Antimicrobial Resistance, Virulence Properties and Genetic Diversity of Salmonella Typhimurium Recovered from Domestic and Imported Seafood
Abstract
1. Introduction
2. Materials and Methods
2.1. Salmonella Isolates
2.2. Antimicrobial Susceptibility Testing
2.3. Detection of Virulence Genes
2.4. Pulsed-Field Gel Electrophoresis of Salmonella
2.5. Statistical Analysis
3. Results
3.1. Prevalence of the Antimicrobial Resistance Phenotypes
3.2. Characterization of Salmonella Isolates Recovered from Seafood for the Presence or Absence of Virulence Genes
3.3. Molecular Characterization of Salmonella Using Pulsed-Field Gel Electrophoresis—PFGE
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Venkat, H.; Matthews, J.; Lumadao, P.; Caballero, B.; Collins, J.; Fowle, N.; Kellis, M.; Tewell, M.; White, S.; Hassan, R.; et al. Salmonella enterica serotype Javiana infections linked to a seafood restaurant in Maricopa county, Arizona, 2016. J. Food. Prot. 2018, 81, 1283–1292. [Google Scholar] [CrossRef]
- Kumar, R.; Datta, T.K.; Lalitha, K.V. Salmonella grows vigorously on seafood and expresses its virulence and stress genes at different temperature exposure. BMC Microbiol. 2015, 15, 254. [Google Scholar] [CrossRef]
- CDC. Salmonella, Salmonella Homepage. Available online: https://www.cdc.gov/salmonella/index.html (accessed on 6 February 2023).
- Iyer, A.; Kumosani, T.; Yousef, J.M.; Barbour, E.; Harakeh, S. Salmonella among humans of Asian countries. J. Life Sci. 2018, 15, 86–91. [Google Scholar]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.-A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States-major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef]
- Lin, Y.; Fong, R.; Leo, J.; Kwan, W.; Ye, A.; Chan, P.; Chong, N.; Lim, G.; Octavia, S.; Lin, M. Distribution of Salmonella spp. along the food chain in Singapore, 2008–2016. Epidemiol. News Bull. 2019, 45, 44–54. [Google Scholar]
- Ferrari, R.G.; Rosario, D.K.A.; Cunha-Neto, A.; Mano, S.B.; Figueiredo, E.E.S.; Conte-Junior, C.A. Worldwide epidemiology of Salmonella serovars in animal-based foods: A Meta-analysis. Appl. Environ. Microbiol. 2019, 85, e00591-19. [Google Scholar] [CrossRef] [PubMed]
- Hassan, R.; Tecle, S.; Adcock, B.; Kellis, M.; Weiss, J.; Saupe, A.; Sorenson, A.; Klos, R.; Blankenship, J.; Blessington, T.; et al. Multistate outbreak of Salmonella Paratyphi B variant L(+) tartrate(+) and Salmonella Weltevreden infections linked to imported frozen raw tuna: USA, March-July 2015. Epidemiol. Infect. 2018, 146, 1461–1467. [Google Scholar] [CrossRef] [PubMed]
- Heinitz, M.L.; Ruble, R.D.; Wagner, D.E.; Tatini, S.R. Incidence of Salmonella in fish and seafood. J. Food. Prot. 2000, 63, 579–592. [Google Scholar] [CrossRef] [PubMed]
- CDC. Surveillance for Foodborne Disease Outbreaks, United States, 2015, Annual Report. Available online: https://www.cdc.gov/foodsafety/pdfs/2015foodborneoutbreaks_508.pdf (accessed on 23 May 2023).
- Cabello, F.C. Heavy use of prophylactic antibiotics in aquaculture: A growing problem for human and animal health and for the environment. Environ. Microbiol. 2006, 8, 1137–1144. [Google Scholar] [CrossRef]
- Elbashir, S.; Parveen, S.; Schwarz, J.; Rippen, T.; Jahncke, M.; DePaola, A. Seafood pathogens and information on antimicrobial resistance: A review. Food. Microbiol. 2018, 70, 85–93. [Google Scholar] [CrossRef]
- CDC. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): Human Isolates Surveillance Report for 2015 (Final Report). Available online: https://www.cdc.gov/narms/pdf/2015-NARMS-Annual-Report-cleared_508.pdf (accessed on 23 May 2023).
- CDC. Antibiotic Resistance Threats in the United States. 2013. Available online: https://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf (accessed on 20 September 2022).
