Genome Analysis of Pseudomonas aeruginosa Strains from Chronically Infected Patients with High Levels of Persister Formation
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Ethical Statement
2.2. Antimicrobial Agents
2.3. Determination of Viable Bacterial Count
2.4. Selection and Quantification of Levofloxacin Persister Cells
2.5. Determination of the MIC of Tested Antibiotics
2.6. The Killing Curve of Bactericidal Antibiotics against Levofloxacin Persisters
2.7. Biofilm Formation Capacity Test
2.8. Genomic DNA (gDNA) Extraction and Whole Genome Sequencing
2.9. Genome Annotation and Phylogeny
2.10. Resistome Analysis
3. Results
3.1. Separation of P. aeruginosa Persister Cells and Determination of Their Antimicrobial Susceptibility
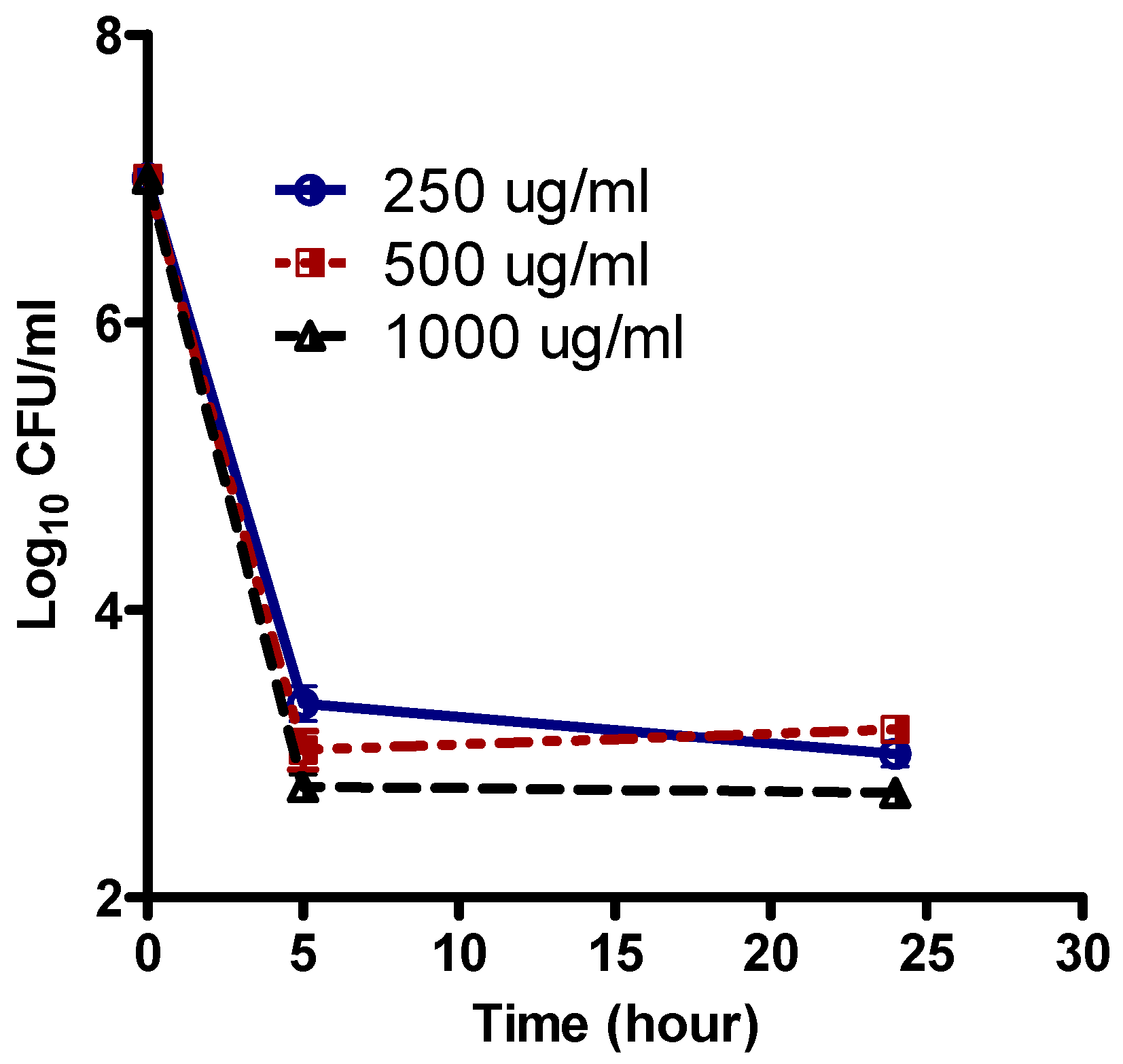
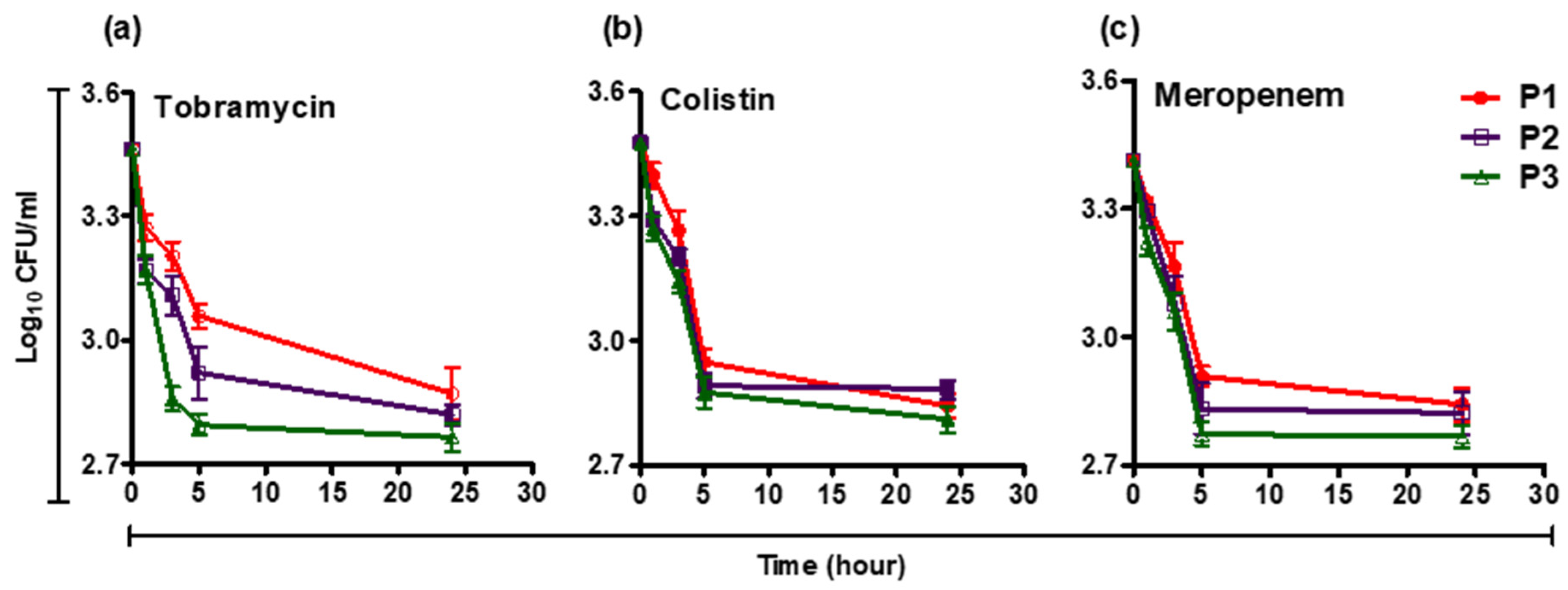
3.2. Survival Curve of Persisters in the Presence of Levofloxacin, Tobramycin, Meropenem, and Colistin
3.3. Biofilm Forming Capacity of Isolates
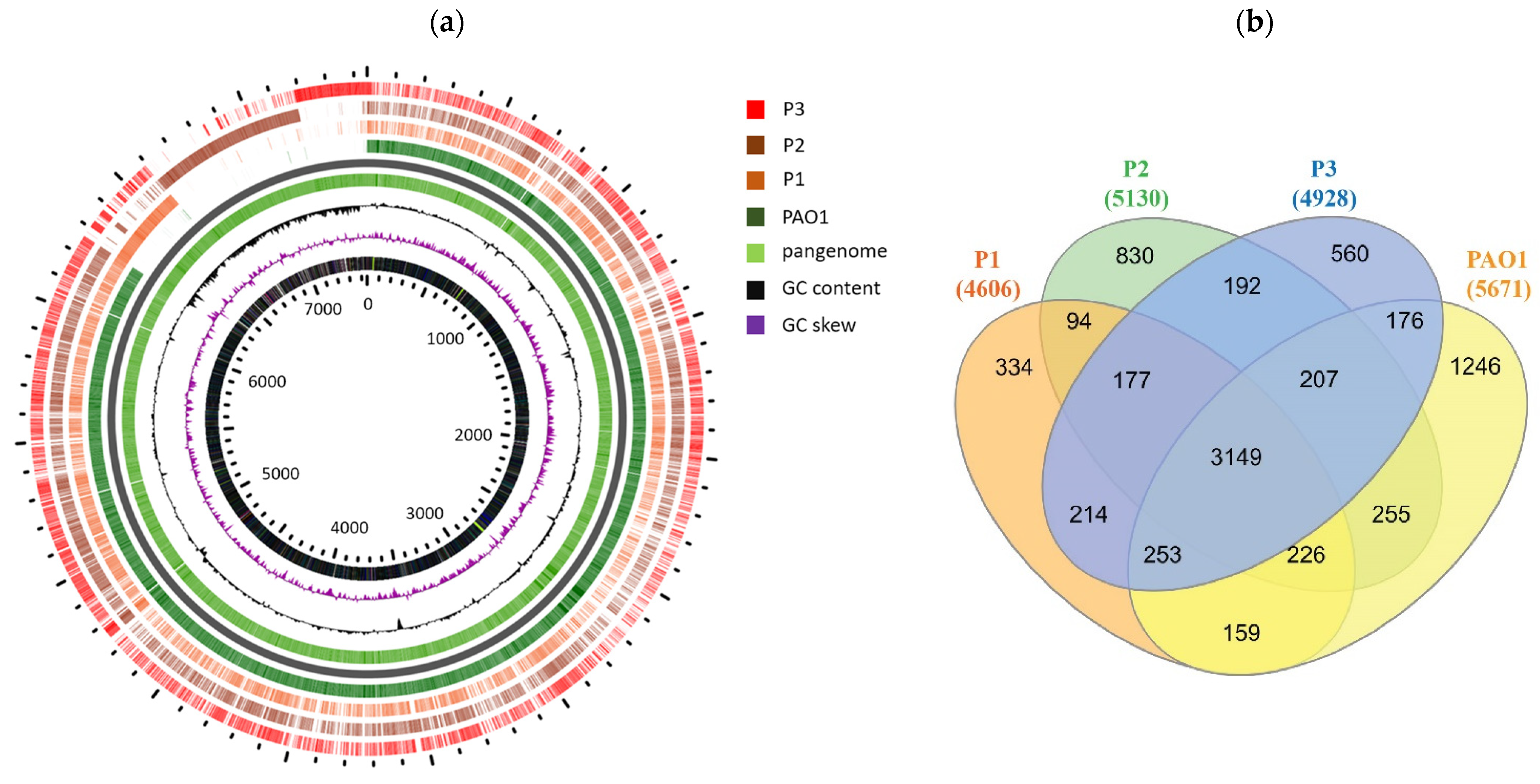
3.4. Whole Genome Sequencing (WGS) Reveals Distinct Features of the Persisters
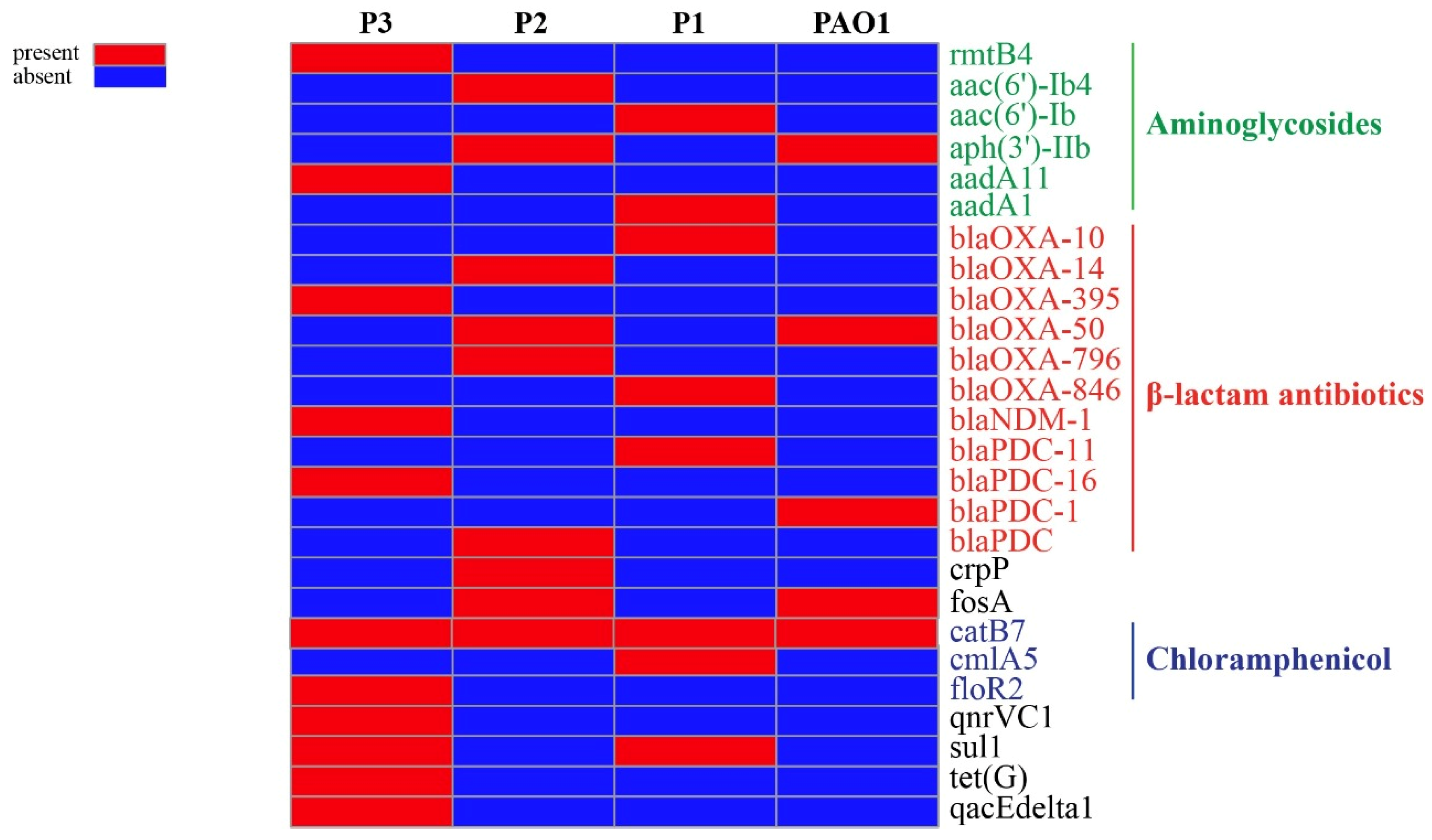
3.5. Antimicrobial Resistance Elements
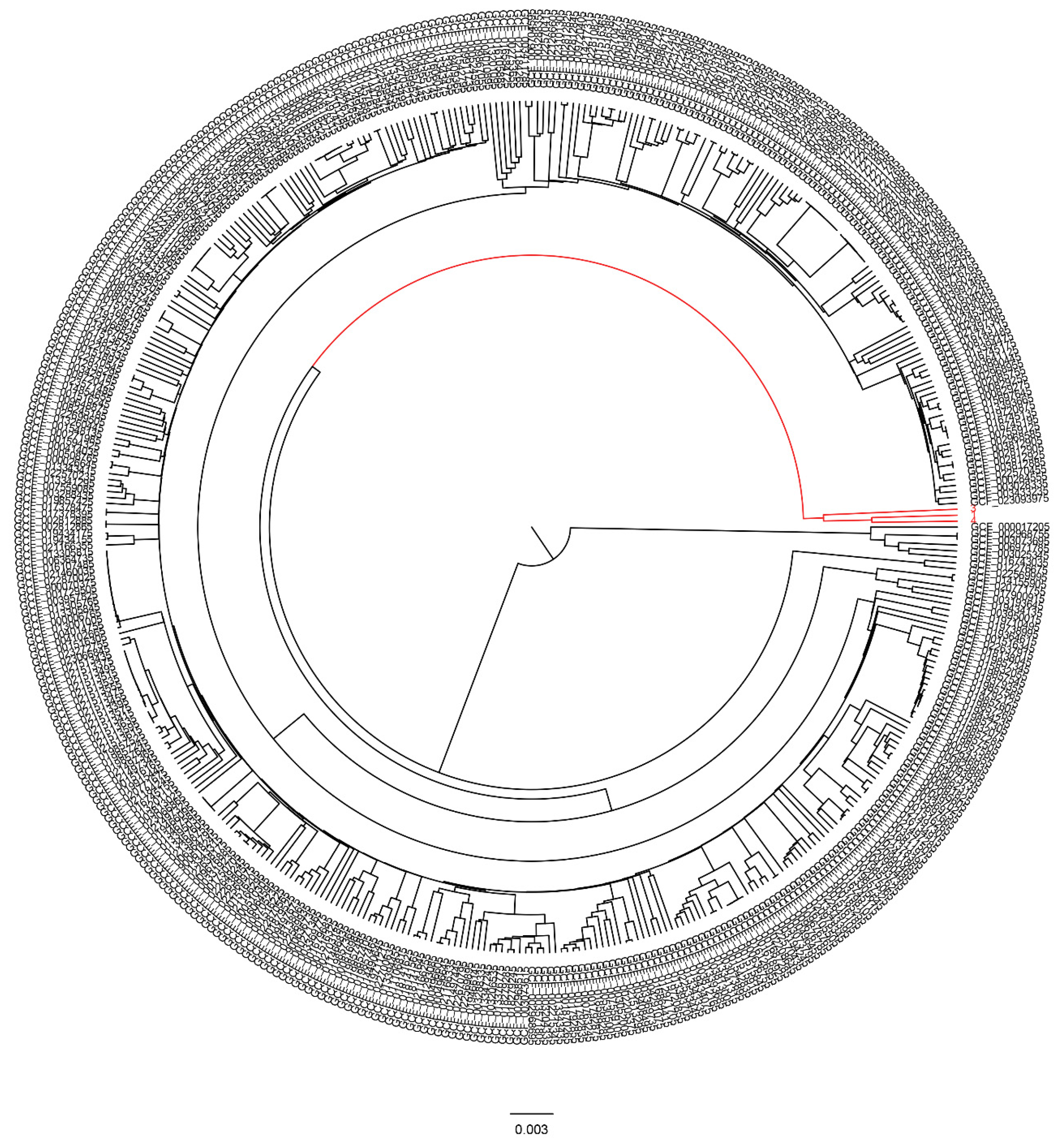
3.6. Global Phylogenetic Analysis of the Persister Isolates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kadry, A.A.; Serry, F.M.; El-Ganiny, A.M.; El-Baz, A.M. Integron occurrence is linked to reduced biocide susceptibility in multidrug resistant Pseudomonas aeruginosa. Br. J. Biomed. Sci. 2017, 74, 78–84. [Google Scholar] [CrossRef]
- Abbas, H.A.; El-Ganiny, A.M.; Kamel, H.A. Phenotypic and genotypic detection of antibiotic resistance of Pseudomonas aeruginosa isolated from urinary tract infections. Afr. Health Sci. 2018, 18, 11–21. [Google Scholar] [CrossRef]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef]
