Development of a Novel Indirect ELISA for the Serological Diagnosis of African Swine Fever Using p11.5 Protein as a Target Antigen
Abstract
1. Introduction
2. Materials and Methods
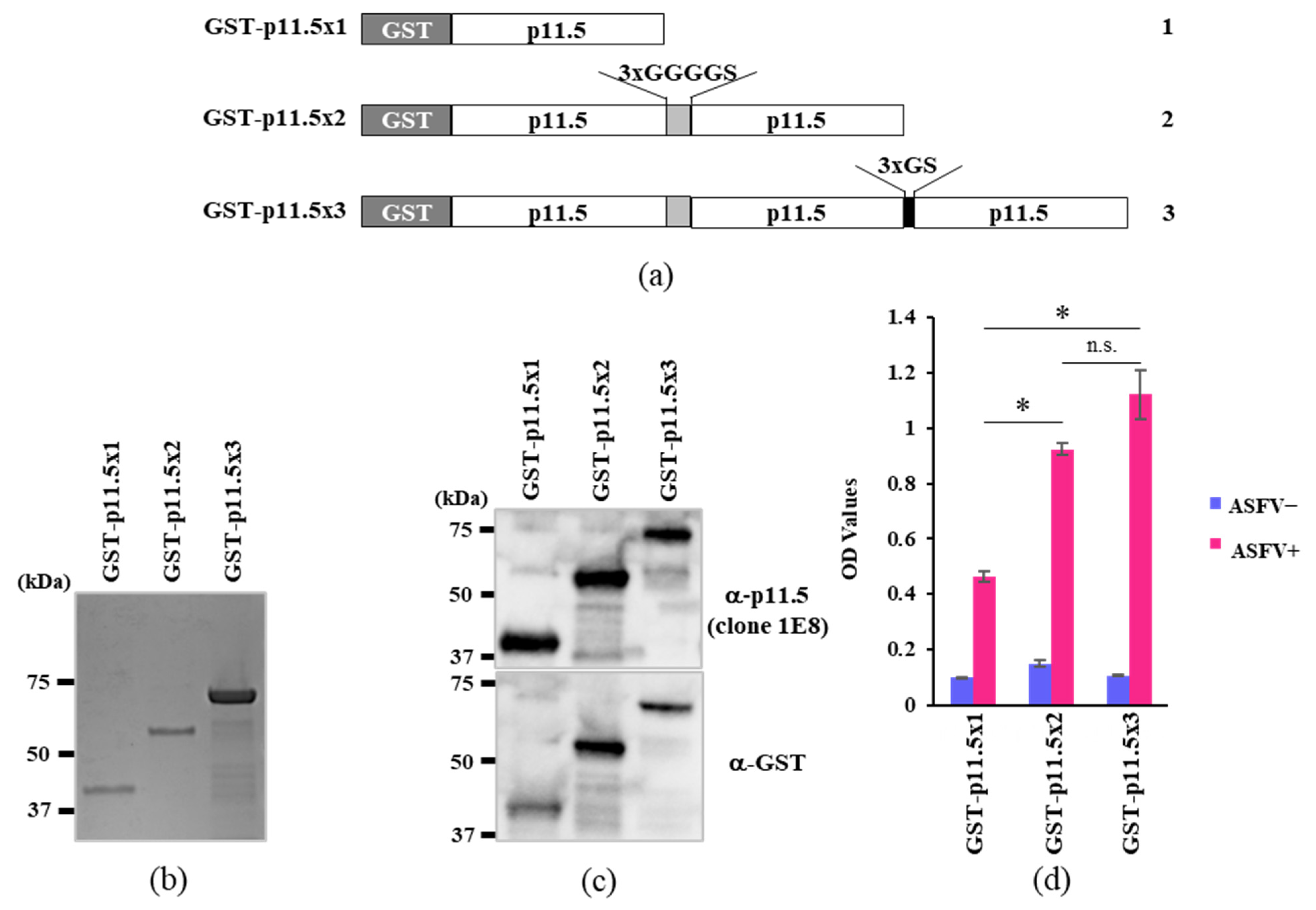
2.1. p11.5 Recombinant DNA Construction
2.2. Recombinant Protein Generation
2.3. Antibodies
2.4. Serum Samples
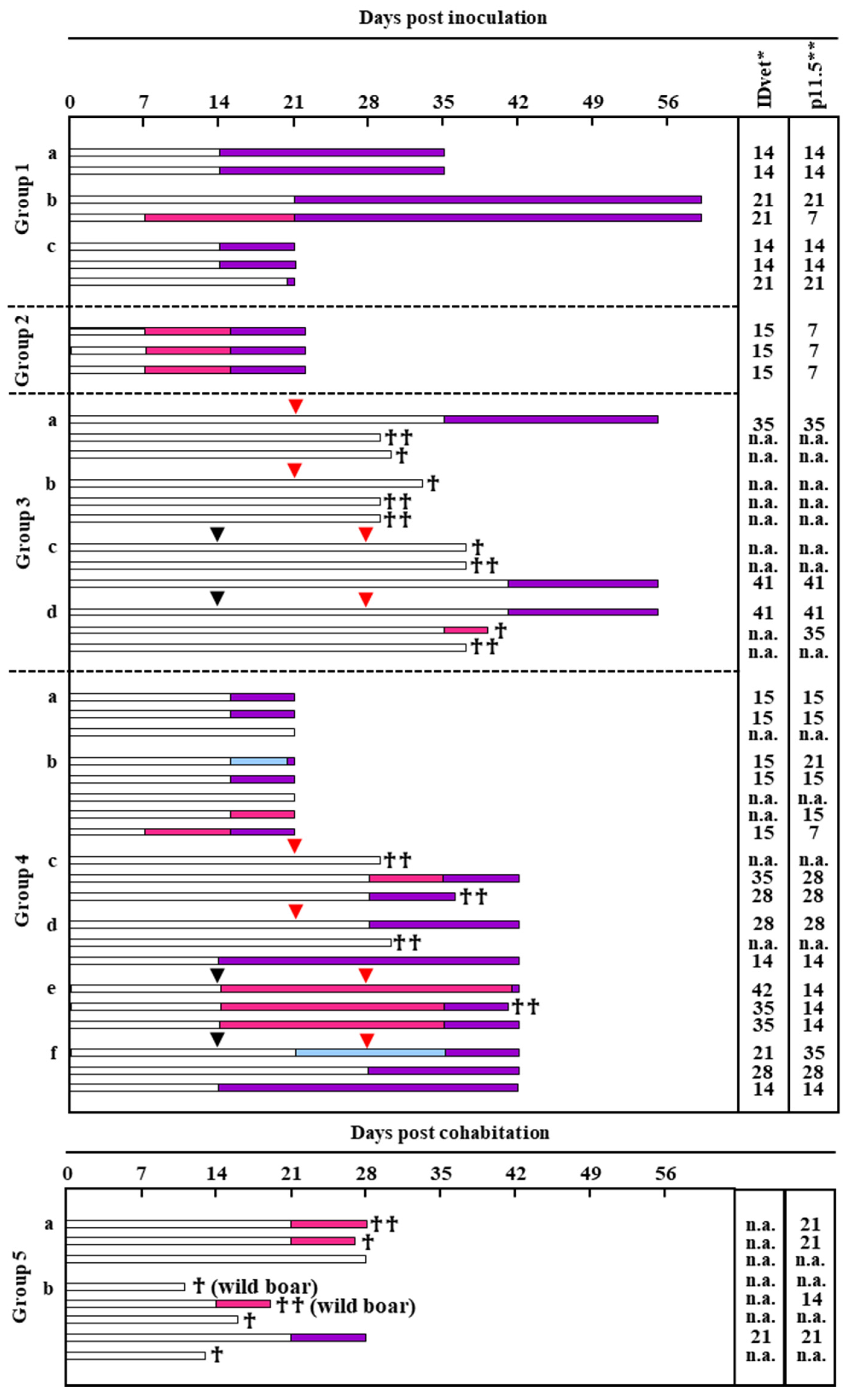
2.4.1. Animal Experiment and Virus Strains
2.4.2. Serum Samples from the Field
2.5. Immunoblotting
2.6. p11.5-Indirect ELISA
2.6.1. Procedure for p11.5-Indirect ELISA
2.6.2. Reproducibility of p11.5-Indirect ELISA
2.6.3. Comparison of p11.5-Indirect ELISA and IDvet Indirect ELISA
2.7. Statistical Analysis
3. Results
3.1. Expression of p11.5 Recombinant Proteins
3.2. Assessment of the Recombinant Proteins as an Indirect ELISA Target Antigen
3.3. Optimization of the Working Conditions of p11.5-Indirect ELISA
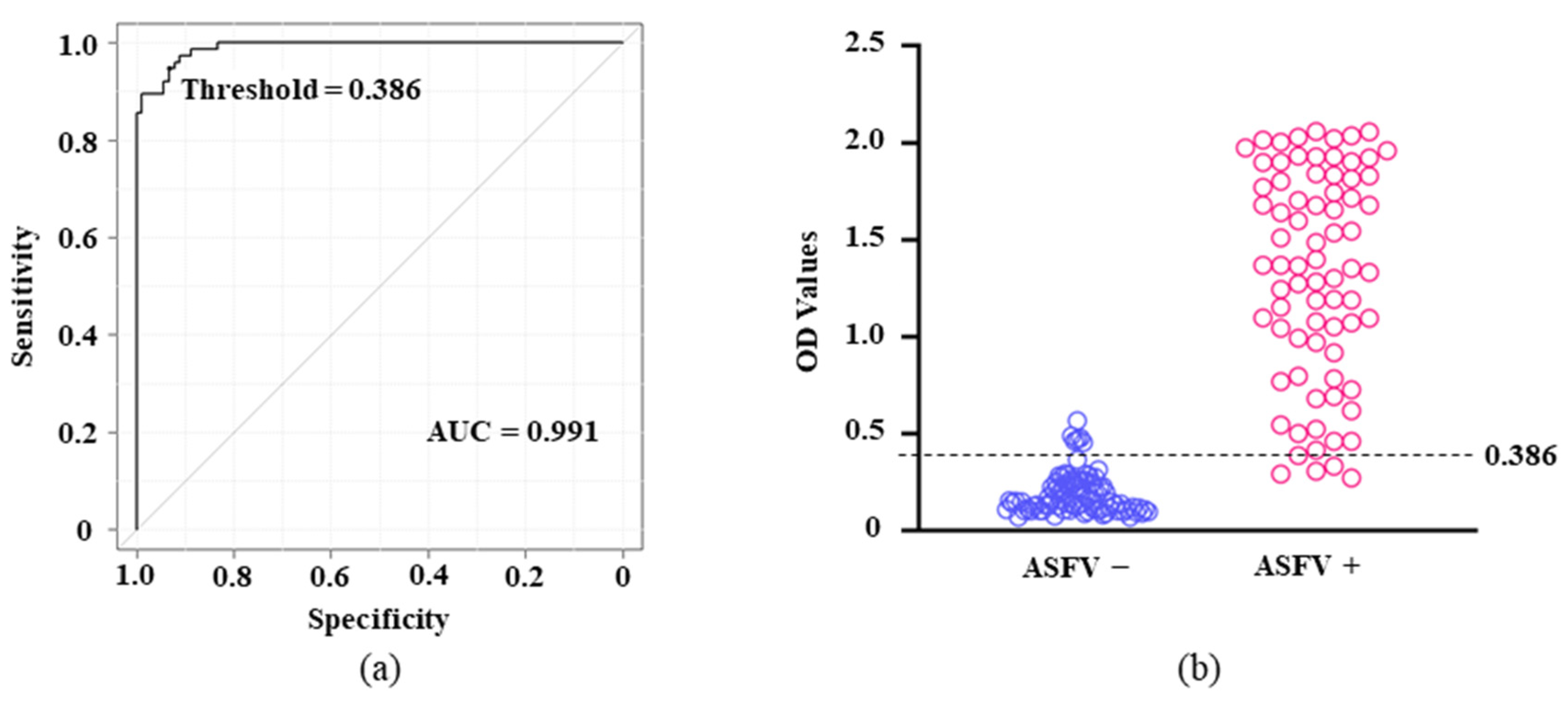
3.4. Standardization and Determination of the Cutoffs in p11.5-Indirect ELISA
3.5. Reproducibility of p11.5-Indirect ELISA
3.6. Comparison of Serological ELISAs for Early Detection of Specific Antibodies in ASFV-Infected Animals
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blome, S.; Franzke, K.; Beer, M. African Swine Fever—A Review of Current Knowledge. Virus Res. 2020, 287, 198099. [Google Scholar] [CrossRef] [PubMed]
