 20-Year Period. Microbiol. Spectr. 2022, 10, e00546-22. [Google Scholar] [CrossRef]
- Safari, D.; Kuo, L.C.; Huang, Y.T.; Liao, C.H.; Sheng, W.H.; Hsueh, P.R. Increase in the rate of azithromycin-resistant Streptococcus pneumoniae isolates carrying the erm(B) and mef(A) genes in Taiwan, 2006–2010. BMC Infect. Dis. 2015, 14, 704. [Google Scholar] [CrossRef]
- Bae, S.; Lee, K. Distribution of capsular serotypes and macrolide resistance mechanisms among macrolide-resistant Streptococcus pneumoniae isolates in Korea. Diagn. Microbiol. Infect. Dis. 2009, 63, 213–216. [Google Scholar] [CrossRef]
- Bogaert, D.; Ha, N.T.; Sluijter, M.; Lemmens, N.; De Groot, R.; Hermans, P.W.M. Molecular epidemiology of pneumococcal carriage among children with upper respiratory tract infections in Hanoi, Vietnam. J. Clin. Microbiol. 2002, 40, 3903–3908. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Parry, C.M.; Duong, N.M.; Zhou, J.; Mai, N.T.H.; Diep, T.S.; Thinh, L.Q.; Wain, J.; Chau, N.V.V.; Griffiths, D.; Day, N.P.J.; et al. Emergence in Vietnam of Streptococcus pneumoniae resistant to multiple antimicrobial agents as a result of dissemination of the multiresistant Spain23F-1 clone. Antimicrob. Agents Chemother. 2002, 46, 3512–3517. [Google Scholar] [CrossRef] [PubMed]
- Bowers, J.R.; Driebe, E.M.; Nibecker, J.L.; Wojack, B.R.; Sarovich, D.S.; Wong, A.H.; Brzoska, P.M.; Hubert, N.; Knadler, A.; Watson, L.M.; et al. Dominance of multidrug resistant CC271 clones in macrolide-resistant Streptococcus pneumoniae in Arizona. BMC Microbiol. 2012, 12, 12. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Fu, J.; Li, L.; Yi, R.; Xu, S.; Chen, J.; Ye, X.; McGrath, E. Molecular epidemiology of Streptococcus pneumoniae isolated from pediatric community-acquired pneumonia in pre-conjugate vaccine era in Western China. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 4. [Google Scholar] [CrossRef]
- Del Grosso, M.; Northwood, J.G.E.; Farrell, D.J.; Pantosti, A. The macrolide resistance genes erm(B) and mef(E) are carried by Tn2010 in dual-gene Streptococcus pneumoniae isolates belonging to clonal complex CC271. Antimicrob. Agents Chemother. 2007, 51, 4184–4186. [Google Scholar] [CrossRef]
- Dzaraly, N.D.; Mohd Desa, M.N.; Muthanna, A.R.; Masri, S.N.; Taib, N.M.; Suhaili, Z.; Sulaiman, N.; Baharin, N.H.Z.; Shuan, C.Y.; Ariffin, Z.; et al. Antimicrobial susceptibility, serotype distribution, virulence profile and molecular typing of piliated clinical isolates of pneumococci from east coast, Peninsular Malaysia. Sci. Rep. 2021, 11, 8220. [Google Scholar] [CrossRef]
- Scherer, E.M.; Beall, B.; Metcalf, B. Serotype-Switch Variant of pneumoniae Sequence Type 271. Emerg. Infect. Dis. 2021, 27, 1689–1692. [Google Scholar] [CrossRef]

| Characteristic | All n (%) | Pneumococcal +ve n (%) | Pneumococcal −ve n (%) | Odds Ratio (95% CI) |
|---|---|---|---|---|
| Total (n) | 1300 | 413 (31.8) | 887 (68.2) | |
| Age | ||||
| 0–1 yr. | 825 (63.5) | 279 (33.8) | 546 (66.2) | 4.03 (1.99–8.16) |
| 2–3 yrs. | 316 (24.3) | 108 (34.2) | 208 (65.8) | 4.10 (1.97–8.51) |
| 4–5 yrs. | 79 (6.1) | 17 (21.5) | 62 (78.5) | 2.16 (0.90–5.20) |
| >5 yrs. | 80 (6.2) | 9 (11.3) | 71 (88.7) | Reference |
| Sex | ||||
| Male | 780 (60.0) | 250 (32.1) | 530 (67.9) | 1.03 (0.81–1.31) |
