Isolation and Identification of Multidrug-Resistant Klebsiella pneumoniae Clones from the Hospital Environment
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Identification of K. pneumoniae Clones
2.2. K. pneumoniae Strains Antibiotic Resistance Test
2.3. Confirmatory Extended-Spectrum β-lactamases (ESBL)
2.4. Molecular Characterization
2.5. Phylogenetic Analysis
3. Results
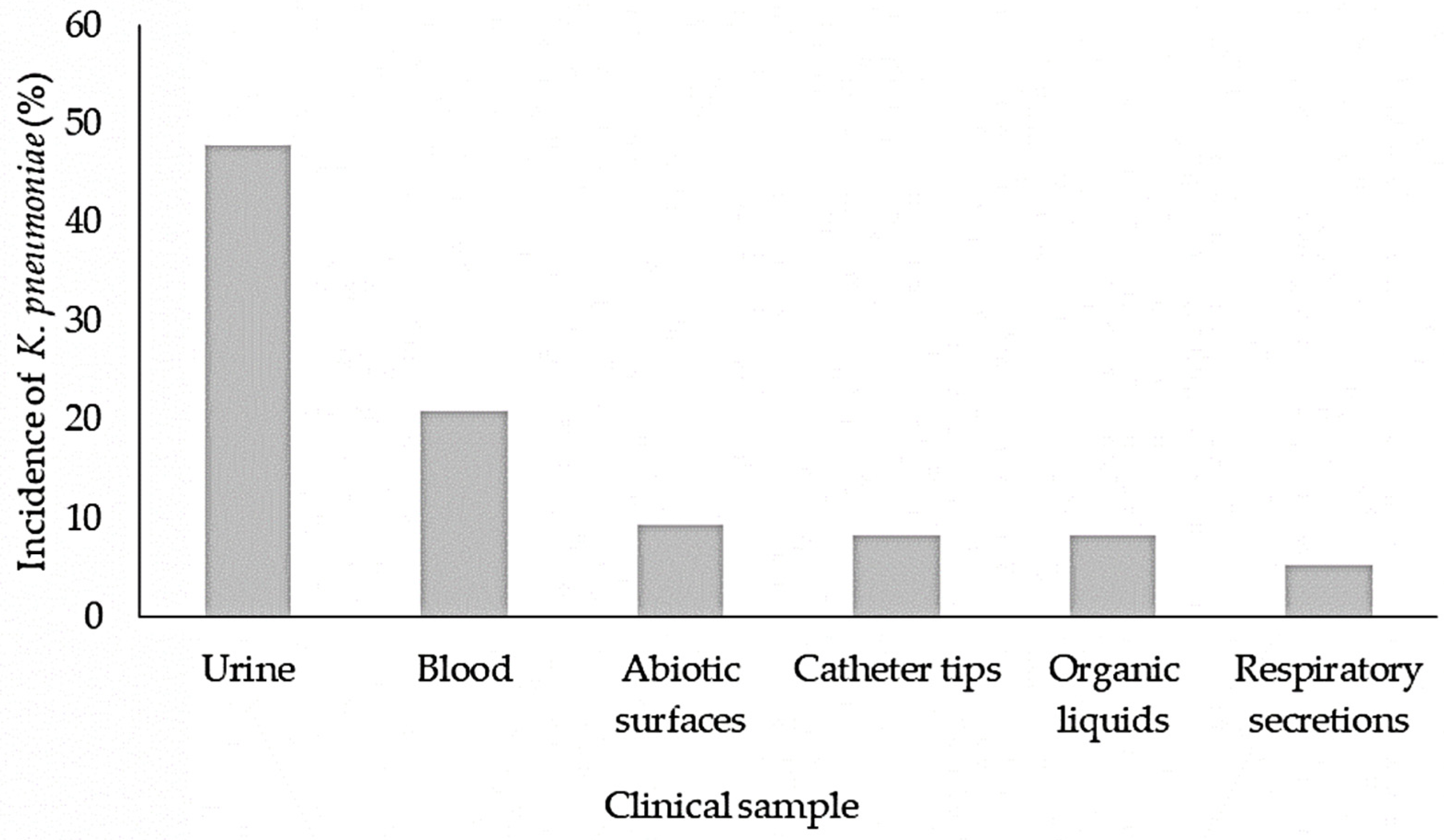
3.1. K. pneumoniae Strains Identification and Susceptibility Test
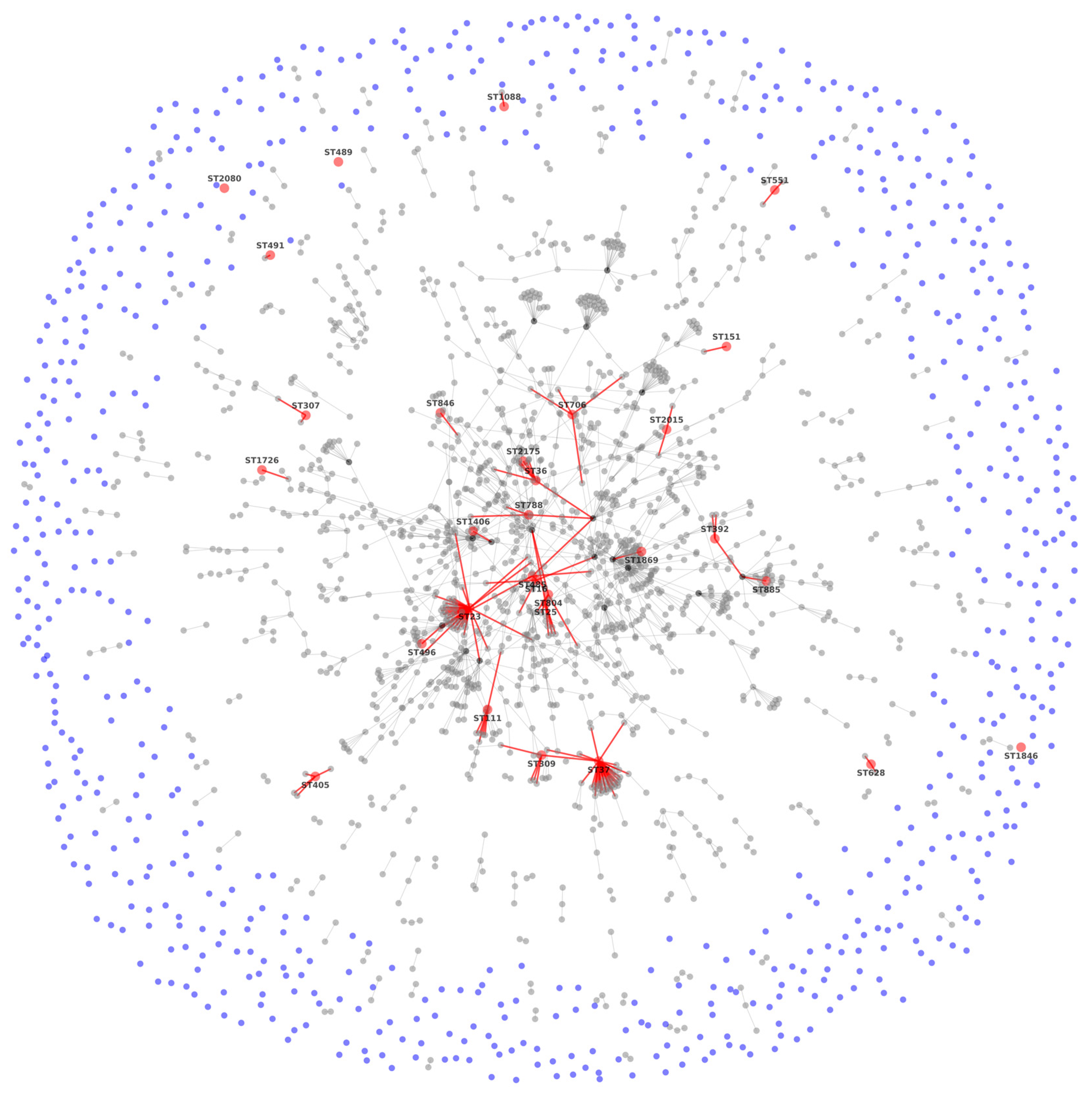
3.2. MLST Results
3.3. Sequence Analysis
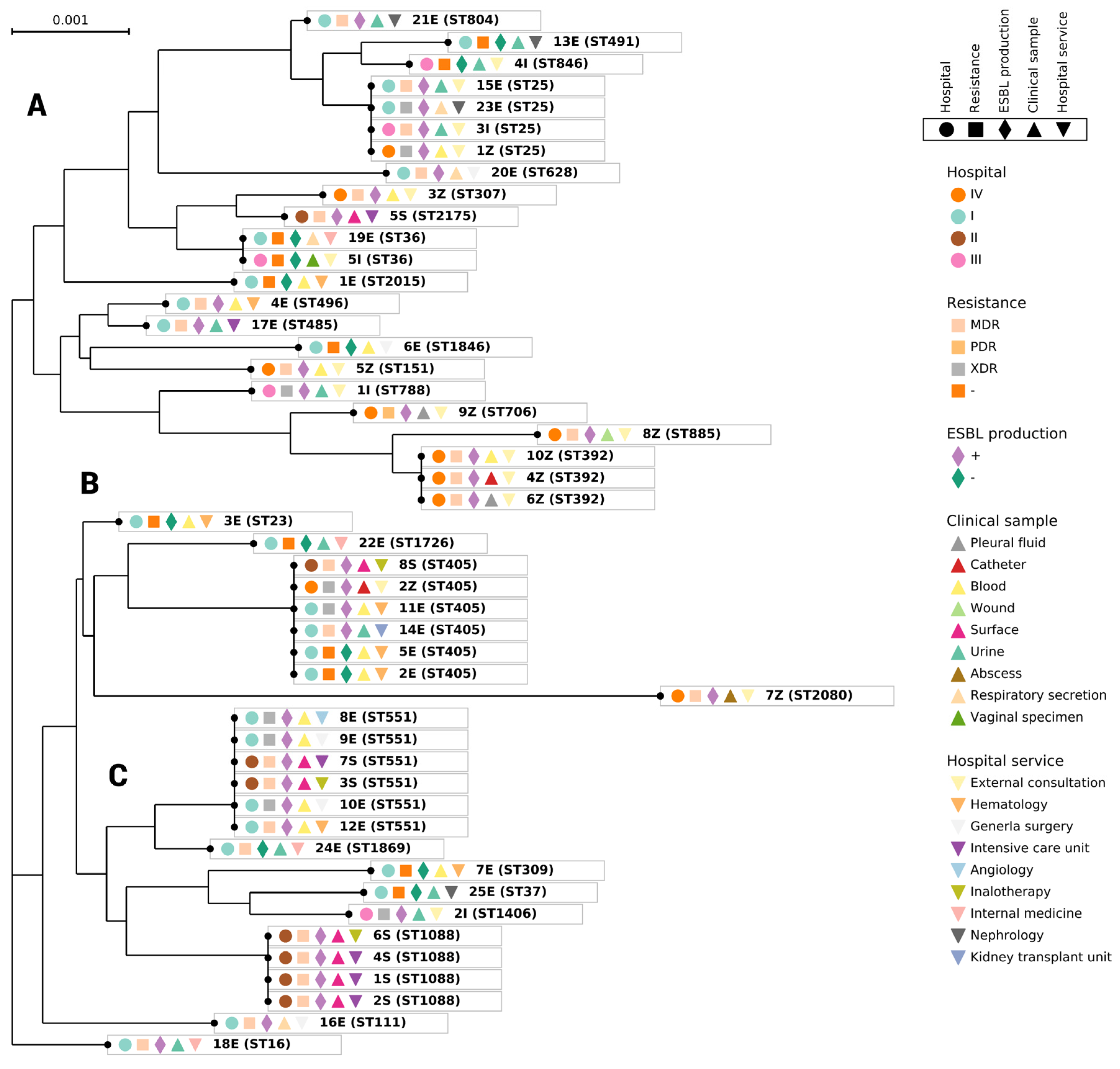
3.4. Phylogenetic Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Miranda-Novales, M.G.; Flores-Moreno, K.; López-Vidal, Y.; Rodríguez-Álvarez, M.; Solórzano-Santos, F.; Hernandez, J.L.S.; De León-Rosales, S.P.; Network, U. Antimicrobial resistance and antibiotic consumption in Mexican hospitals. Salud Pública de México 2019, 62, 42–49. [Google Scholar] [CrossRef]
- López Vargas, J.A.; Echeverri Toro, L.M. K. pneumoniae: ¿la nueva “superbacteria”? Patogenicidad, epidemiología y mecanismos de resistencia. Iatreia 2010, 23, 157–165. [Google Scholar] [CrossRef]
- Bialek-Davenet, S.; Criscuolo, A.; Ailloud, F.; Passet, V.; Jones, L.; Delannoy-Vieillard, A.-S.; Garin, B.; Le Hello, S.; Arlet, G.; Nicolas-Chanoine, M.-H.; et al. Genomic Definition of Hypervirulent and Multidrug-Resistant Klebsiella pneumoniae clonal groups. Emerg. Infect. Dis. 2014, 20, 1812–1820. [Google Scholar] [CrossRef] [PubMed]