- Cameron, D.R.; Shan, Y.; Zalis, E.A.; Isabella, V.; Lewis, K. A genetic determinant of persister cell formation in bacterial pathogens. J. Bacteriol. 2018, 200, e00303–e00318. [Google Scholar] [CrossRef]
- Beharry, Z.; Palzkill, T. Functional analysis of active site residues of the fosfomycin resistance enzyme FosA from Pseudomonas aeruginosa. J. Biol. Chem. 2005, 280, 17786–17791. [Google Scholar] [CrossRef]
- Willenborg, J.; Willms, D.; Bertram, R.; Goethe, R.; Valentin-Weigand, P. Characterization of multi-drug tolerant persister cells in Streptococcus suis. BMC Microbiol. 2014, 14, 120. [Google Scholar] [CrossRef]
- Bigger, J.W. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet 1944, 244, 497–500. [Google Scholar] [CrossRef]
- Moyed, H.S.; Bertrand, K.P. HipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 1983, 155, 768–775. [Google Scholar] [CrossRef]
- Fisher, R.A.; Gollan, B.; Helaine, S. Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 2017, 15, 453–564. [Google Scholar] [CrossRef]
- Pu, Y.; Zhao, Z.; Li, Y.; Zou, J.; Ma, Q.; Zhao, Y.; Ke, Y.; Zhu, Y.; Chen, H.; Baker, M.A.; et al. Enhanced efflux activity facilitates drug tolerance in dormant bacterial cells. Mol. Cell 2016, 62, 284–294. [Google Scholar] [CrossRef]
- Thi, M.T.T.; Wibowo, D.; Rehm, B.H.A. Pseudomonas aeruginosa Biofilms. Int. J. Mol. Sci. 2020, 21, 8671. [Google Scholar] [CrossRef]
- Seleem, N.M.; Abd El Latif, H.K.; Shaldam, M.A.; El-Ganiny, A.M. Drugs with new lease of life as quorum sensing inhibitors: For combating MDR Acinetobacter baumannii infections. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1687–1702. [Google Scholar] [CrossRef]
- Ahmed, F.; Mirani, Z.A.; Mirani, P.N.; Imdad, M.J.; Khan, F.Z.; Khan, M.N.; Khan, A.B.; Li, Y.; Zhao, Y. Pseudomonas aeruginosa response to acidic stress and imipenem resistance. Appl. Sci. 2022, 12, 8357. [Google Scholar] [CrossRef]
