Effective Methods of Estimation of Pathogen Prevalence in Pooled Ticks
Abstract
1. Introduction
2. Materials and Methods
2.1. Search Strategy, Study Selection and Data Analysis
2.2. Tested Dataset and Statistical Analysis
3. Results
3.1. Summary of Literature Data
3.2. Tested Dataset and Statistical Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Estrada-Peña, A.; Ayllón, N.; de la Fuente, J. Impact of climate trends on tick-borne pathogen transmission. Front. Physiol. 2012, 3, 64. [Google Scholar] [CrossRef]
- Sonenshine, D.E. Ranse expansion of tick disease vectors in North America: Implications for Spread of tick-borne disease. Int. J. Environ. Res. Public Health 2018, 15, 478. [Google Scholar] [CrossRef] [PubMed]
- Diuk-Wasser, M.A.; VanAcker, M.C.; Fernandez, M.P. Impact of land use changes and habitat fragmentation on the eco-epidemiology of tick-borne diseases. J. Med. Entomol. 2021, 58, 1546–1564. [Google Scholar] [CrossRef] [PubMed]
- Eisen, R.J.; Paddock, C.D. Tick and tick-borne pathogen surveillance as a public health tool in the United States. J. Med. Entomol. 2021, 58, 1490–1502. [Google Scholar] [CrossRef] [PubMed]
- Dorfman, R. The detection of defective members of large populations. Ann. Math. Stat. 1943, 14, 436–440. [Google Scholar] [CrossRef]
- Farrington, C.P. Estimating prevalence by group testing using generalized linear models. Stat. Med. 1992, 11, 1591–1597. [Google Scholar] [CrossRef]
- Mendoza-Blanco, J.R.; Tu, X.M.; Iyengar, S. Bayesian inference on prevalence using a missing-data approach with simulation-based techniques: Applications to HIV screening. Stat. Med. 1996, 15, 2161–2176. [Google Scholar] [CrossRef]
- Cowling, D.W.; Gardner, I.A.; Johnson, W.O. Comparison of methods for estimation of individual-level prevalence based on pooled samples. Prev. Vet. Med. 1999, 39, 211–225. [Google Scholar] [CrossRef]
- Martín-Sánchez, J.; Gállego, M.; Barón, S.; Castillejo, S.; Morillas-Marquez, F. Pool screen PCR for estimating the prevalence of Leishmania infantum infection in sandflies (Diptera: Nematocera, Phlebotomidae). Trans. R Soc. Trop. Med. Hyg. 2006, 100, 527–532. [Google Scholar] [CrossRef]
- Randolph, S.E. Ticks are not insects: Consequences of contrasting vector biology for transmission potential. Parasitol. Today 1998, 14, 186–192. [Google Scholar] [CrossRef]
- Randolph, S.E.; Storey, K. Impact of microclimate on immature tick-rodent host interactions (Acari: Ixodidae): Implications for parasite transmission. J. Med. Entomol. 1999, 36, 741–748. [Google Scholar] [CrossRef] [PubMed]
- Mancini, G.; Montarsi, F.; Calzolari, M.; Capelli, G.; Dottori, M.; Ravagnan, S.; Lelli, D.; Chiari, M.; Santilli, A.; Quaglia, M.; et al. Mosquito species involved in the circulation of West Nile and Usutu viruses in Italy. Vet. Ital. 2017, 53, 97–110. [Google Scholar] [CrossRef] [PubMed]
- Bertola, M.; Montarsi, F.; Obber, F.; Da Rold, G.; Carlin, S.; Toniolo, F.; Porcellato, E.; Falcaro, C.; Mondardini, V.; Ormelli, S.; et al. Occurrence and identification of Ixodes ricinus borne pathogens in Northeastern Italy. Pathogens 2021, 10, 1181. [Google Scholar] [CrossRef]