- Galindo, I.; Alonso, C. African Swine Fever Virus: A Review. Viruses 2017, 9, 103. [Google Scholar] [CrossRef] [PubMed]
- Eustace Montgomery, R. On a Form of Swine Fever Occurring in British East Africa (Kenya Colony). J. Comp. Pathol. Ther. 1921, 34, 159–191. [Google Scholar] [CrossRef]
- Sánchez-Cordón, P.J.; Montoya, M.; Reis, A.L.; Dixon, L.K. African Swine Fever: A Re-Emerging Viral Disease Threatening the Global Pig Industry. Vet. J. 2018, 233, 41–48. [Google Scholar] [CrossRef]
- Gogin, A.; Gerasimov, V.; Malogolovkin, A.; Kolbasov, D. African Swine Fever in the North Caucasus Region and the Russian Federation in Years 2007–2012. Virus Res 2013, 173, 198–203. [Google Scholar] [CrossRef]
- Urbano, A.C.; Ferreira, F. African Swine Fever Control and Prevention: An Update on Vaccine Development. Emerg. Microbes. Infect. 2022, 11, 2021–2033. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhao, S.; Zhang, H.; Qin, Z.; Shan, H.; Cai, X. Vaccines for African Swine Fever: An Update. Front. Microbiol. 2023, 14, 1139494. [Google Scholar] [CrossRef] [PubMed]
- World Organisation for Animal Health. Chapter 15.1. Infection with African Swine Fever Virus; World Organisation for Animal Health: Paris, France, 2022. [Google Scholar]