- Garza-González, E.; Morfín-Otero, R.; Mendoza-Olazarán, S.; Bocanegra-Ibarias, P.; Flores-Treviño, S.; Rodríguez-Noriega, E.; Ponce-De-León, A.; Sanchez-Francia, D.; Franco-Cendejas, R.; Arroyo-Escalante, S.; et al. A snapshot of antimicrobial resistance in Mexico. Results from 47 centers from 20 states during a six-month period. PLoS ONE 2019, 14, e0209865. [Google Scholar] [CrossRef] [PubMed]
- Vera-Leiva, A.; Barría-Loaiza, C.; Carrasco-Anabalón, S.; Lima, C.; Aguayo-Reyes, A.; Domínguez, M.; Bello-Toledo, H.; González-Rocha, G. KPC: Klebsiella pneumoniae carbapenemasa, principal carbapenemasa en enterobacterias. Rev. Chilena Infectol. 2017, 34, 476–484. [Google Scholar] [CrossRef]
- Wang, Q.; Li, B.; Tsang, A.K.L.; Yi, Y.; Woo, P.C.Y.; Liu, C.H. Genotypic Analysis of Klebsiella pneumoniae Isolates in a Beijing Hospital Reveals High Genetic Diversity and Clonal Population Structure of Drug-Resistant Isolates. PLoS ONE 2013, 8, e57091. [Google Scholar] [CrossRef]
- Gomez-Simmonds, A.; Uhlemann, A.-C. Clinical Implications of Genomic Adaptation and Evolution of Carbapenem-Resistant Klebsiella pneumoniae. J. Infect. Dis. 2017, 215, S18–S27. [Google Scholar] [CrossRef]
- Gijón, D.; Tedim, A.P.; Valverde, A.; Rodríguez, I.; Morosini, M.-I.; Coque, T.M.; Manrique, M.; Pareja, E.; Tobes, R.; Ruiz-Garbajosa, P.; et al. Early OXA-48-Producing Enterobacterales Isolates Recovered in a Spanish Hospital Reveal a Complex Introduction Dominated by Sequence Type 11 (ST11) and ST405 Klebsiella pneumoniae Clones. Msphere 2020, 5, e00080-20. [Google Scholar] [CrossRef]
- Koneman, E.W.; Allen, S.; Janda, W.M.; Schereckenberger, P.C.; Win Jr, W.C. Microbiological diagnosis: Text and Color Atlas, 6th ed.; Editorial Médica Panamericana: Buenos Aires, Argentina, 2008. [Google Scholar]