- Soares, A.; Alexandre, K.; Etienne, M. Tolerance and persistence of Pseudomonas aeruginosa in biofilms exposed to antibiotics: Molecular mechanisms, antibiotic strategies and therapeutic perspectives. Front. Microbiol. 2020, 11, 2057. [Google Scholar] [CrossRef]
- Wiuff, C.; Zappala, R.M.; Regoes, R.R.; Garner, K.N.; Baquero, F.; Levin, B.R. Phenotypic tolerance: Antibiotic enrichment of noninherited resistance in bacterial populations. Antimicrob. Agents Chemother. 2005, 49, 1483–1494. [Google Scholar] [CrossRef]
- Kwan, B.W.; Valenta, J.A.; Benedik, M.J.; Wood, T.K. Arrested protein synthesis increases persister-like cell formation. Antimicrob. Agents Chemother. 2013, 57, 1468–1473. [Google Scholar] [CrossRef]
- Lewis, K. Persister cells. Annu. Rev. Microbiol. 2010, 64, 357–372. [Google Scholar] [CrossRef]
- Kussell, E.; Leibler, S. Phenotypic diversity, population growth, and information in fluctuating environments. Science 2005, 309, 2075–2078. [Google Scholar] [CrossRef]
- Kaspy, I.; Rotem, E.; Weiss, N.; Ronin, I.; Balaban, N.Q.; Glaser, G. HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nat. Commun. 2013, 4, 3001. [Google Scholar] [CrossRef]
- Germain, E.; Castro-Roa, D.; Zenkin, N.; Gerdes, K. Molecular mechanism of bacterial persistence by HipA. Mol. cell. 2013, 52, 248–254. [Google Scholar] [CrossRef]
- Pacios, O.; Blasco, L.; Bleriot, I.; Fernandez-Garcia, L.; Ambroa, A.; López, M.; Bou, G.; Cantón, R.; Garcia-Contreras, R.; Wood, T.K.; et al. (p) ppGpp and its role in bacterial persistence: New challenges. Antimicrob. Agents Chemother. 2020, 64, e01283-20. [Google Scholar] [CrossRef]
- Maisonneuve, E.; Castro-Camargo, M.; Gerdes, K. (p) ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 2013, 154, 1140–1150. [Google Scholar] [CrossRef]
- Wang, T.; El Meouche, I.; Dunlop, M.J. Bacterial persistence induced by salicylate via reactive oxygen species. Sci. Rep. 2017, 7, 43839. [Google Scholar] [CrossRef] [PubMed]