- Ott, D.; Ulrich, K.; Ginsbach, P.; Öhme, R.; Bock-Hensley, O.; Falk, U.; Teinert, M.; Lenhard, T. Tick-borne encephalitis virus (TBEV) prevalence in field-collected ticks (Ixodes ricinus) and phylogenetic, structural and virulence analysis in a TBE high-risk endemic area in Southwestern Germany. Parasit. Vectors 2020, 13, 303. [Google Scholar] [CrossRef] [PubMed]
- Cutler, S.J.; Vayssier-Taussat, M.; Estrada-Peña, A.; Potkonjak, A.; Mihalca, A.D.; Zeller, H. Tick-borne diseases and co-infection: Current considerations. Ticks Tick Borne Dis. 2021, 12, 101607. [Google Scholar] [CrossRef]
- Pfäffle, M.; Littwin, N.; Muders, S.V.; Petney, T.N. The ecology of tick-borne diseases. Int. J. Parasitol. 2013, 43, 1059–1077. [Google Scholar] [CrossRef]
- Wang, S.; Hua, X.; Cui, L. Characterization of microbiota diversity of engorged ticks collected from dogs in China. J. Vet. Sci. 2021, 22, e37. [Google Scholar] [CrossRef]
- Pautienius, A.; Armonaite, A.; Simkute, E.; Zagrabskaite, R.; Buitkuviene, J.; Alpizar-Jara, R.; Grigas, J.; Zakiene, I.; Zienius, D.; Salomskas, A.; et al. Cross-sectional study on the prevalence and factors influencing occurrence of tick-borne encephalitis in horses in Lithuania. Pathogens 2021, 10, 140. [Google Scholar] [CrossRef]
- Madder, M.; Day, M.; Schunack, B.; Fourie, J.; Labuschange, M.; van der Westhuizen, W.; Johnson, S.; Githigia, S.M.; Akande, F.A.; Nzalawahe, J.S.; et al. A community approach for pathogens and their arthropod vectors (ticks and fleas) in cats of sub-Saharan Africa. Parasit. Vectors 2022, 15, 321. [Google Scholar] [CrossRef]
- Ondruš, J.; Balážová, A.; Baláž, V.; Zechmeisterová, K.; Novobilský, A.; Široký, P. Candidatus Neoehrlichia mikurensis is widespread in questing Ixodes ricinus ticks in the Czech Republic. Ticks Tick Borne Dis. 2020, 11, 101371. [Google Scholar] [CrossRef]
- Lakew, B.T.; Kheravii, S.K.; Wu, S.B.; Eastwood, S.; Andrew, N.R.; Jenkins, C.; Walkden-Brown, S.W. Endemic infection of cattle with multiple genotypes of Theileria orientalis on the Northern Tablelands of New South Wales despite limited presence of ticks. Ticks Tick Borne Dis. 2021, 12, 101645. [Google Scholar] [CrossRef] [PubMed]
- Krishnamoorthy, P.; Sudhagar, S.; Goudar, A.L.; Jacob, S.S.; Suresh, K.P. Molecular survey and phylogenetic analysis of tick-borne pathogens in ticks infesting cattle from two South Indian states. Vet. Parasitol. Reg. Stud. Reports 2021, 25, 100595. [Google Scholar] [CrossRef]
- Li, X.; Ji, H.; Wang, D.; Che, L.; Zhang, L.; Li, L.; Yin, Q.; Liu, Q.; Wei, F.; Wang, Z. Molecular detection and phylogenetic analysis of tick-borne encephalitis virus in ticks in Northeastern China. J. Med. Virol. 2022, 94, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Furstenau, T.N.; Cocking, J.H.; Hepp, C.M.; Fofanov, V.Y. Sample pooling methods for efficient pathogen screening: Practical implications. PLoS ONE 2020, 15, e0236849. [Google Scholar] [CrossRef]