- Cerezo, S.G.; Gutierrez, A.E.; Gomez, J.L.V. Diagnostico de laboratorio de infecciones gastrointestinales. Secretaria de Salud: Madrid, Spain, 1994. [Google Scholar]
- Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement. CLSI Document M100-S24. 2014, 34.
- Cao, M.D.; Ganesamoorthy, D.; Elliott, A.G.; Zhang, H.; Cooper, M.A.; Coin, L.J. Streaming algorithms for identification of pathogens and antibiotic resistance potential from real-time MinIONTM sequencing. Gigascience 2016, 5, 32. [Google Scholar] [CrossRef]
- Diancourt, L.; Passet, V.; Verhoef, J.; Grimont, P.A.D.; Brisse, S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 2005, 43, 4178–4182. [Google Scholar] [CrossRef] [PubMed]
- Feil, E.J.; Li, B.C.; Aanensen, D.M.; Hanage, W.P.; Spratt, B.G. eBURST: Inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 2004, 186, 1518–1530. [Google Scholar] [CrossRef]
- Jolley, K.A.; Feil, E.J.; Chan, M.-S.; Maiden, M.C.J. Sequence type analysis and recombinational tests (START). Bioinformatics 2001, 17, 1230–1231. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Holten, D.D.; Van Wijk, J.J. Force-directed edge bundling for graph visualization. Comput. Graph. Forum 2009, 28, 983–990. [Google Scholar] [CrossRef]
- Lemoine, F.; Correia, D.; Lefort, V.; Doppelt-Azeroual, O.; Mareuil, F.; Cohen-Boulakia, S.; Gascuel, O. NGPhylogeny.fr: New generation phylogenetic services for non-specialists. Nucleic Acids Res. 2019, 47, W260–W265. [Google Scholar] [CrossRef] [PubMed]