- Helms, M.; Vastrup, P.; Gerner-Smidt, P.; Mølbak, K. Excess mortality associated with antimicrobial drug-resistant Salmonella Typhimurium. Emerg. Infect. Dis. 2002, 8, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.H.Y.; Yan, M.; Chan, E.W.C.; Biao, K.; Chen, S. Emergence of clinical Salmonella enterica serovar Typhimurium isolates with concurrent resistance to ciprofloxacin, ceftriaxone, and azithromycin. Antimicrob. Agents Chemother. 2014, 58, 3752–3756. [Google Scholar] [CrossRef]
- Beshiru, A.; Igbinosa, I.H.; Igbinosa, E.O. Prevalence of antimicrobial resistance and virulence gene elements of Salmonella serovars from Ready-to-Eat (RTE) shrimps. Front. Microbiol. 2019, 10, 01613. [Google Scholar] [CrossRef]
- Martinez-Urtaza, J.; Liebana, E.; Garcia-Migura, L.; Perez-Piñeiro, P.; Saco, M. Characterization of Salmonella enterica serovar typhimurium from marine environments in coastal waters of Galicia (Spain). Appl. Environ. Microbiol. 2004, 70, 4030–4034. [Google Scholar] [CrossRef]
- National Marine Fisheries Service. Fisheries of the United States. 2019. Available online: https://www.fisheries.noaa.gov/national/sustainable-fisheries/fisheries-united-states (accessed on 18 October 2021).
- Gould, L.H.; Kline, J.; Monahan, C.; Vierk, K. Outbreaks of disease associated with food imported into the United States, 1996–2014. Emerg. Infect. Dis. 2017, 23, 525–528. [Google Scholar] [CrossRef] [PubMed]
- Hounmanou, Y.M.G.; Dalsgaard, A.; Sopacua, T.F.; Uddin, G.M.N.; Leekitcharoenphon, P.; Hendriksen, R.S.; Olsen, J.E.; Larsen, M.H. Molecular characteristics and zoonotic potential of Salmonella Weltevreden from cultured shrimp and tilapia in Vietnam and China. Front. Microbiol. 2020, 11, 01985. [Google Scholar] [CrossRef] [PubMed]
- Budiati, T.; Rusul, G.; Wan-Abdullah, W.N.; Chuah, L.O.; Ahmad, R.; Thong, K.L. Geneticrelatedness of Salmonella serovars isolated from catfish (Clarias gariepinus) and tilapia (Tilapia mossambica) obtained from wet markets and ponds in Penang, Malaysia. J. Food. Prot. 2016, 79, 659–665. [Google Scholar] [CrossRef]
- Friesema, I.; de Jong, A.; Hofhuis, A.; Heck, M.; van den Kerkhof, H.; de Jonge, R.; Hameryck, D.; Nagel, K.; van Vilsteren, G.; van Beek, P.; et al. Large outbreak of Salmonella Thompson related to smoked salmon in the Netherlands, August to December 2012. Euro Surveill. 2014, 19, 20918. [Google Scholar] [CrossRef]
- Chaudhary, J.H.; Nayak, J.B.; Brahmbhatt, M.N.; Makwana, P.P. Virulence genes detection of Salmonella serovars isolated from pork and slaughterhouse environment in Ahmedabad, Gujarat. Vet. World 2015, 8, 121–124. [Google Scholar] [CrossRef]
- Tekale, A.; Savalia, C.; Kshirsagar, D.; Brahmbhatt, M.; Chatur, Y. Detection and virulence gene characterization of Salmonella isolates from fish by conventional and molecular methods. J. Vet. Public Health 2015, 13, 43–46. [Google Scholar]
- Kumar, P.; Agarwal, R.; Avinash, R.; Prasannavadhana, A.; Thomas, P.; Kumar, A.; Kataria, J.; Kerketta, P. Loop mediated isothermal amplification (LAMP) test for rapid detection of Salmonella in the chicken meat. J. Vet. Public Health 2014, 12, 7–12. [Google Scholar]
- Akiyama, T.; Khan, A.A.; Cheng, C.M.; Stefanova, R. Molecular characterization of Salmonella enterica serovar Saintpaul isolated from imported seafood, pepper, environmental and clinical samples. Food. Microbiol. 2011, 28, 1124–1128. [Google Scholar] [CrossRef] [PubMed]
- Bhatta, D.R.; Bangtrakulnonth, A.; Tishyadhigama, P.; Saroj, S.D.; Bandekar, J.R.; Hendriksen, R.S.; Kapadnis, B.P. Serotyping, PCR, phage-typing and antibiotic sensitivity testing of Salmonella serovars isolated from urban drinking water supply systems of Nepal. Lett. Appl. Microbiol. 2007, 44, 588–594. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, S.D.d.; Rodenbusch, C.R.; Michael, G.B.; Cardoso, M.I.R.; Canal, C.W.; Brandelli, A. Detection of virulence genes in Salmonella Enteritidis isolated from different sources. Braz. J. Microbiol. 2003, 34, 123–124. [Google Scholar] [CrossRef]
- Bhowmick, P.P.; Srikumar, S.; Devegowda, D.; Shekar, M.; Darshanee Ruwandeepika, H.A.; Karunasagar, I. Serotyping & molecular characterization for study of genetic diversity among seafood associated nontyphoidal Salmonella serovars. Indian J. Med. Res. 2012, 135, 371–381. [Google Scholar]
- Rodriguez-Castro, A.; Ansede-Bermejo, J.; Blanco-Abad, V.; Varela-Pet, J.; Garcia-Martin, O.; Martinez-Urtaza, J. Prevalence and genetic diversity of pathogenic populations of Vibrio parahaemolyticus in coastal waters of Galicia, Spain. Environ. Microbiol. Rep. 2010, 2, 58–66. [Google Scholar] [CrossRef]