- Blome, S.; Gabriel, C.; Beer, M. Pathogenesis of African Swine Fever in Domestic Pigs and European Wild Boar. Virus Res. 2013, 173, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Salguero, F.J. Comparative Pathology and Pathogenesis of African Swine Fever Infection in Swine. Front. Vet. Sci. 2020, 7, 282. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Cordón, P.J.; Nunez, A.; Neimanis, A.; Wikström-Lassa, E.; Montoya, M.; Crooke, H.; Gavier-Widén, D. African Swine Fever: Disease Dynamics in Wild Boar Experimentally Infected with ASFV Isolates Belonging to Genotype I and II. Viruses 2019, 11, 852. [Google Scholar] [CrossRef] [PubMed]
- Kameyama, K.; Kitamura, T.; Okadera, K.; Ikezawa, M.; Masujin, K.; Kokuho, T. Usability of Immortalized Porcine Kidney Macrophage Cultures for the Isolation of ASFV without Affecting Virulence. Viruses 2022, 14, 1794. [Google Scholar] [CrossRef] [PubMed]
- Kanazawa, I.; Karaki, H.; Washitani, I.; Hirohashi, S.; Asashima, M.; Osumi, N.; Taniguchi, M.; Nomoto, A.; Miyashita, Y.; Yano, H.; et al. Guidelines for Proper Conduct of Animal Experiments; Science Council of Japan: Tokyo, Japan, 2006. [Google Scholar]
- Kilkenny, C.; Browne, W.J.; Cuthill, I.C.; Emerson, M.; Altman, D.G. Improving Bioscience Research Reporting: The ARRIVE Guidelines for Reporting Animal Research. Osteoarthr. Cartil. 2012, 20, 256–260. [Google Scholar] [CrossRef] [PubMed]