| Female | 520 (40.0) | 163 (31.4) | 357 (68.6) | Reference |
| Day-care or school attendance | ||||
| Yes | 671 (51.6) | 251 (37.4) | 420 (62.6) | 1.72 (1.36–2.18) |
| No | 629 (48.4) | 162 (25.8) | 467 (74.2) | Reference |
| Prior antibiotic use a | ||||
| Yes | 950 (73.1) | 317 (33.4) | 633 (66.6) | 1.32 (1.01–1.74) |
| No | 350 (26.9) | 96 (27.4) | 254 (72.6 | Reference |
| From a big family (>4 members) | ||||
| Yes | 692 (53.2) | 211 (30.5) | 481 (69.5) | 0.88 (0.69–1.11) |
| No | 608 (46.8) | 202 (33.2) | 406 (66.8) | Reference |
| Smoker(s) in the household (n = 1273) | ||||
| Yes | 403 (31.7) | 275 (31.6) | 595 (68.4) | 1.03 (0.94–1.12) |
| No | 870 (68.3) | 128 (31.8) | 275 (68.2) | Reference |
| Wheeze | ||||
| Yes | 666 (51.2) | 206 (30.9) | 460 (69.1) | 0.92 (0.73–1.17) |
| No | 634 (48.8) | 207 (32.6) | 427 (67.4) | Reference |
| Cough * | ||||
| Yes | 1298 (99.8) | 412 (31.7) | 886 (68.2) | 0.47 (0.29–7.45) |
| No | 2 (0.2) | 1 (50.0) | 1 (50.0) | Reference |
| Crackle upon lung examination | ||||
| Yes | 284 (21.8) | 88 (31.0) | 196 (69.0) | 0.95 (0.72–1.27) |
| No | 1016 (78.2) | 325 (31.9) | 691 (68.0) | Reference |
| Danger signs b | ||||
| Yes | 27 (2.1) | 10 (37.0) | 17 (63.0) | 1.27 (0.59–2.75) |
| No | 1273 (97.9) | 403 (31.7) | 870 (68.3) | Reference |
| Pneumonia | ||||
| Yes | 116 (8.9) | 34 (29.3) | 82 (70.7) | 0.88 (0.58–1.33) |
| No | 1184 (91.1) | 379 (32.0) | 805 (68.0) | Reference |
| Difficulty breathing c | ||||
| Yes | 129 (9.9) | 38 (29.5) | 91 (70.5) | 0.89 (0.59–1.32) |
| No | 1171 (90.1) | 375 (32.0) | 796 (68.0) | Reference |
| Any virus detection d | ||||
| Yes | 851 (65.5) | 276 (32.4) | 575 (67.6) | 1.09 (0.85–1.39) |
| No | 449 (34.5) | 137 (30.5) | 312 (69.5) | Reference |
| Duration of hospitalization | ||||
| Range (min–max) | 0–22 days | |||
| 0–5 days | 894 (68.8) | 289 (32.3) | 605 (67.7) | Reference |
| 6–10 days | 374 (28.8) | 118 (31.6) | 256 (68.4) | 0.96 (0.74–1.25) |
| >10 days | 32 (2.4) | 6 (18.8) | 26 (81.2) | 0.48 (0.19–1.19) |
| Antimicrobials | Susceptible n (%) | Intermediate n (%) | Resistance n (%) | MIC (µg/mL) Breakpoints S-R |
|---|---|---|---|---|
| * Penicillin (parenteral non-meningitis) | 394 (98.7) | 4 (1) | 1 (0.3) | ≤2–≥8 |
| Amoxicillin (non-meningitis) | 271 (67.9) | 66 (16.5) | 62 (15.5) | ≤2–≥8 |
| Ampicillin | 42 (10.5) | 281 (70.4) | 76 (19.0) | ≤0.25–≥8 |
| Amoxicillin/Clavulanate | 290 (72.7) | 66 (16.5) | 43 (10.8) | ≤2/1–≥8/4 |
| Cefuroxime (parenteral) | 74 (18.5) | 38 (9.5) | 287 (71.9) | ≤0.5–≥2 |
| * Cefotaxime (non-meningitis) | 368 (92.2) | 26 (6.5) | 5 (1.3) | ≤1–≥4 |
| Imipenem | 232 (58.1) | 126 (31.6) | 41 (10.3) | ≤0.12–≥1 |
| Erythromycin | 55 (13.8) | 5 (1.3) | 339 (85.0) | ≤0.25–≥1 |
| Clarithromycin | 48 (12.0) | 5 (1.3) | 346 (86.7) | ≤0.25–≥1 |
| Chloramphenicol | 270 (67.7) | 0 | 129 (32.3) | ≤4–≥8 |
| Clindamycin | 84 (21.1) | 3 (0.8) | 312 (78.2) | ≤0.25–≥1 |
| Tetracycline | 74 (18.5) | 21 (5.3) | 304 (76.2) | ≤1–≥4 |
| Ciprofloxacin | 28 (7.0) | 369 (92.5) | 2 (0.5) | ≤0.25–≥4 |
| CC/Singletons | No. of Isolates (%) | No. of STs | Sequence Types (STs) | PMEN | Resistance Patterns * | Serotypes |
|---|---|---|---|---|---|---|
| CC (9) | ||||||