- Park, J.W.; Lee, S.H.; Lee, G.S.; Seo, J.J.; Chung, J.K. Epidemiological characteristics of field tick-borne pathogens in Gwang-ju Metropolitan Area, South Korea, from 2014 to 2018. Osong. Public Health Res. Perspect. 2020, 11, 177–184. [Google Scholar] [CrossRef]
- Vitek, C.J.; Richards, S.L.; Mores, C.N.; Day, J.F.; Lord, C.C. Arbovirus transmission by Culex nigripalpus in Florida, 2005. J. Med. Entomol. 2008, 45, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Orshan, L.; Bin, H.; Schnur, H.; Kaufman, A.; Valinsky, A.; Shulman, L.; Weiss, L.; Mendelson, E.; Pener, H. Mosquito vectors of West Nile Fever in Israel. J. Med. Entomol. 2008, 45, 939–947. [Google Scholar] [CrossRef] [PubMed]
- Andreassen, A.; Jore, S.; Cuber, P.; Dudman, S.; Tengs, T.; Isaksen, K.; Hygen, H.O.; Viljugrein, H.; Anestad, G.; Ottesen, P.; et al. Prevalence of tick-borne encephalitis virus in tick nymphs in relation to climatic factors on the southern coast of Norway. Parasit Vectors 2012, 5, 177. [Google Scholar] [CrossRef]
- Morozkin, E.S.; Makenov, M.T.; Zhurenkova, O.B.; Kholodilov, I.S.; Belova, O.A.; Radyuk, E.V.; Fyodorova, M.V.; Grigoreva, Y.E.; Litov, A.G.; Valdokhina, A.V.; et al. Integrated jingmenvirus polymerase gene in Ixodes ricinus genome. Viruses 2022, 14, 1908. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 28 February 2023).
- McLure, A.; O’Neill, B.; Mayfield, H.; Lau, C.; McPherson, B. PoolTestR: An R package for estimating prevalence and regression modelling for molecular xenomonitoring and other applications with pooled samples. Environ. Model Softw. 2021, 145, 105158. [Google Scholar] [CrossRef]
- Biggerstaff, B.J. PooledInfRate, Version 4.0: A Microsoft® Office Excel© Add-In to Compute Prevalence Estimates from Pooled Samples; Centers for Disease Control and Prevention: Fort Collins, CO, USA, 2009. [Google Scholar]
- Sergeant, E.S.G. Epitools Epidemiological Calculators. Ausvet Animal Health Services and Australian Biosecurity Cooperative Research Centre for Emerging Infectious Disease. Available online: http://epitools.ausvet.com.au (accessed on 28 February 2023).
- Centers for Disease Control and Prevention (CDC). Available online: https://www.cdc.gov/epiinfo/index.html (accessed on 28 February 2023).
- Rózsa, L.; Reiczigel, J.; Majoros, G. Quantifying parasites in samples of hosts. J. Parasitol. 2000, 86, 228–232. [Google Scholar] [CrossRef]
- Reiczigel, J.; Rózsa, L. Quantitative Parasitology 3.0. Budapest. 2005. Available online: http://www.zoologia.hu/qp/qp.html (accessed on 28 February 2023).
- Hauck, W.W. Confidence intervals for seroprevalence determined from pooled sera. Ann. Epidemiol. 1991, 1, 277–281. [Google Scholar] [CrossRef]
- Krebs, C.J. Ecological Methodology, 2nd ed.; Harlow: Menlo Park, CA, USA, 1999; p. 620. [Google Scholar]
- Devleesschauwer, B.; Torgerson, P.; Charlier, J.; Levecke, B.; Praet, N.; Roelandt, S.; Smit, S.; Dorny, P.; Berkvens, D.; Speybroeck, N. Prevalence: Tools for Prevalence Assessment Studies. R Package Version 0.4.0. 2015. Available online: https://cran.r-project.org/package=prevalence (accessed on 28 February 2023).