- Zhao, S.; Qaiyumi, S.; Friedman, S.; Singh, R.; Foley, S.L.; White, D.G.; McDermott, P.F.; Donkar, T.; Bolin, C.; Munro, S.; et al. Characterization of Salmonella enterica serotype Newport isolated from humans and food animals. J. Clin. Microbiol. 2003, 41, 5366–5371. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, B.; Barrett, T.J.; Hunter, S.B.; Tauxe, R.V. PulseNet: The molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 2001, 7, 382–389. [Google Scholar] [CrossRef]
- Elbashir, S.; Jahncke, M.; DePaola, A.; Bowers, J.; Schwarz, J.; Punchihewage-Don, A.J.; Min, B.; Rippen, T.; Parveen, S. Prevalence and abundance of bacterial pathogens of concern in shrimp, catfish and tilapia obtained at retail stores in Maryland, USA. Pathogens 2023, 12, 187. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Methods for Dilutions Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. In Approved Standard M7–A7, 7th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2006. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; 20th Informational Supplement (June 2010 Update); CLSI Document M100-S20-U; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2010. [Google Scholar]
- Mohamed, T.; Zhao, S.; White, D.G.; Parveen, S. Molecular characterization of antibiotic resistant Salmonella Typhimurium and Salmonella Kentucky isolated from pre- and post-chill whole broilers carcasses. Food Microbiol. 2014, 38, 6–15. [Google Scholar] [CrossRef]
- Olah, P.A.; Sherwood, J.S.; Logue, C.M. Molecular analysis of Salmonella isolates recovered from processed Turkey carcasses. J. Food Prot. 2005, 68, 845–849. [Google Scholar] [CrossRef]
- Rahn, K.; De Grandis, S.A.; Clarke, R.C.; McEwen, S.A.; Galán, J.E.; Ginocchio, C.; Curtiss, R., 3rd; Gyles, C.L. Amplification of an invA gene sequence of Salmonella Typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol. Cell. Probes. 1992, 6, 271–279. [Google Scholar] [CrossRef]
- Lampel, K.A.; Orlandi, P.A.; Kornegay, L. Improved template preparation for PCR-based assays for detection of food-borne bacterial pathogens. Appl. Environ. Microbiol. 2000, 66, 4539–4542. [Google Scholar] [CrossRef]
- Galán, J.E.; Curtiss, R., 3rd. Cloning and molecular characterization of genes whose products allow Salmonella Typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. USA 1989, 86, 6383–6387. [Google Scholar] [CrossRef] [PubMed]
- Gulig, P.A.; Danbara, H.; Guiney, D.G.; Lax, A.J.; Norel, F.; Rhen, M. Molecular analysis of spv virulence genes of the Salmonella virulence plasmids. Mol. Microbiol. 1993, 7, 825–830. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Broughton, E.I.; Walker, D.G. Prevalence of antibiotic-resistant Salmonella in fish in Guangdong, China. Foodborne Pathog. Dis. 2009, 6, 519–521. [Google Scholar] [CrossRef] [PubMed]
- Das, P.K.; Mandal, A.; Rahman, M.M.; Sarkar, S.L.; Jahid, I.K.; Hossain, M.A.; Alam, A.S.M.R.U.; Roy, P.C. Salmonella enterica serovar Typhimurium and Enteritidis isolated from raw shrimp in Bangladesh: An investigation based on molecular characteristics, survival, virulence, antibiotic resistance, and biofilm formation attributes. J. Food Qual. 2022, 2022, 3420364. [Google Scholar] [CrossRef]
- Karp, B.E.; Leeper, M.M.; Chen, J.C.; Tagg, K.A.; Francois Watkins, L.K.; Friedman, C.R. Multidrug-resistant Salmonella serotype Anatum in travelers and seafood from Asia, United States. Emerg. Infect. Dis. 2020, 26, 1030–1033. [Google Scholar] [CrossRef]
- Obaidat, M.M.; Bani Salman, A.E. Antimicrobial resistance percentages of Salmonella and Shigella in seafood imported to Jordan: Higher percentages and more diverse profiles in Shigella. J. Food. Prot. 2017, 80, 414–419. [Google Scholar] [CrossRef]
- Wang, F.; Jiang, L.; Yang, Q.; Han, F.; Chen, S.; Pu, S.; Vance, A.; Ge, B. Prevalence and antimicrobial susceptibility of major foodborne pathogens in imported seafood. J. Food Prot. 2011, 74, 1451–1461. [Google Scholar] [CrossRef]
- Zhao, S.; McDermott, P.F.; Friedman, S.; Qaiyumi, S.; Abbott, J.; Kiessling, C.; Ayers, S.; Singh, R.; Hubert, S.; Sofos, J.; et al. Characterization of antimicrobial-resistant Salmonella isolated from imported foods. J. Food Prot. 2006, 69, 500–507. [Google Scholar] [CrossRef] [PubMed]
- Braak, I.K.V. Opportunities to Improve Quality of Exports of Indian Seafood. Available online: https://www.rvo.nl/sites/default/files/2022-04/Opportunities%20to%20improve%20quality%20of%20exports%20of%20Indian%20Seafood.pdf (accessed on 8 May 2023).