- King, D.P.; Reid, S.M.; Hutchings, G.H.; Grierson, S.S.; Wilkinson, P.J.; Dixon, L.K.; Bastos, A.D.S.; Drew, T.W. Development of a TaqMan® PCR Assay with Internal Amplification Control for the Detection of African Swine Fever Virus. J. Virol. Methods 2003, 107, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Nishi, T.; Okadera, K.; Fukai, K.; Yoshizaki, M.; Nakasuji, A.; Yoneyama, S.; Kokuho, T. Establishment of a Direct PCR Assay for Simultaneous Differential Diagnosis of African Swine Fever and Classical Swine Fever Using Crude Tissue Samples. Viruses 2022, 14, 498. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, T.; Masujin, K.; Yamazoe, R.; Kameyama, K.; Watanabe, M.; Ikezawa, M.; Yamada, M.; Kokuho, T. A Spontaneously Occurring African Swine Fever Virus with 11 Gene Deletions Partially Protects Pigs Challenged with the Parental Strain. Viruses 2023, 15, 311. [Google Scholar] [CrossRef] [PubMed]
- Boinas, F.S.; Hutchings, G.H.; Dixon, L.K.; Wilkinson, P.J. Characterization of Pathogenic and Non-Pathogenic African Swine Fever Virus Isolates from Ornithodoros erraticus Inhabiting Pig Premises in Portugal. J. Gen. Virol. 2004, 85, 2177–2187. [Google Scholar] [CrossRef] [PubMed]
- Masujin, K.; Kitamura, T.; Kameyama, K.I.; Okadera, K.; Nishi, T.; Takenouchi, T.; Kitani, H.; Kokuho, T. An Immortalized Porcine Macrophage Cell Line Competent for the Isolation of African Swine Fever Virus. Sci. Rep. 2021, 11, 4759. [Google Scholar] [CrossRef] [PubMed]