- Kazutaka, K.; Misakwa, K.; Kei-ichi, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Criscuolo, A.; Gribaldo, S. BMGE (Block Mapping and Gathering with Entropy): A new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 2010, 10, 210. [Google Scholar] [CrossRef]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A Comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef]
- Azam, M.; Gaind, R.; Yadav, G.; Sharma, A.; Upmanyu, K.; Jain, M.; Singh, R. Colistin resistance among multiple sequence types of Klebsiella pneumoniae is associated with diverse resistance mechanisms: A report from India. Front. Microbiol. 2021, 12, 609840. [Google Scholar] [CrossRef] [PubMed]
- Eger, E.; Schwabe, M.; Schulig, L.; Hübner, N.-O.; Bohnert, J.A.; Bornscheuer, U.T.; Heiden, S.E.; Müller, J.U.; Adnan, F.; Becker, K.; et al. Extensively drug-resistant Klebsiella pneumoniae counteracts fitness and virulence costs that accompanied ceftazidime-avibactam resistance acquisition. Microbiol. Spectr. 2022, 10, e00148-22. [Google Scholar] [CrossRef] [PubMed]
- Fumagalli, O.; Tall, B.D.; Schipper, C.; A Oelschlaeger, T. N-glycosylated proteins are involved in efficient internalization of Klebsiella pneumoniae by cultured human epithelial cells. Infect. Immun. 1997, 65, 4445–4451. [Google Scholar] [CrossRef] [PubMed]
- Secretaria de Salud, G.d.M. Boletín Infecciones Asociadas a la Atención de la Salud (IAAS) Red Hospitalaria de Vigilancia Epidemiológica (RHOVE). 2022, 23. Available online: https://www.gob.mx/cms/uploads/attachment/file/773554/BOLET_N_RHOVE_SEPTIEMBRE_2022_Final_31102022.pdf (accessed on 15 June 2022).