- Henriksson, P.J.G.; Rico, A.; Troell, M.; Klinger, D.H.; Buschmann, A.H.; Saksida, S.; Chadag, M.V.; Zhang, W. Unpacking factors influencing antimicrobial use in global aquaculture and their implication for management: A review from a systems perspective. Sustain. Sci. 2018, 13, 1105–1120. [Google Scholar] [CrossRef]
- Guidi, L.R.; Santos, F.A.; Ribeiro, A.C.S.R.; Fernandes, C.; Silva, L.H.M.; Gloria, M.B.A. Quinolones and tetracyclines in aquaculture fish by a simple and rapid LC-MS/MS method. Food Chem. 2018, 245, 1232–1238. [Google Scholar] [CrossRef] [PubMed]
- Adesiji, Y.O.; Kogaluru Shivakumaraswamy, S.; Kumar Deekshit, V.; Shivani Kallappa, G.; Karunasagar, I. Molecular characterization of antimicrobial multi-drug resistance in non-typhoidal Salmonellae from chicken and clam in Mangalore, India. J. Biomed. Res. 2017, 32, 237–244. [Google Scholar] [CrossRef]
- Nolan, L.K.; Giddings, C.W.; Brown, J. The distribution of invA, pagC and spvC genes among Salmonella isolates from animals. Vet. Res. Commun. 1995, 19, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Soto, S.M.; Rodríguez, I.; Rodicio, M.R.; Vila, J.; Mendoza, M.C. Detection of virulence determinants in clinical strains of Salmonella enterica serovar Enteritidis and mapping on macrorestriction profiles. J. Med. Microbiol. 2006, 55, 365–373. [Google Scholar] [CrossRef]

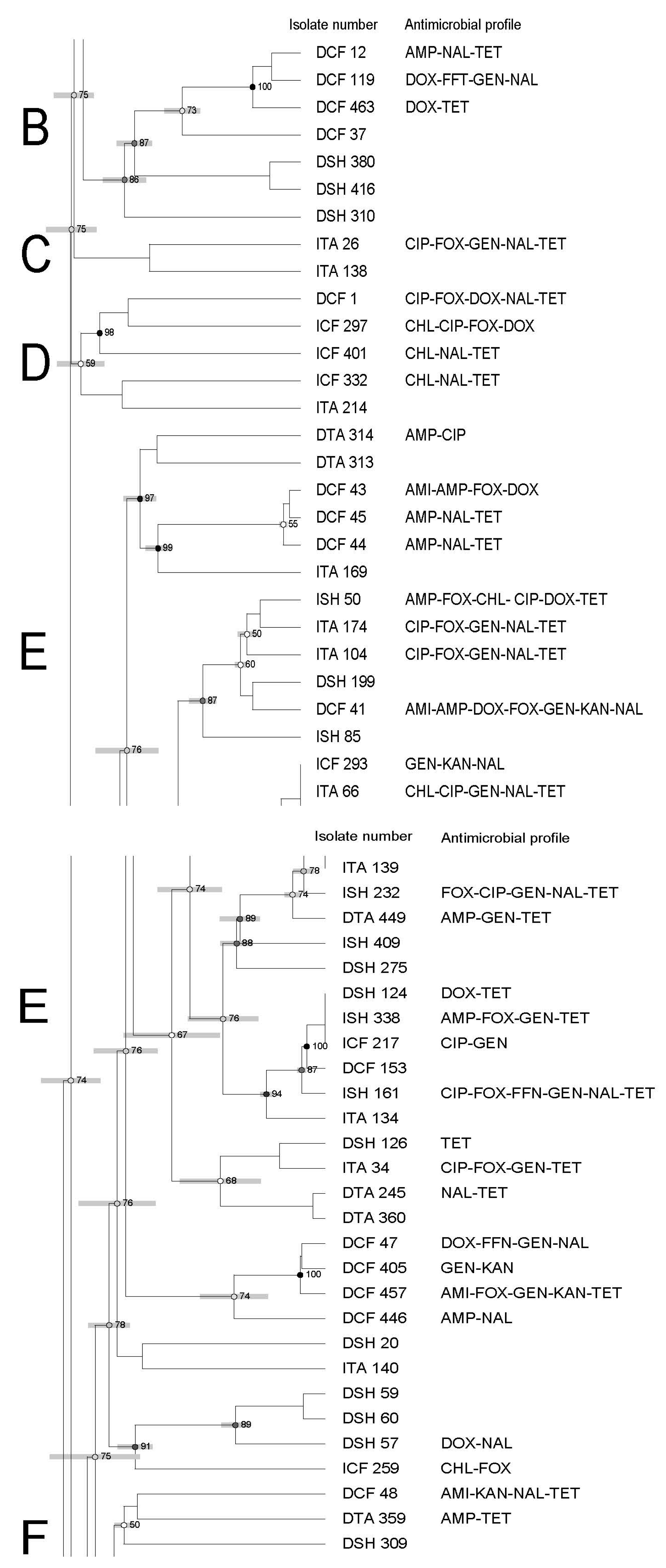


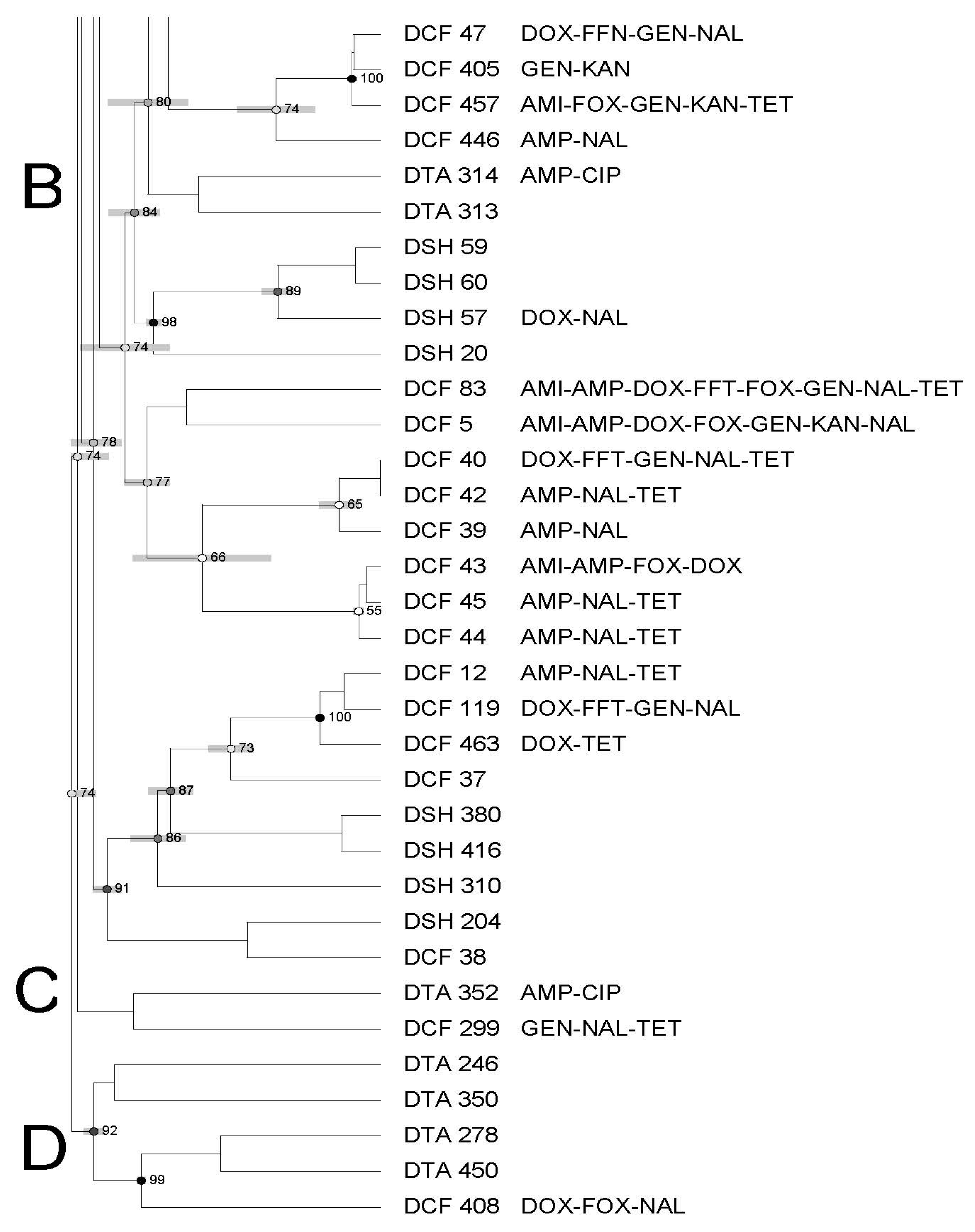

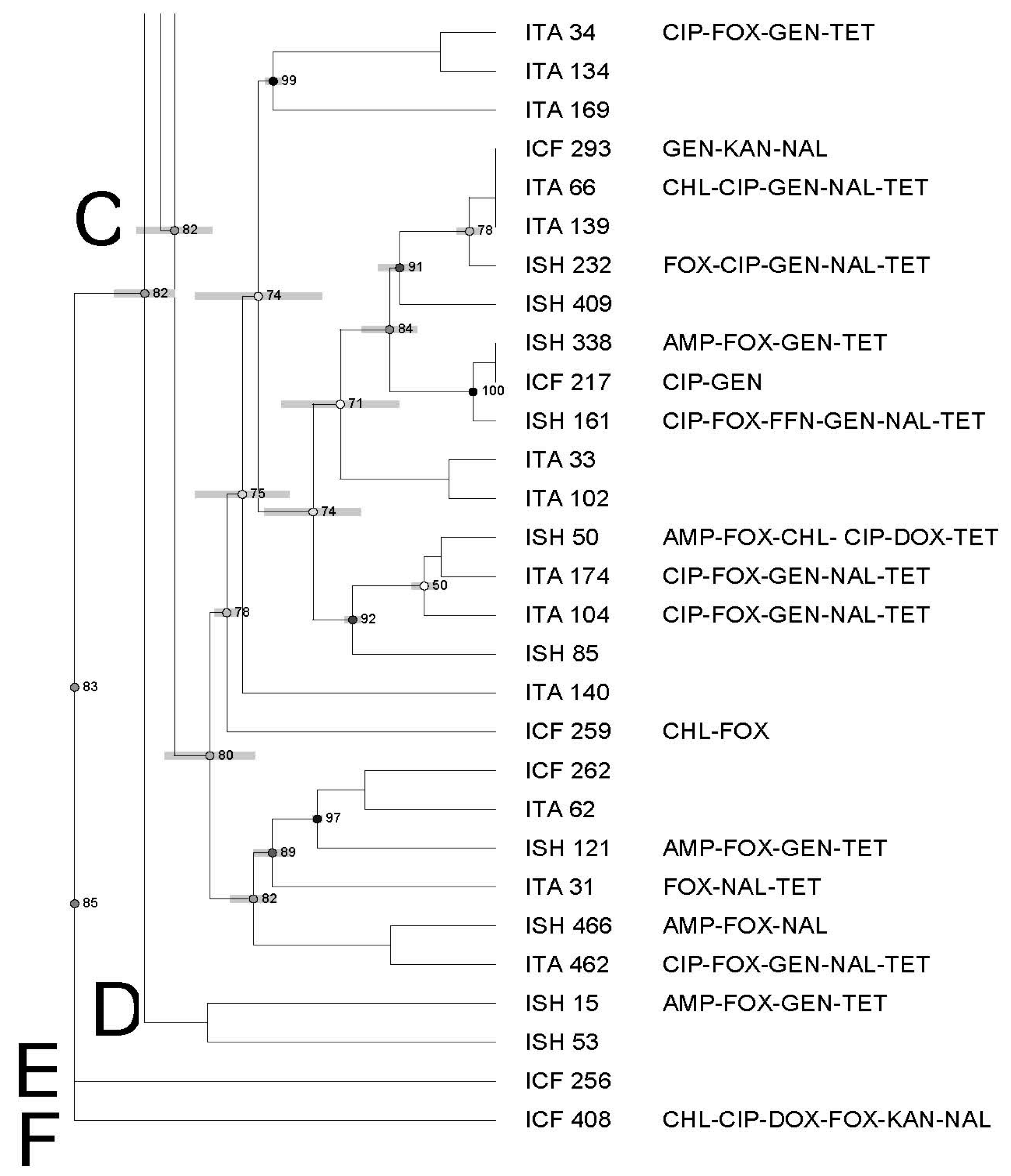
| Antimicrobial | Symbol | Concentration mg/mL | Break Points |
|---|---|---|---|
| Amikacin | AMI | 0.25–32 | ≤8–≥16 |
| Amoxicillin/Clavulanic Acid 2:1 | A.U.G2 | 8/4–32/16 | ≤8/4–≥32/16 |
| Ampicillin | AMP | 2–16 | ≤8–≥8 |