- Kanda, Y. Investigation of the Freely Available Easy-to-Use Software ‘EZR’ for Medical Statistics. Bone Marrow Transpl. 2013, 48, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Sun, Y.; Huang, S.; Qiu, H.-J. Multifaceted Immune Responses to African Swine Fever Virus: Implications for Vaccine Development. Vet. Microbiol. 2020, 249, 108832. [Google Scholar] [CrossRef] [PubMed]
- Gladue, D.P.; Ramirez-Medina, E.; Vuono, E.; Silva, E.; Rai, A.; Pruitt, S.; Espinoza, N.; Velazquez-Salinas, L.; Borca, M.V. Deletion of the A137R Gene from the Pandemic Strain of African Swine Fever Virus Attenuates the Strain and Offers Protection against the Virulent Pandemic Virus. J. Virol. 2021, 95, e01139-21. [Google Scholar] [CrossRef] [PubMed]
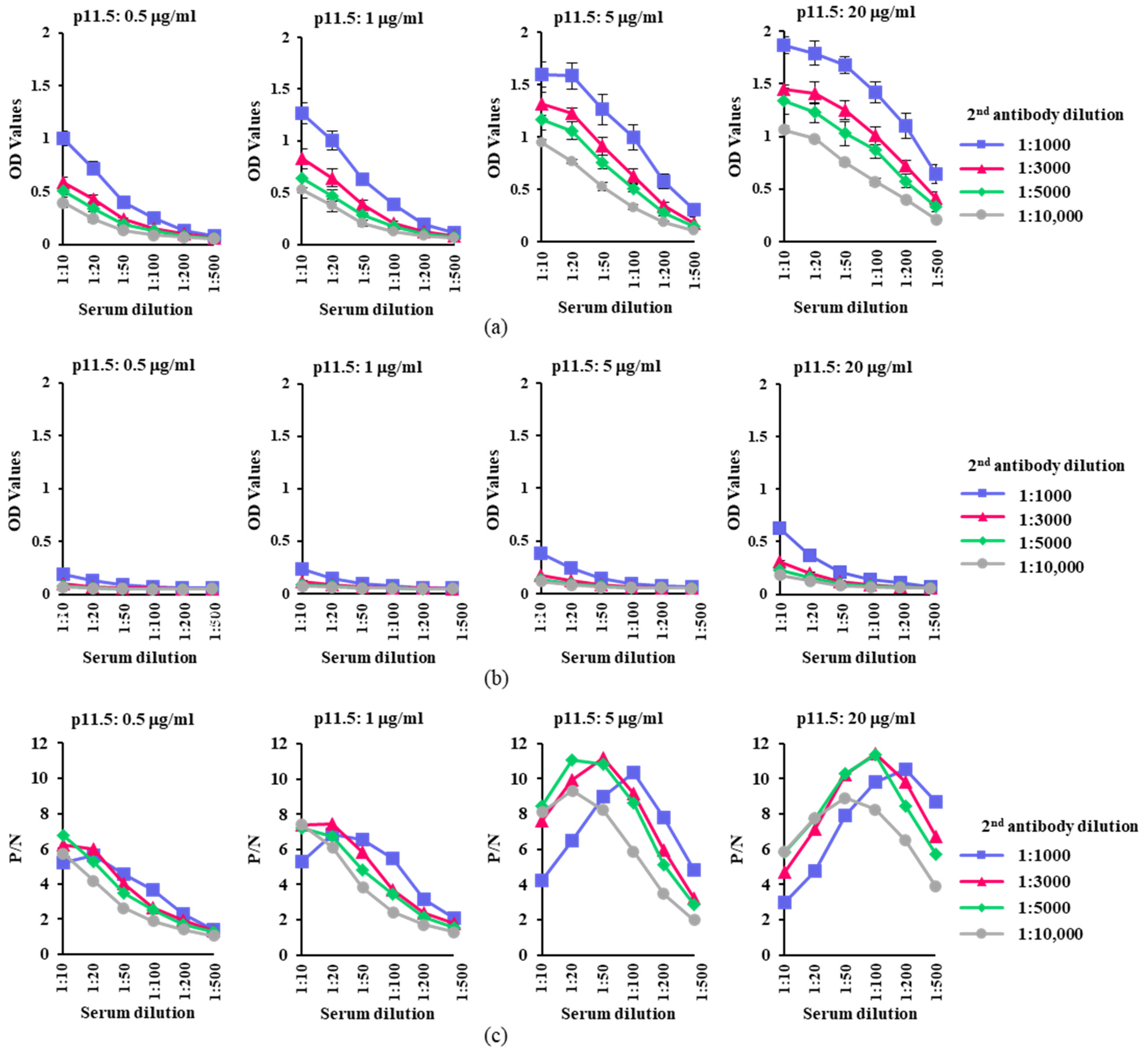
| Antigen Concentration | Serum Dilution | ||||||
|---|---|---|---|---|---|---|---|
| 1:10 | 1:20 | 1:50 | 1:100 | 1:200 | 1:500 | ||
| 20 μg/mL | P | 1.336 | 1.226 | 1.028 | 0.876 | 0.577 | 0.331 |
| N | 0.230 | 0.159 | 0.100 | 0.077 | 0.068 | 0.058 | |
| P/N | 5.81 | 7.73 | 10.31 | 11.38 | 8.44 | 5.74 | |
| 5 μg/mL | P | 1.171 | 1.062 | 0.761 | 0.510 | 0.277 | 0.152 |
| N | 0.139 | 0.096 | 0.070 | 0.059 | 0.054 | 0.053 | |
| P/N | 8.42 | 11.06 | 10.82 | 8.60 | 5.10 | 2.88 | |
| 1 μg/mL | P | 0.637 | 0.470 | 0.285 | 0.177 | 0.107 | 0.071 |
| N | 0.089 | 0.070 | 0.059 | 0.051 | 0.049 | 0.046 | |