- McV Messam, L.L.; Branscum, A.J.; Collins, M.T.; Gardner, I.A. Frequentist and Bayesian approaches to prevalence estimation using examples from Johne’s disease. Anim. Health Res. Rev. 2008, 9, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.J.; Moffitt, C.M. Estimation of pathogen prevalence in pooled samples using maximum likelihood methods and open-source software. J. Aquat. Anim. Health 2005, 17, 386–391. [Google Scholar] [CrossRef]
- Daniel, W.W. Biostatistics: A Foundation for Analysis in the Health Sciences, 7th ed.; John Wiley & Sons, Inc.: New York, NY, USA, 1999; p. 956. [Google Scholar]
- Ebert, T.A.; Brlansky, R.; Rogers, M. Reexamining the pooled sampling approach for estimating prevalence of infected insect vectors. Ann. Entomol. Soc. Am. 2010, 103, 827–837. [Google Scholar] [CrossRef]
- Pettersson, J.H.; Golovljova, I.; Vene, S.; Jaenson, T.G. Prevalence of tick-borne encephalitis virus in Ixodes ricinus ticks in Northern Europe with particular reference to Southern Sweden. Parasit. Vectors 2014, 7, 102. [Google Scholar] [CrossRef]
- Walter, S.D.; Hildreth, S.W.; Beaty, B.J. Estimation of infection rates in populations of organisms using pools of variable size. Am. J. Epidemiol. 1980, 112, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.L.; Reeves, W.C. Statistical estimation of virus infection rates in mosquito vector populations. Am. J. Hyg. 1962, 75, 377–391. [Google Scholar] [CrossRef]
| Prevalence Index | Papers Using the Method/Total Papers | Associated with Confidence Interval (CI) 95% | Specific Methods/Software/Package | Association between Different Indexes |
|---|---|---|---|---|
| Pool positivity rate (PPR) | 106/185 * (57%) | - | - | PPR could be associated with MIR (n = 17) |
| Minimum infection rate (MIR) | 64/185 ** (35%) | 18/184 (10%) | MIR could be associated (n = 4) with maximum infection rate (MaxIR) index, which was calculated by considering that all the ticks present in a positive pool are infected. | |
| Estimate pool prevalence (EPP) | 35/185 (19%) | 26/35 (74%) | EPP could be associated with MIR (n = 4) or PPR (n = 1) |