| Cefoxitin | FOX | 2–16 | ≤8–≥32 |
| Ceftiofur | XNL | 0.12–8 | ≤2–≥8 |
| Ceftriaxone | AXO | 1–8 | ≤1–≥4 |
| Chloramphenicol | CHL | 0.5–16 | ≤8–≥32 |
| Ciprofloxacin | CIP | 0.12–4 | ≤1–≥4 |
| Doxycycline | DOX | 0.5–8 | ≤4–≥16 |
| Florfenicol | FFN | 0.5–16 | ≤2–≥8 |
| Gentamicin | GEN | 1–8 | ≤4–≥16 |
| Imipenem | IMI | 1–8 | ≤2–≥8 |
| Kanamycin | KAN | 8–64 | ≤16–≥64 |
| Nalidixic Acid | NAL | 1–8 | ≤16–≥32 |
| Nitrofurantoin | NIT | 0.25–16 | ≤2–≥128 |
| Tetracycline | TET | 0.5–8 | ≤4–≥16 |
| Trimethoprim/sulfamethoxazole | SXT | 2/38–4/76 | ≤2/38–≥4/76 |
| Target | Forward Primer (5′ → 3′) | Reverse Primer (5′ → 3′) |
|---|---|---|
| invA | GTGAAATTATCGCCACGTTCGGGCAA | TCATCGCACCGTCAAAGGAACC |
| pagC | TATGAGGATCACTCTCCGGTA | ATTCTCCAGCGGATTCATCTA |
| spvC | ACTCCTTGCACAACCAAATGCGGA | TGTCTTCTGCATTTCGCCACCATCA |
| spvR | CAGGTTCCTTCAGTATCGCA | TTTGGCCGGAAATGGTCAGT |
| Gene | Initial Temp (°C) | Annealing Temp (°C) | Annealing Time (s) | Final Temp (°C) |
|---|---|---|---|---|
| invA | 94 | 55 | 72 | 72 |
| pagC | 94 | 55 | 72 | 72 |
| spvC | 94 | 59 | 72 | 72 |
| spvR | 94 | 60 | 72 | 72 |
| Antimicrobial | Domestic # (%) | Imported # (%) | ||||
|---|---|---|---|---|---|---|
| S | I | R | S | I | R | |
| Amikacin | 58(94) | 4(6) | 0(0) | 65(100) | 0(0) | 0(0) |
| Amoxicillin/Clavulanic Acid 2:1 | 62(100) | 0 | 0 | 65(100) | 0 | 0 |
| Ampicillin | 46(74) | 0 | 16(26) | 58(89) | 0 | 7(11) |
| Cefoxitin | 58(94) | 0 | 4(6) | 52(80) | 0 | 13(20) |
| /Ceftiofur | 62(100) | 0 | 0 | 65(100) | 0 | 0 |
| Ceftriaxone | 62(100) | 0 | 0 | 65(100) | 0 | 0 |
| Chloramphenicol | 62(100) | 0 | 0 | 51(78) | 0 | 14(22) |
| Ciprofloxacin | 52(84) | 5(8) | 5(8) | 44(68) | 0 | 21(32) |