| P/N | 7.19 | 6.75 | 4.85 | 3.48 | 2.17 | 1.54 | |
| 0.5 μg/mL | P | 0.506 | 0.341 | 0.188 | 0.128 | 0.081 | 0.059 |
| N | 0.075 | 0.064 | 0.054 | 0.050 | 0.048 | 0.047 | |
| P/N | 6.75 | 5.30 | 3.48 | 2.54 | 1.71 | 1.26 | |
| p11.5-Indirect ELISA | IDvet-Indirect ELISA | ||
|---|---|---|---|
| + | − | Total | |
| + | 71 | 5 | 76 |
| − | 5 | 85 | 90 |
| Total | 76 | 90 | 166 |
| Intra-Assay | Inter-Assay | ||||||
|---|---|---|---|---|---|---|---|
| Samples | Mean OD Value | SD | CV% | Mean OD Value | SD | CV% | |
| Negative | No.1 | 0.141 | 0.009 | 6.39 | 0.142 | 0.003 | 2.01 |
| No.2 | 0.107 | 0.002 | 1.58 | 0.103 | 0.009 | 8.7 | |
| No.3 | 0.109 | 0.003 | 2.64 | 0.120 | 0.011 | 8.79 | |
| No.4 | 0.084 | 0 | 0 | 0.098 | 0.002 | 2.09 | |
| No.5 | 0.087 | 0.002 | 2.48 | 0.092 | 0.004 | 4.57 | |
| Positive | Group 1 | 1.440 | 0.045 | 3.09 | 1.440 | 0.028 | 1.95 |
| Group 2 | 1.390 | 0.068 | 4.86 | 1.630 | 0.09 | 5.54 | |
| Group 3 | 0.919 | 0.081 | 8.81 | 0.908 | 0.024 | 2.59 | |
| Group 4 | 1.430 | 0.073 | 5.06 | 1.460 | 0.081 | 5.55 | |
| Group 5 | 0.665 | 0.014 | 2.09 | 0.780 | 0.068 | 8.75 | |
| Genotype I | Genotype II | Genotype X | |||
|---|---|---|---|---|---|
| Group 1 | Group 2 | Group 3 | Group 4 | Group 5 | |
| IDvet-indirect ELISA | 7 (17) | 3 (15) | 3 (39) | 15 (23.7) | 1 (21) |
| p11.5-indirect ELISA | 7 (15) | 3 (7) | 4 (38) | 16 (19.1) | 3 (21), 1 * (14) |