| Dataset | PPR (CI) | MIR (CI) | EPP (CI) |
|---|---|---|---|
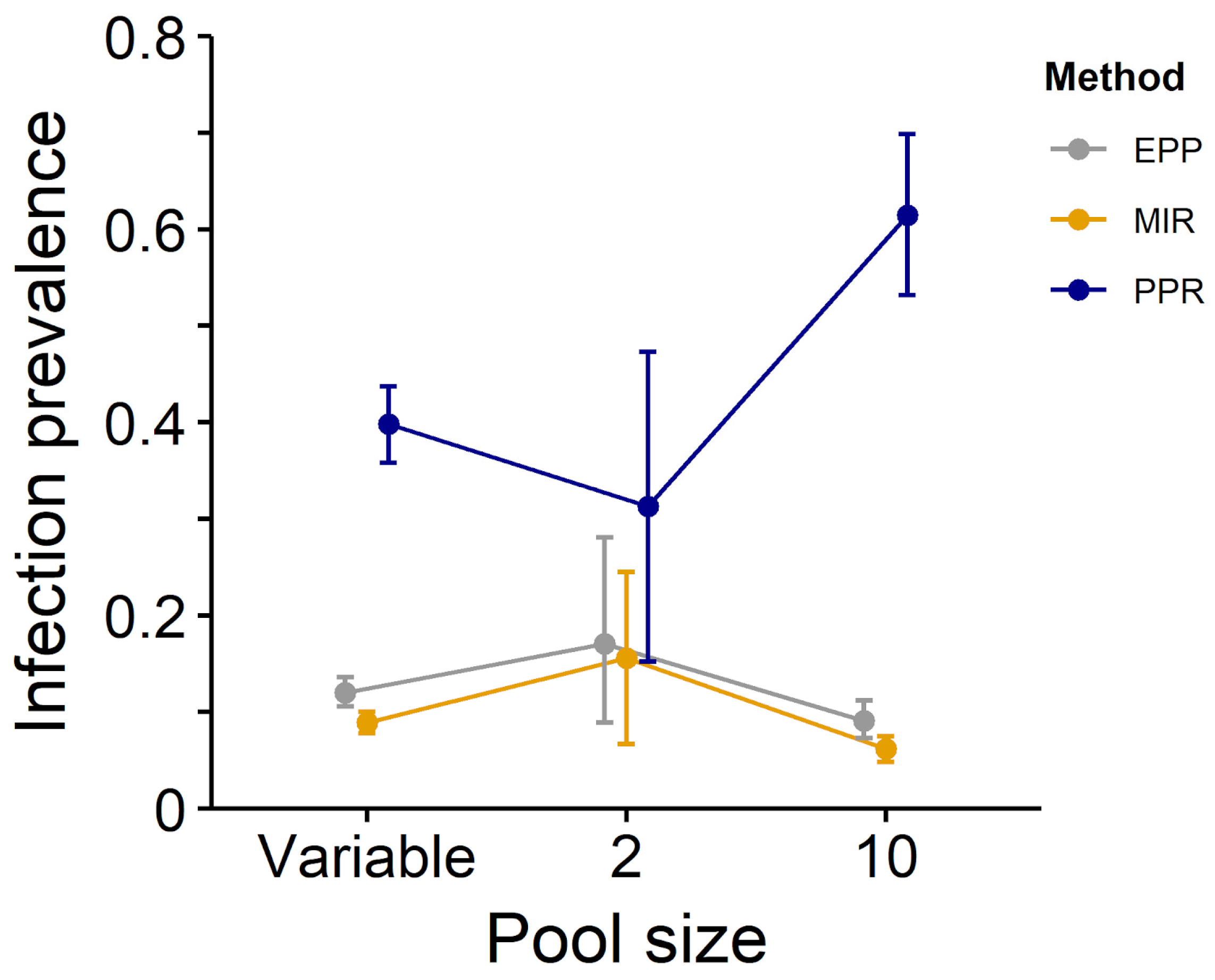
| All diseases, unequal pools | 0.398 (0.358–0.437) * | 0.089 (0.078–0.100) * | 0.120 (0.106–0.136) |
| All diseases, pool = 2 | 0.313 (0.152–0.473) | 0.156 (0.067–0.245) | 0.171 (0.089–0.281) |
| All diseases, pool = 10 | 0.615 (0.532–0.699) | 0.062 (0.048–0.075) | 0.091 (0.073–0.112) |
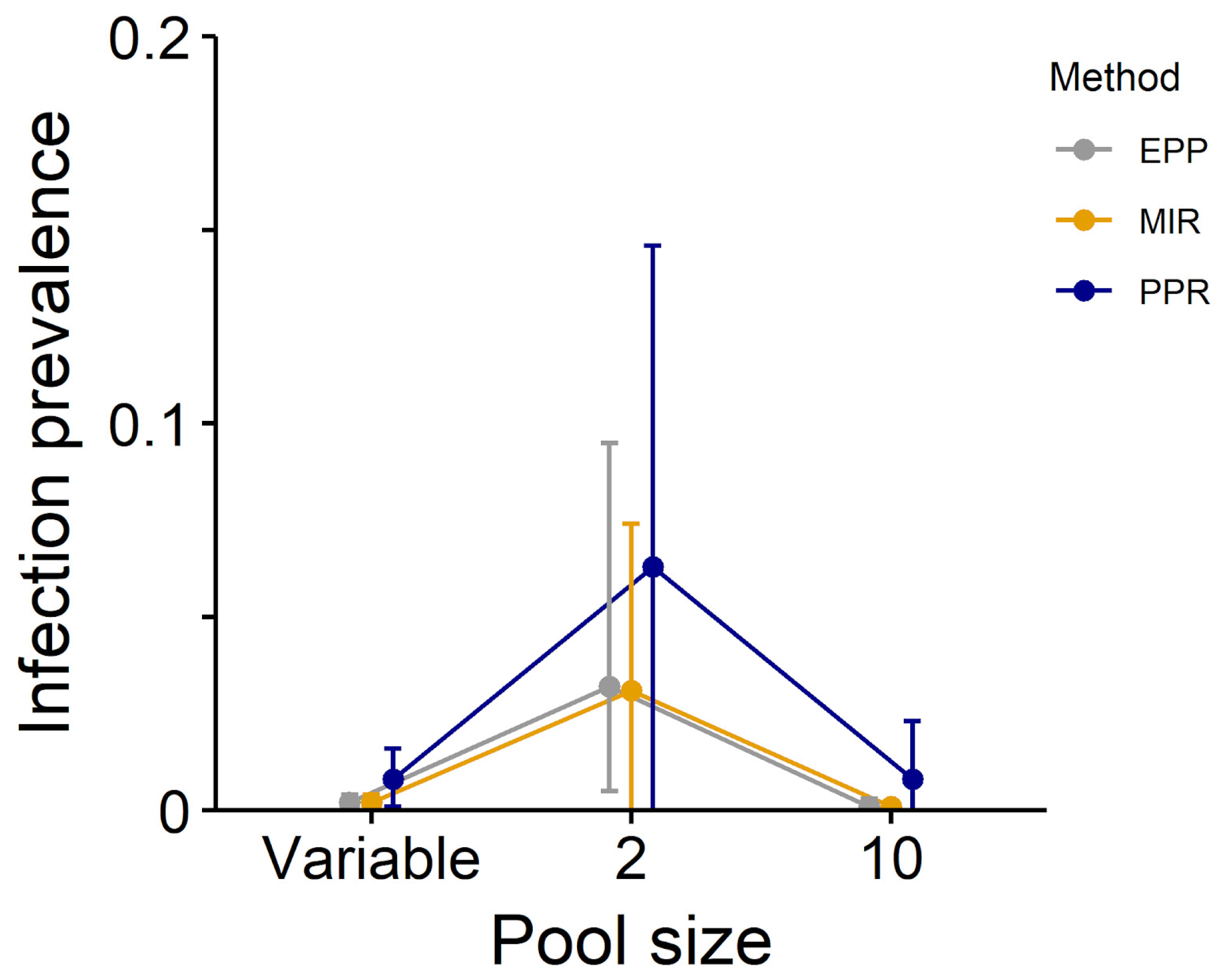
| Low prevalence, unequal pools | 0.008 (0.001–0.016) * | 0.002 (0.000–0.004) * | 0.002 (0.001–0.004) |
| Low prevalence, pool = 2 a | 0.063 (0.000–0.146) | 0.031 (0.000–0.074) | 0.032 (0.005–0.095) |
| Low prevalence, pool = 10 | 0.008 (0.000–0.023) | 0.001 (0.000–0.002) | 0.001 (0.000–0.003) |
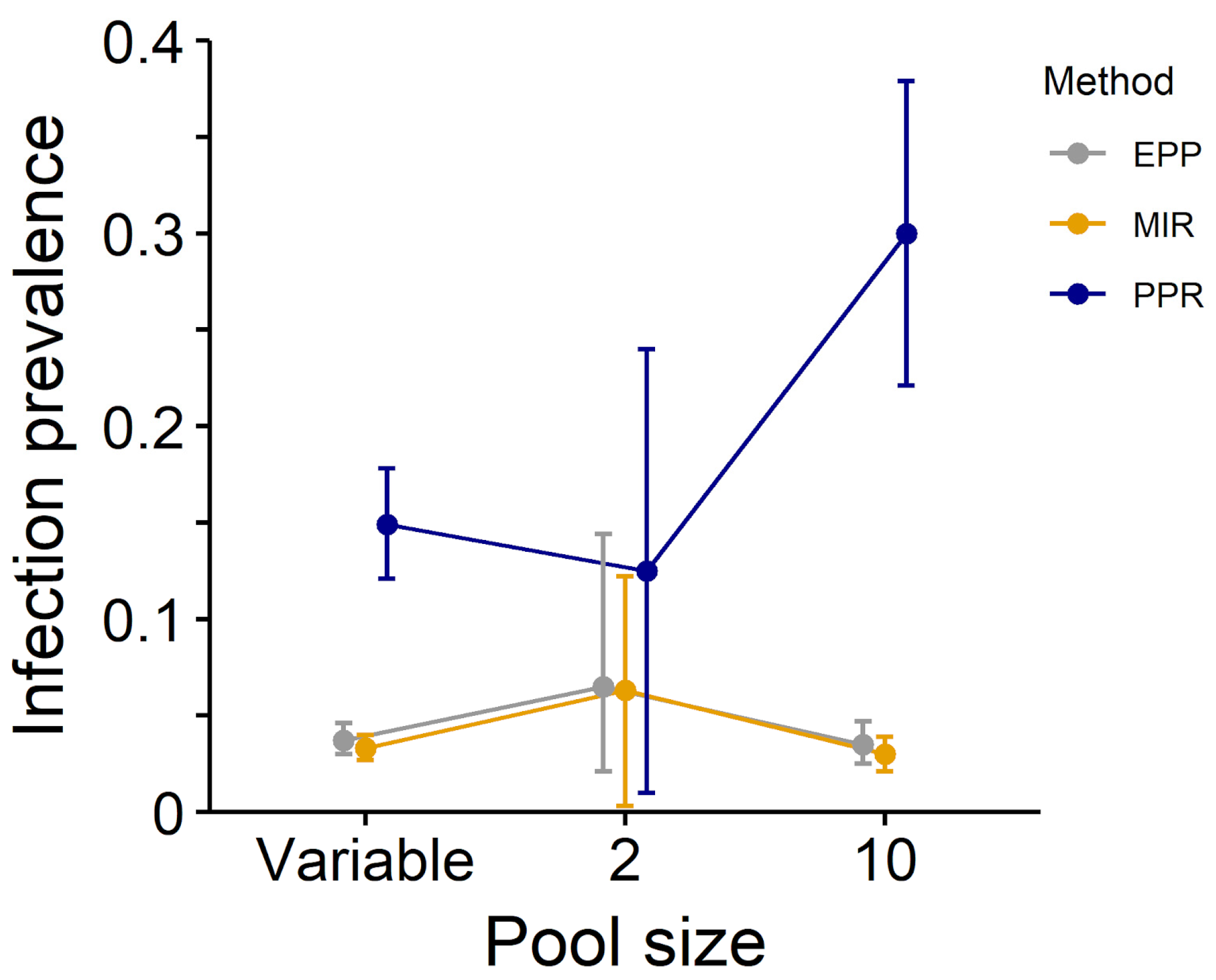
| High prevalence, unequal pools | 0.166 (0.136–0.196) * | 0.037 (0.030–0.044) * | 0.042 (0.035–0.051) |
| High prevalence, pool = 2 | 0.125 (0.010–0.240) | 0.063 (0.003–0.122) | 0.065 (0.021–0.144) |