| Doxycycline | 53(85) | 0 | 9(15) | 57(88) | 0 | 7(12) |
| Florfenicol | 59(95) | 0 | 3(5) | 61(94) | 0 | 4(6) |
| Gentamicin | 48(77) | 0 | 14(23) | 50(81) | 0 | 12(19) |
| Imipenem | 62(100) | 0 | 0 | 65(100) | 0 | 0 |
| Kanamycin | 58(94) | 0 | 4(6) | 64(98) | 0 | 1(2) |
| Nalidixic Acid | 48(77) | 0 | 14(23) | 40(65) | 0 | 25(35) |
| Nitrofurantoin | 62(100) | 0 | 0 | 65(100) | 0 | 0 |
| Tetracycline | 40(65) | 0 | 22(35) | 39(60) | 0 | 26(40) |
| Trimethoprim/sulfamethoxazole | 62(100) | 0 | 0 | 65(100) | 0 | 0 |
| Antimicrobial Resistance Profile | Number of Isolates | |
|---|---|---|
| Domestic (n = 62) | Imported (n = 65) | |
| AMI-AMP-FOX-DOX-FFN-GEN*-NAL*-TET | 1 | 0 |
| AMI**-AMP**-FOX**-DOX*-GEN**-KAN**-NAL-TET | 6 | 0 |
| AMP-CIP | 2 | 0 |
| AMP-FOX-CHL-CIP-DOX-FFN-GEN-NAL-TET | 0 | 2 |
| AMP-FOX-GEN**-TET | 0 | 4 |
| AMP-FOX-NAL | 0 | 2 |
| AMP-GEN-TET | 2 | 0 |
| AMP-NAL-TET | 4 | 0 |
| AMP-NAL | 1 | 0 |
| AMP-TET | 1 | 0 |
| CHL-NAL-TET | 0 | 3 |
| CIP-DOX-FFN-GEN-NAL-TET | 0 | 2 |
| CIP*-DOX*-FFN*-GEN*-TET* | 1 | 0 |
| CIP-GEN | 0 | 3 |
| DOX**-FFN**-GEN**-NAL**-TET** | 4 | 0 |
| DOX-NAL | 2 | 0 |
| DOX-TET | 4 | 0 |
| FOX-CIP-GEN-NAL-TET | 0 | 6 |
| FOX-CIP-GEN-TET | 0 | 2 |
| FOX**-CHL**-CIP**DOX-KAN**-NAL** | 0 | 5 |
| FOX-NAL-TET | 0 | 2 |
| GEN-KAN-NAL-TET | 0 | 3 |
| NAL-TET | 1 | 0 |
| TET | 1 | 3 |
| Gene | Size (bp) | % of Positive Isolates/Type and Source | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| DSH (n = 21) | ISH (n = 20) | DCF (n = 28) | ICF (n = 14) | DTA (n = 16) | ITA (n = 28) | Domestic (n = 65) | Imported (n = 62) | Total | ||
| invA | 284 | 90 | 100 | 86 | 100 | 100 | 100 | 91 | 100 | 95 |