- Salud, S.d. Boletín Infecciones Asociadas a la Atención de la Salud (IAAS) Red Hospitalaria de Vigilancia Epidemiológica (RHOVE). 2016, 116. Available online: https://epidemiologia.salud.gob.mx/gobmx/salud/documentos/manuales/28_Manual_RHoVE.pdf (accessed on 15 June 2022).
- Rodríguez Salgado, M. Frecuencia de infecciones asociadas a la atención de la salud en los principales sistemas de información de México. Boletín CONAMED-OPS 2018, 3, 17. Available online: http://www.conamed.gob.mx/gobmx/boletin/pdf/boletin17/frecuencia_infecciones.pdf (accessed on 15 June 2022).
- Nájera-Bello, J.A.; Villanueva-Pastrana, N.; Barlandas-Rendón, N.R.E.; Quintana-Ponce, S.; Cruz-Navarrete, E.; Maya-Rodríguez, P.A. Bacterial drug resistance of priority pathogens isolated in Chilpancingo, Guerrero, Mexico. Revista Mexicana de Patología Clínica y Medicina de Laboratorio 2022, 68, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Karlowsky, J.A.; Lob, S.H.; DeRyke, C.A.; Siddiqui, F.; Young, K.; Motyl, M.R.; Sahm, D.F. Prevalence of ESBL non-CRE Escherichia coli and Klebsiella pneumoniae among clinical isolates collected by the SMART global surveillance programme from 2015 to 2019. Int. J. Antimicrob. Agents 2022, 59, 106535. [Google Scholar] [CrossRef]
- Garza-Ramos, U.; Barrios, H.; Reyna-Flores, F.; Sánchez-Pérez, A.; Tamayo-Legorreta, E.; Ibarra-Pacheco, A.; Salazar-Salinas, J.; Núñez-Ceballos, R.; Silva-Sanchez, J. Characteristics of KPC-2–producing Klebsiella pneumoniae (ST258) clinical isolates from outbreaks in 2 Mexican medical centers. Diagn. Microbiol. Infect. Dis. 2014, 79, 483–485. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Zulueta, P.; Silva-Sánchez, J.; Barrios, H.; Reyes-Mar, J.; Vélez-Pérez, F.; Arroyo-Escalante, S.; Ochoa-Carrera, L.; Delgado-Sapien, G.; Morales-Espinoza Mdel, R.; Tamayo-Legorreta, E.; et al. First outbreak of KPC-3-producing Klebsiella pneumoniae (ST258) clinical isolates in a Mexican Medical Center. Antimicrob. Agents Chemother. 2013, 57, 4086–4088. [Google Scholar] [CrossRef] [PubMed]
- Aquino-Andrade, A.; Merida-Vieyra, J.; De La Garza, E.A.; Arzate-Barbosa, P.; Ranero, A.D.C. Carbapenemase-producing Enterobacteriaceae in Mexico: Report of seven non-clonal cases in a pediatric hospital. BMC Microbiol. 2018, 18, 38. [Google Scholar] [CrossRef] [PubMed]
- Torres-Gonzalez, P.; Cervera-Hernandez, M.; Niembro-Ortega, M.D.; Leal-Vega, F.; Cruz-Hervert, L.P.; García-García, L.; Galindo-Fraga, A.; Martinez-Gamboa, A.; Valle, M.B.-D.; Sifuentes-Osornio, J.; et al. Factors associated to prevalence and incidence of carbapenem-resistant enterobacteriaceae fecal carriage: A cohort study in a mexican tertiary care hospital. PLoS One 2015, 10, e0139883. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Anes, J.; Devineau, S.; Fanning, S. Klebsiella pneumoniae: Prevalence, reservoirs, antimicrobial resistance, pathogenicity, and infection: A hitherto unrecognized zoonotic bacterium. Foodborne Pathog. Dis. 2021, 18, 63–84. [Google Scholar] [CrossRef]