- Hazan, R.; Maura, D.; Que, Y.; Rahme, L.G. Assessing Pseudomonas aeruginosa persister/antibiotic tolerant Cell. In Pseudomonas Methods and Protocols. Methods in Molecular Biology; Filloux, A., Ramos, J.L., Eds.; Humana: New York, NY, USA, 2014; Volume 1149, pp. 699–707. [Google Scholar] [CrossRef]
- Wood, T.K.; Knabel, S.J.; Kwan, B.W. Bacterial persister cell formation and dormancy. Appl. Environ. Microbiol. 2013, 79, 7116–7121. [Google Scholar] [CrossRef] [PubMed]
- Žiemytė, M.; Carda-Diéguez, M.; Rodríguez-Díaz, J.C.; Ventero, M.P.; Mira, A.; Ferrer, M.D. Real-time monitoring of Pseudomonas aeruginosa biofilm growth dynamics and persister cells’ eradication. Emerg. Microbes Infect. 2021, 10, 2062–2075. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.H. Determination of viable cell counts: Bacterial growth curves. In Experiments in Molecular Genetics; Miller, J.H., Ed.; Cold Spring Harbor: New York, NY, USA, 1972; pp. 31–36. [Google Scholar]
- De Groote, V.N.; Fauvart, M.; Kint, C.I.; Verstraeten, N.; Jans, A.; Cornelis, P.; Michiels, J. Pseudomonas aeruginosa fosfomycin resistance mechanisms affect non-inherited fluoroquinolone tolerance. J. Med. Microbiol. 2011, 60, 329–336. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; 27th Informational Supplement; M100–S28; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Narayanaswamy, V.P.; Keagy, L.L.; Duris, K.; Wiesmann, W.; Loughran, A.J.; Townsend, S.M.; Baker, S. Novel Glycopolymer eradicates antibiotic- and CCCP-induced persister cells in Pseudomonas aeruginosa. Front. Microbiol. 2018, 9, 1724. [Google Scholar] [CrossRef]
- Stepanović, S.; Vuković, D.; Hola, V.; Di Bonaventura, G.; Djukić, S.; Cirković, I.; Ruzicka, F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 2007, 115, 891–899. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef] [PubMed]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef]