| Number of pigs used in the experiment. | 7 | 3 | 12 | 20 | 6, 2 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Watanabe, M.; Kitamura, T.; Nagata, K.; Ikezawa, M.; Kameyama, K.-i.; Masujin, K.; Kokuho, T. Development of a Novel Indirect ELISA for the Serological Diagnosis of African Swine Fever Using p11.5 Protein as a Target Antigen. Pathogens 2023, 12, 774. https://doi.org/10.3390/pathogens12060774
Watanabe M, Kitamura T, Nagata K, Ikezawa M, Kameyama K-i, Masujin K, Kokuho T. Development of a Novel Indirect ELISA for the Serological Diagnosis of African Swine Fever Using p11.5 Protein as a Target Antigen. Pathogens. 2023; 12(6):774. https://doi.org/10.3390/pathogens12060774
Chicago/Turabian StyleWatanabe, Mizuki, Tomoya Kitamura, Koji Nagata, Mitsutaka Ikezawa, Ken-ichiro Kameyama, Kentaro Masujin, and Takehiro Kokuho. 2023. "Development of a Novel Indirect ELISA for the Serological Diagnosis of African Swine Fever Using p11.5 Protein as a Target Antigen" Pathogens 12, no. 6: 774. https://doi.org/10.3390/pathogens12060774
APA StyleWatanabe, M., Kitamura, T., Nagata, K., Ikezawa, M., Kameyama, K.-i., Masujin, K., & Kokuho, T. (2023). Development of a Novel Indirect ELISA for the Serological Diagnosis of African Swine Fever Using p11.5 Protein as a Target Antigen. Pathogens, 12(6), 774. https://doi.org/10.3390/pathogens12060774