- Ostria-Hernandez, M.L.; la Rosa, K.C.J.-D.; Arzate-Barbosa, P.; Lara-Hernández, A.; Sakai, F.; Ibarra, J.A.; Castro-Escarpulli, G.; Vidal, J.E. Nosocomial, multidrug-resistant Klebsiella pneumoniae strains isolated from mexico city produce robust biofilms on abiotic surfaces but not on human lung cells. Microb. Drug Resist. 2018, 24, 422–433. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.J. Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol. Rev. 2017, 42, fux053. [Google Scholar] [CrossRef]
- Sleiman, A.; Awada, B.; Mocadie, M.; Sherri, N.; Haraoui, L.-P.; Baby, V.; Araj, G.F.; Kanj, S.S.; Rizk, N.; Matar, G.M.; et al. An unequivocal superbug: PDR Klebsiella pneumoniae with an arsenal of resistance and virulence factor genes. J. Infect. Dev. Ctries. 2021, 15, 404–414. [Google Scholar] [CrossRef]
- Ramírez-Alfaro, C.; Villalobos-Vindas, J. Análisis de las bacteremias por Klebsiella pneumoniae en pacientes del Hospital México. Acta Med. Costarric. 2016, 58, 62–68. [Google Scholar]
- Higashino, M.; Murata, M.; Morinaga, Y.; Akamatsu, N.; Matsuda, J.; Takeda, K.; Kaku, N.; Kosai, K.; Uno, N.; Hasegawa, H.; et al. Fluoroquinolone resistance in extended-spectrum β-lactamase-producing Klebsiella pneumoniae in a Japanese tertiary hospital: Silent shifting to CTX-M-15-producing K. pneumoniae. J. Med. Microbiol. 2017, 66, 1476–1482. [Google Scholar] [CrossRef] [PubMed]
- Bocanegra-Ibarias, P.; Garza-González, E.; Morfín-Otero, R.; Barrios, H.; Villarreal-Treviño, L.; Rodríguez-Noriega, E.; Garza-Ramos, U.; Petersen-Morfin, S.; Silva-Sanchez, J. Molecular and microbiological report of a hospital outbreak of NDM-1-carrying Enterobacteriaceae in Mexico. PLoS ONE 2017, 12, e0179651. [Google Scholar] [CrossRef] [PubMed]
- Lowe, M.; Kock, M.M.; Coetzee, J.; Hoosien, E.; Peirano, G.; Strydom, K.-A.; Ehlers, M.M.; Mbelle, N.M.; Shashkina, E.; Haslam, D.B.; et al. Klebsiella pneumoniae ST307 with blaOXA-181, South Africa, 2014–2016. Emerg. Infect. Dis. 2019, 25, 739–747. [Google Scholar] [CrossRef]
- López-Camacho, E.; Paño-Pardo, J.R.; Ruiz-Carrascoso, G.; Wesselink, J.-J.; Lusa-Bernal, S.; Ramos-Ruiz, R.; Ovalle, S.; Gómez-Gil, R.; Pérez-Blanco, V.; Pérez-Vázquez, M.; et al. Population structure of OXA-48-producing Klebsiella pneumoniae ST405 isolates during a hospital outbreak characterised by genomic typing. J. Glob. Antimicrob. Resist. 2018, 15, 48–54. [Google Scholar] [CrossRef]
- Del Franco, M.; Paone, L.; Novati, R.; Giacomazzi, C.G.; Bagattini, M.; Galotto, C.; Montanera, P.G.; Triassi, M.; Zarrilli, R. Molecular epidemiology of carbapenem resistant Enterobacteriaceae in Valle d’Aosta region, Italy, shows the emergence of KPC-2 producing Klebsiella pneumoniae clonal complex 101 (ST101 and ST1789). BMC Microbiol. 2015, 15, 260. [Google Scholar] [CrossRef]