- Tonkin-Hill, G.; MacAlasdair, N.; Ruis, C.; Weimann, A.; Horesh, G.; Lees, J.A.; Gladstone, R.A.; Lo, S.; Beaudoin, C.; Floto, R.A.; et al. Producing polished prokaryotic pangenomes with the panaroo pipeline. Genome Biol. 2020, 21, 180. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef]
- Alikhan, N.-F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genomics. 2011, 12, 402. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef]
- Cohen, N.R.; Lobritz, M.A.; Collins, J.J. Microbial persistence and the road to drug resistance. Cell Host Microbe. 2013, 13, 632–642. [Google Scholar] [CrossRef]
- Orman, M.A.; Brynildsen, M.P. Establishment of a method to rapidly assay bacterial persister metabolism. Antimicrob. Agents Chemother. 2013, 57, 4398–4409. [Google Scholar] [CrossRef]
- Mulcahy, L.R.; Burns, J.L.; Lory, S.; Lewis, K. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J. Bacteriol. 2010, 192, 6191–6199. [Google Scholar] [CrossRef] [PubMed]
- Bartell, J.A.; Cameron, D.R.; Mojsoska, B.; Haagensen, J.A.; Pressler, T.; Sommer, L.M.; Lewis, K.; Molin, S.; Johansen, H.K. Bacterial persisters in long-term infection: Emergence and fitness in a complex host environment. PLoS Pathogens 2020, 16, e1009112. [Google Scholar] [CrossRef] [PubMed]
- Lechner, S.; Lewis, K.; Bertram, R. Staphylococcus aureus persisters tolerant to bactericidal antibiotics. J. Mol. Microbiol. Biotechnol. 2012, 22, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Guérillot, R.; Kostoulias, X.; Donovan, L.; Li, L.; Carter, G.P.; Hachani, A.; Vandelannoote, K.; Giulieri, S.; Monk, I.R.; Kunimoto, M.; et al. Unstable chromosome rearrangements in Staphylococcus aureus cause phenotype switching associated with persistent infections. Proc. Natl. Acad. Sci. USA 2019, 116, 20135–20140. [Google Scholar] [CrossRef] [PubMed]