| 90 | 23 (8.7) | 5 | 8465, 1624, 90, 9332, 95 | Spain6B-2 | ERY + CXM + TE + C | 6A/B |
| 1106 | 6 (2.2) | 5 | 17855, 17854, 1106, 14138, 17853 | ERY + CXM + TE | 23F(1), 11A/D (1), 16F(1), NT (3) | |
| 271 | 65 (24.6) | 5 | 236, 283, 320, 4467271 | Taiwan19F-14 | ERY + CXM + TE | 19F(62), 14(1), 19A(1), 6A/B(1) |
| 81 | 29 (11) | 4 | 2395, 81, 6796, 83 | Spain23F-1 | ERY + CXM | 23F(26), 15B/C (3) |
| 880 | 15 (5.7) | 4 | 3176, 880, 17847, 9116 | ERY + CXM + TE | 23F(13), 19F(1) 11A/D(1) | |
| 17849 | 8 (3) | 4 | 17849, 17846, 1593, 15730 | ERY + CXM + TE | 19F(1), 6A/B(1) NT (6) | |
| 63 | 12 (4.5) | 4 | 17850, 2675, 63, 782 | Sweden15A-25 | ERY + TE | 14(6), 15A/F(6) |
| 13223 | 52 (19.7) | 2 | 13223, 9650 | ERY + CXM + TE + C | 6A/B | |
| 1961 | 2 (0.8) | 2 | 1961, 17848 | TE | 15B/C | |
| Singletons (17) | ||||||
| 319 | 4 (1.5) | 1 | singleton | ERY | 19A | |
| 8966 | 1 (0.4) | 1 | singleton | ERY + CXM + TE | 22F/A | |
| 17845 | 9 (3.4) | 1 | singleton | ERY + CXM + TE | 14 | |
| 17856 | 1 (0.4) | 1 | singleton | ERY + CXM + TE | 6A/B | |
| 17857 | 1 (0.4) | 1 | singleton | 0 | 15B/C | |
| 17851 | 2 (0.8) | 1 | singleton | CXM + TE | NT | |
| 4852 | 1 (0.4) | 1 | singleton | ERY + CXM + TE | 19A | |
| 166 | 7 (2.7) | 1 | singleton | ERY + CXM + TE | 11A/D | |
| 855 | 4 (1.5) | 1 | singleton | ERY + CXM | 6A/B | |
| 448 | 2 (0.8) | 1 | singleton | ERY + CXM | 14 (1), 16F (1) | |
| 4417 | 5 (1.9) | 1 | singleton | ERY + CXM + TE | 6A/B | |
| 17852 | 6 (2.3) | 1 | singleton | ERY + CXM + TE | NT(4), 6A/B(2) | |
| 1518 | 2 (0.8) | 1 | singleton | ERY + CXM | 6A/B | |
| 1545 | 1 (0.4) | 1 | singleton | 0 | 15B/C | |
| 17859 | 2 (0.8) | 1 | singleton | ERY + CXM + TE | 11A/D, NT | |
| 558 | 1 (0.4) | 1 | singleton | CXM + TE | 35B | |
| 17860 | 3 (1.1) | 1 | singleton | ERY + CXM + TE + AMP | 6A/B(1), NT(2) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wambugu, P.; Shah, M.-M.; Nguyen, H.-A.; Le, K.-A.; Le, H.-H.; Vo, H.-M.; Toizumi, M.; Bui, M.-X.; Dang, D.-A.; Yoshida, L.-M. Molecular Epidemiology of Streptococcus pneumoniae Detected in Hospitalized Pediatric Acute Respiratory Infection Cases in Central Vietnam. Pathogens 2023, 12, 943. https://doi.org/10.3390/pathogens12070943
Wambugu P, Shah M-M, Nguyen H-A, Le K-A, Le H-H, Vo H-M, Toizumi M, Bui M-X, Dang D-A, Yoshida L-M. Molecular Epidemiology of Streptococcus pneumoniae Detected in Hospitalized Pediatric Acute Respiratory Infection Cases in Central Vietnam. Pathogens. 2023; 12(7):943. https://doi.org/10.3390/pathogens12070943
Chicago/Turabian StyleWambugu, Peris, Mohammad-Monir Shah, Hien-Anh Nguyen, Kim-Anh Le, Huy-Hoang Le, Hien-Minh Vo, Michiko Toizumi, Minh-Xuan Bui, Duc-Anh Dang, and Lay-Myint Yoshida. 2023. "Molecular Epidemiology of Streptococcus pneumoniae Detected in Hospitalized Pediatric Acute Respiratory Infection Cases in Central Vietnam" Pathogens 12, no. 7: 943. https://doi.org/10.3390/pathogens12070943
APA StyleWambugu, P., Shah, M.-M., Nguyen, H.-A., Le, K.-A., Le, H.-H., Vo, H.-M., Toizumi, M., Bui, M.-X., Dang, D.-A., & Yoshida, L.-M. (2023). Molecular Epidemiology of Streptococcus pneumoniae Detected in Hospitalized Pediatric Acute Respiratory Infection Cases in Central Vietnam. Pathogens, 12(7), 943. https://doi.org/10.3390/pathogens12070943