| pagC | 318 | 52 | 95 | 46 | 43 | 69 | 89 | 54 | 81 | 67 |
| spvC | 571 | 29 | 50 | 36 | 57 | 63 | 39 | 40 | 47 | 43 |
| spvR | 310 | 29 | 50 | 36 | 57 | 63 | 39 | 40 | 47 | 43 |
| Antimicrobial | Resistance % | Remarks | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Our Study | S1 | S2 | S3 | S4 | S5 | S6 | S7 | |||
| D | I | D | D | D | D | I | I | I | ||
| AMI | 8 | 0 | N/A | N/A | + | N/A | N/A | N/A | N/A | |
| A.U.G2 | 0 | 0 | 72.7 | N/A | N/A | + | 52.2 | N/A | N/A | |
| AMP | 26 | 11 | 90.9 | 20 | N/A | + | 44.8 | T | N/A | S6/ND |
| FOX | 6 | 20 | N/A | N/A | N/A | + | N/A | T | N/A | S6/ND |
| XNL | 0 | 0 | N/A | N/A | N/A | + | N/A | N/A | N/A | |
| AXO | 0 | 0 | N/A | N/A | N/A | + | 0 | N/A | N/A | |
| CHL | 0 | 22 | 18 | 20 | N/A | + | 4.5 | T | + | S6/ND |
| CIP | 8 | 32 | 55 | 20 | + | + | 1.5 | T | + | S6/ND |
| DOX | 15 | 12 | 63.6 | N/A | N/A | N/A | N/A | N/A | N/A | |
| FFN | 5 | 6 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| GEN | 22 | 34 | 27.3 | 20 | N/A | + | 6 | T | N/A | S6/ND |
| IMI | 0 | 0 | 27 | N/A | N/A | N/A | N/A | N/A | N/A | |
| KAN | 6 | 2 | 9.1 | N/A | + | N/A | 10.4 | N/A | + | |
| NAL | 23 | 37 | N/A | 40 | N/A | N/A | 4.5 | N/A | + | |
| NIT | 0 | 0 | N/A | 20 | N/A | N/A | N/A | N/A | N/A | |
| TET | 36 | 40 | 55 | N/A | N/A | + | 9 | T | + | S6/ND |
| SXT | 0 | 0 | N/A | N/A | N/A | + | 4.5 | T | + | S6/ND |
| Others | 0 | 0 | + | + | + | + | N/A | T | + | Varies |
| Gene | % of Positive Isolates | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Our Study | S1 | S2 | S3 | S4 | S5 | S6 | S7 | S8 | S9 | |
| invA | 95 | 100 | 100 | 100 | 100 | 100 | 100 | 94.2 | N/A | 100 |
| pagC | 67 | 100 | N/A | N/A | N/A | N/A | N/A | 99 | N/A | N/A |