- Salcedo-Sora, J.E.; Kell, D.B. A Quantitative survey of bacterial persistence in the presence of antibiotics: Towards antipersister antimicrobial discovery. Antibiotics 2020, 9, 508. [Google Scholar] [CrossRef]
- Coates, J.; Park, B.R.; Le, D.; Şimşek, E.; Chaudhry, W.; Kim, M. Antibiotic-induced population fluctuations and stochastic clearance of bacteria. Elife 2018, 7, e32976. [Google Scholar] [CrossRef]
- Datar, R.; Coello Pelegrin, A.; Orenga, S.; Chalansonnet, V.; Mirande, C.; Dombrecht, J.; Perry, J.D.; Perry, A.; Goossens, H.; van Belkum, A. Phenotypic and genomic variability of serial peri-lung transplantation Pseudomonas aeruginosa isolates from cystic fibrosis patients. Front. Microbiol. 2021, 12, 604555. [Google Scholar] [CrossRef]
- Karatuna, O.; Yagci, A. Analysis of quorum sensing-dependent virulence factor production and its relationship with antimicrobial susceptibility in Ps. aeruginosa respiratory isolates. Clin. Microbiol. Infect. 2010, 16, 1770–1775. [Google Scholar] [CrossRef]
- Asbury, J.; Jazayeri, J.A. The significance of Pseudomonas aeruginosa infection and biofilms to cystic fibrosis patients: The need for the development of new therapies. Adv. Biotechnol. Microbiol. 2018, 8, 555742. [Google Scholar] [CrossRef]
- Drenkard, E.; Ausubel, F.M. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 2002, 416, 740–743. [Google Scholar] [CrossRef]
- Jurado-Martín, I.; Sainz-Mejías, M.; McClean, S. Pseudomonas aeruginosa: An audacious pathogen with an adaptable arsenal of virulence factors. Int. J. Mol. Sci. 2021, 22, 3128. [Google Scholar] [CrossRef]
- Stover, C.K.; Pham, X.Q.; Erwin, A.L.; Mizoguchi, S.D.; Warrener, P.; Hickey, M.J.; Brinkman, F.S.; Hufnagle, W.O.; Kowalik, D.J.; Lagrou, M.; et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 2000, 406, 959–964. [Google Scholar] [CrossRef] [PubMed]