| High prevalence, pool = 10 | 0.315 (0.236–0.395) | 0.032 (0.022–0.041) | 0.037 (0.027–0.050) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fracasso, G.; Grillini, M.; Grassi, L.; Gradoni, F.; Rold, G.d.; Bertola, M. Effective Methods of Estimation of Pathogen Prevalence in Pooled Ticks. Pathogens 2023, 12, 557. https://doi.org/10.3390/pathogens12040557
Fracasso G, Grillini M, Grassi L, Gradoni F, Rold Gd, Bertola M. Effective Methods of Estimation of Pathogen Prevalence in Pooled Ticks. Pathogens. 2023; 12(4):557. https://doi.org/10.3390/pathogens12040557
Chicago/Turabian StyleFracasso, Gerardo, Marika Grillini, Laura Grassi, Francesco Gradoni, Graziana da Rold, and Michela Bertola. 2023. "Effective Methods of Estimation of Pathogen Prevalence in Pooled Ticks" Pathogens 12, no. 4: 557. https://doi.org/10.3390/pathogens12040557
APA StyleFracasso, G., Grillini, M., Grassi, L., Gradoni, F., Rold, G. d., & Bertola, M. (2023). Effective Methods of Estimation of Pathogen Prevalence in Pooled Ticks. Pathogens, 12(4), 557. https://doi.org/10.3390/pathogens12040557