- Villa, L.; Feudi, C.; Fortini, D.; Brisse, S.; Passet, V.; Bonura, C.; Endimiani, A.; Mammina, C.; Ocampo, A.M.; Jiménez, J.N.; et al. Diversity, virulence, and antimicrobial resistance of the KPC-producing Klebsiella pneumoniae ST307 clone. Microb. Genom. 2017, 3, e000110. [Google Scholar] [CrossRef]
- Zaha, D.C.; Bungau, S.; Aleya, S.; Tit, D.M.; Vesa, C.M.; Popa, A.R.; Pantis, C.; Maghiar, O.A.; Bratu, O.G.; Furau, C.; et al. What antibiotics for what pathogens? The sensitivity spectrum of isolated strains in an intensive care unit. Sci. Total Environ. 2019, 687, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Llaca-Díaz, J.M.; Mendoza-Olazarán, S.; Camacho-Ortiz, A.; Flores, S.; Garza-González, E. One-Year Surveillance of ESKAPE Pathogens in an Intensive Care Unit of Monterrey, Mexico. Chemotherapy 2012, 58, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Salazar, C.; Giménez, M.; Riera, N.; Parada, A.; Puig, J.; Galiana, A.; Grill, F.; Vieytes, M.; Mason, C.E.; Antelo, V.; et al. Human microbiota drives hospital-associated antimicrobial resistance dissemination in the urban environment and mirrors patient case rates. Microbiome 2022, 10, 208. [Google Scholar] [CrossRef] [PubMed]

| Allelic Profile | ||||||||
|---|---|---|---|---|---|---|---|---|
| Strain | gapA | rpoB | mdh | pgi | phoE | InfB | tonB | ST |
| K. quasipneumoniae ATCC 700603 | 18 | 52 | 26 | 71 | 98 | 22 | 51 | 489 |
| 1E | 2 | 1 | 1 | 3 | 8 | 1 | 15 | 2015 |
| 2E | 2 | 1 | 62 | 3 | 10 | 4 | 110 | 405 |
| 3E | 2 | 1 | 1 | 1 | 9 | 4 | 12 | 23 |
| 4E | 2 | 1 | 1 | 6 | 7 | 4 | 12 | 496 |
| 5E | 2 | 1 | 62 | 3 | 10 | 4 | 110 | 405 |
| 6E | 2 | 1 | 164 | 1 | 7 | 4 | 303 | 1846 |
| 7E | 2 | 9 | 2 | 1 | 13 | 1 | 10 | 309 |
| 8E | 3 | 1 | 1 | 1 | 9 | 4 | 135 | 551 |
| 9E | 3 | 1 | 1 | 1 | 9 | 4 | 135 | 551 |
| 10E | 3 | 1 | 1 | 1 | 9 | 4 | 135 | 551 |
| 11E | 2 | 1 | 62 | 3 | 10 | 4 | 110 | 405 |
| 12E | 3 | 1 | 1 | 1 | 9 | 4 | 135 | 551 |
| 13E | 51 | 1 | 5 | 1 | 9 | 4 | 13 | 491 |
| 14E | 2 | 1 | 62 | 3 | 10 | 4 | 110 | 405 |
| 15E | 2 | 1 | 1 | 1 | 10 | 4 | 13 | 25 |
| 16E | 2 | 1 | 5 | 1 | 17 | 4 | 42 | 111 |
| 17E | 2 | 1 | 1 | 1 | 7 | 1 | 12 | 485 |
| 18E | 2 | 1 | 2 | 1 | 4 | 4 | 4 | 16 |
| 19E | 2 | 1 | 2 | 1 | 7 | 1 | 7 | 36 |
| 20E | 2 | 60 | 11 | 1 | 4 | 8 | 24 | 628 |
| 21E | 2 | 1 | 2 | 1 | 1 | 4 | 13 | 804 |
| 22E | 2 | 1 | 1 | 117 | 10 | 4 | 18 | 1726 |
| 23E | 2 | 1 | 1 | 1 | 10 | 4 | 13 | 25 |
| 24E | 3 | 3 | 1 | 1 | 9 | 1 | 4 | 1869 |
| 25E | 2 | 9 | 2 | 1 | 13 | 1 | 16 | 37 |
| 1S | 2 | 1 | 1 | 10 | 1 | 1 | 76 | 1088 |
| 2S | 2 | 1 | 1 | 10 | 1 | 1 | 76 | 1088 |
| 3S | 3 | 1 | 1 | 1 | 9 | 4 | 135 | 551 |
| 4S | 2 | 1 | 1 | 10 | 1 | 1 | 76 | 1088 |
| 5S | 2 | 1 | 2 | 1 | 213 | 1 | 7 | 2175 |
| 6S | 2 | 1 | 1 | 10 | 1 | 1 | 76 | 1088 |
| 7S | 3 | 1 | 1 | 1 | 9 | 4 | 135 | 551 |
| 8S | 2 | 1 | 162 | 3 | 10 | 4 | 110 | 405 |
| 1I | 2 | 4 | 2 | 1 | 7 | 1 | 12 | 788 |
| 2I | 3 | 3 | 1 | 1 | 13 | 1 | 79 | 1406 |
| 3I | 2 | 1 | 1 | 1 | 10 | 4 | 13 | 25 |
| 4I | 2 | 1 | 97 | 1 | 9 | 4 | 13 | 846 |