- Mathee, K.; Narasimhan, G.; Valdes, C.; Qiu, X.; Matewish, J.M.; Koehrsen, M.; Rokas, A.; Yandava, C.N.; Engels, R.; Zeng, E.; et al. Dynamics of Pseudomonas aeruginosa genome evolution. Proc. Natl. Acad. Sci. USA 2008, 105, 3100–3105. [Google Scholar] [CrossRef] [PubMed]
- Alcalde-Rico, M.; Olivares-Pacheco, J.; Alvarez-Ortega, C.; Cámara, M.; Martínez, J.L. Role of the multidrug resistance efflux pump MexCD-OprJ in the Pseudomonas aeruginosa quorum sensing response. Front. Microbiolol. 2018, 9, 2752. [Google Scholar] [CrossRef]
- Bhandari, S.; Adhikari, S.; Karki, D.; Chand, A.B.; Sapkota, S.; Dhungel, B.; Banjara, M.R.; Joshi, P.; Lekhak, B.; Rijal, K.R. Antibiotic resistance, biofilm formation and detection of mexA/mexB efflux-pump genes among clinical isolates of Pseudomonas aeruginosa in a tertiary care hospital, Nepal. Front. Trop. Dis. 2022, 2, 810863. [Google Scholar] [CrossRef]
- Adewoye, L.; Sutherland, A.; Srikumar, R.; Poole, K. The mexR repressor of the mexAB-oprM multidrug efflux operon in Pseudomonas aeruginosa: Characterization of mutations compromising activity. J. Bacteriol. 2002, 184, 4308–4312. [Google Scholar] [CrossRef]
- Sobel, M.L.; Hocquet, D.; Cao, L.; Plesiat, P.; Poole, K. Mutations in PA3574 (nalD) lead to increased MexAB-OprM expression and multidrug resistance in laboratory and clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2005, 49, 1782–1786. [Google Scholar] [CrossRef]
- Suresh, M.; Nithya, N.; Jayasree, P.R.; Vimal, K.P.; Manish Kumar, P.R. Mutational analyses of regulatory genes, mexR, nalC, nalD and mexZ of mexAB-oprM and mexXY operons, in efflux pump hyperexpressing multidrug-resistant clinical isolates of Pseudomonas aeruginosa. World J. Microbiol. Biotechnol. 2018, 34, 83. [Google Scholar] [CrossRef] [PubMed]