| spvC | 43 | N/A | N/A | 100 | ND | N/A | 90.2 | P | 97 | ND |
| spvR | 43 | N/A | N/A | N/A | 81 | N/A | 91.2 | N/A | N/A | ND |
| # of Other tested genes | N/A | 15 | 5 | N/A | 2 | 3 | N/A | N/A | 15 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elbashir, S.M.; Adnan, A.M.; Bowers, J.; DePaola, A.; Jahncke, M.; Punchihewage-Don, A.J.; Da Silva, L.V.; Hashem, F.; Parveen, S. Antimicrobial Resistance, Virulence Properties and Genetic Diversity of Salmonella Typhimurium Recovered from Domestic and Imported Seafood. Pathogens 2023, 12, 897. https://doi.org/10.3390/pathogens12070897
Elbashir SM, Adnan AM, Bowers J, DePaola A, Jahncke M, Punchihewage-Don AJ, Da Silva LV, Hashem F, Parveen S. Antimicrobial Resistance, Virulence Properties and Genetic Diversity of Salmonella Typhimurium Recovered from Domestic and Imported Seafood. Pathogens. 2023; 12(7):897. https://doi.org/10.3390/pathogens12070897
Chicago/Turabian StyleElbashir, Salah M., Adib M. Adnan, John Bowers, Angelo DePaola, Michael Jahncke, Anuradha J. Punchihewage-Don, Ligia V. Da Silva, Fawzy Hashem, and Salina Parveen. 2023. "Antimicrobial Resistance, Virulence Properties and Genetic Diversity of Salmonella Typhimurium Recovered from Domestic and Imported Seafood" Pathogens 12, no. 7: 897. https://doi.org/10.3390/pathogens12070897
APA StyleElbashir, S. M., Adnan, A. M., Bowers, J., DePaola, A., Jahncke, M., Punchihewage-Don, A. J., Da Silva, L. V., Hashem, F., & Parveen, S. (2023). Antimicrobial Resistance, Virulence Properties and Genetic Diversity of Salmonella Typhimurium Recovered from Domestic and Imported Seafood. Pathogens, 12(7), 897. https://doi.org/10.3390/pathogens12070897