| 5I | 2 | 1 | 2 | 1 | 7 | 1 | 7 | 36 |
| 1Z | 2 | 1 | 1 | 1 | 10 | 4 | 13 | 25 |
| 2Z | 2 | 1 | 62 | 3 | 10 | 4 | 110 | 405 |
| 3Z | 4 | 1 | 2 | 52 | 1 | 1 | 7 | 307 |
| 4Z | 3 | 4 | 6 | 1 | 7 | 4 | 40 | 392 |
| 5Z | 4 | 1 | 32 | 1 | 7 | 4 | 10 | 151 |
| 6Z | 3 | 4 | 6 | 1 | 7 | 4 | 40 | 392 |
| 7Z | 4 | 5 | 88 | 1 | 1 | 94 | 23 | 2080 |
| 8Z | 3 | 4 | 6 | 1 | 7 | 4 | 13 | 885 |
| 9Z | 2 | 4 | 1 | 1 | 7 | 4 | 4 | 706 |
| 10Z | 3 | 4 | 6 | 1 | 7 | 4 | 40 | 392 |
| Gene | PCR (bp) | Haplotype | Polymorphic Sites | Total Mutations | π | θ | G + C | dN | dS | dN/dS |
|---|---|---|---|---|---|---|---|---|---|---|
| gapA | 450 | 4 | 3 | 3 | 0.02794 | 0.00364 | 0.5615 | 0.0000 | 0.0132 | 0.0000 |
| InfB | 318 | 6 | 7 | 7 | 0.00797 | 0.00964 | 0.6140 | 0.0097 | 0.0037 | 2.6548 |
| mdh | 477 | 10 | 21 | 21 | 0.00526 | 0.00684 | 0.5577 | 0.0086 | 0.0145 | 0.5944 |
| pgi | 432 | 6 | 6 | 6 | 0.00463 | 0.00608 | 0.5733 | 0.0042 | 0.0056 | 0.7531 |
| phoE | 420 | 9 | 9 | 9 | 0.00728 | 0.00788 | 0.5585 | 0.0000 | 0.0311 | 0.0000 |
| rpoB | 501 | 4 | 13 | 13 | 0.01331 | 0.01331 | 0.5415 | 0.0128 | 0.0147 | 0.8713 |
| tonB | 414 | 17 | 16 | 16 | 0.01027 | 0.01143 | 0.6466 | 0.0050 | 0.0257 | 0.1961 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Córdova-Espinoza, M.G.; Giono-Cerezo, S.; Sierra-Atanacio, E.G.; Escamilla-Gutiérrez, A.; Carrillo-Tapia, E.; Carrillo-Vázquez, L.I.; Mendoza-Pérez, F.; Leyte-Lugo, M.; González-Vázquez, R.; Mayorga-Reyes, L.; et al. Isolation and Identification of Multidrug-Resistant Klebsiella pneumoniae Clones from the Hospital Environment. Pathogens 2023, 12, 634. https://doi.org/10.3390/pathogens12050634
Córdova-Espinoza MG, Giono-Cerezo S, Sierra-Atanacio EG, Escamilla-Gutiérrez A, Carrillo-Tapia E, Carrillo-Vázquez LI, Mendoza-Pérez F, Leyte-Lugo M, González-Vázquez R, Mayorga-Reyes L, et al. Isolation and Identification of Multidrug-Resistant Klebsiella pneumoniae Clones from the Hospital Environment. Pathogens. 2023; 12(5):634. https://doi.org/10.3390/pathogens12050634
Chicago/Turabian StyleCórdova-Espinoza, María Guadalupe, Silvia Giono-Cerezo, Erika Gabriela Sierra-Atanacio, Alejandro Escamilla-Gutiérrez, Eduardo Carrillo-Tapia, Laura Isabel Carrillo-Vázquez, Felipe Mendoza-Pérez, Martha Leyte-Lugo, Raquel González-Vázquez, Lino Mayorga-Reyes, and et al. 2023. "Isolation and Identification of Multidrug-Resistant Klebsiella pneumoniae Clones from the Hospital Environment" Pathogens 12, no. 5: 634. https://doi.org/10.3390/pathogens12050634
APA StyleCórdova-Espinoza, M. G., Giono-Cerezo, S., Sierra-Atanacio, E. G., Escamilla-Gutiérrez, A., Carrillo-Tapia, E., Carrillo-Vázquez, L. I., Mendoza-Pérez, F., Leyte-Lugo, M., González-Vázquez, R., Mayorga-Reyes, L., & González-Vázquez, R. (2023). Isolation and Identification of Multidrug-Resistant Klebsiella pneumoniae Clones from the Hospital Environment. Pathogens, 12(5), 634. https://doi.org/10.3390/pathogens12050634