- Amsalu, A.; Sapula, S.A.; De Barros Lopes, M.; Hart, B.J.; Nguyen, A.H.; Drigo, B.; Turnidge, J.; Leong, L.E.; Venter, H. Efflux pump-driven antibiotic and biocide cross-resistance in Pseudomonas aeruginosa isolated from different ecological niches: A case study in the development of multidrug resistance in environmental hotspots. Microorganisms 2020, 8, 1647. [Google Scholar] [CrossRef] [PubMed]

| Strain Code | MICs (µg/mL) | MDR | |||||||
|---|---|---|---|---|---|---|---|---|---|
| CHL | AK | LEV | CEP | FEP | MEM | CT | TOB | ||
| PAO1 | 2 (S) | 4 (S) | 0.5 (S) | 4 (S) | 1 (S) | 0.5 (S) | 1 (S) | 2 (S) | - |
| P1 | 256 (R) | 128 (R) | 8 (R) | 256 (R) | 64 (R) | 64 (R) | 16 (R) | 128 (R) | 5 |
| P2 | 512 (R) | 512 (R) | 8 (R) | 512 (R) | 128 (R) | 16 (R) | 32 (R) | 128 (R) | 5 |
| P3 | 256 (R) | 128 (R) | 16 (R) | 512 (R) | 64 (R) | 16 (R) | 16 (R) | 512 (R) | 5 |
| Isolate | Absorbance (OD) at 570 nm | Biofilm Forming Capacity |
|---|---|---|
| P1 | 0.967 | S |
| P2 | 0.831 | S |
| P3 | 0.872 | S |
| PAO1 | 0.584 | S |
| NP | 0.547 | S |
| ODc | 0.1 | Control (no biofilm) |
| Persister Number | Efflux Pump Component | Efflux Pump | Mutation in MexAB-OprM Efflux Pump Regulatory Proteins | ||
|---|---|---|---|---|---|
| MFP | RND | OMP | |||
| P1 | MexA | MexB | OprM | MexAB-OprM | MexR (V126E), NalC (S209R, G71E) |
| MexC | MexD | OprJ | MexCD-OprJ | ||
| MexJ | MexK | OpmH | MexJK-OpmH | ||
| MexM | MexN | OprM | MexMN-OprM | ||
| MexP | MexQ | - | |||
| MexV | MexW | OprM | MexVW-OprM | ||
| P2 | MexA | MexB | OprM | MexAB-OprM | MexR (V126E), NalC (S209R, G71E) |
| MexC | MexD | OprJ | MexCD-OprJ | ||
| MexE | MexF | OprN | MexEF-OprN | ||
| MexG | MexH,I | - | |||
| MexJ MexM | MexK MexN | OpmH OprM | MexJK-OpmH MexMN-OprM | ||
| MexP | MexQ | - | |||
| MexV | MexW | OprM | MexVW-OprM | ||
| P3 | MexA | MexB | OprM | MexAB-OprM | MexR (V126E), NalC (S209R, G71E) |
| MexC | MexD | OprJ | MexCD-OprJ | ||
| MexE | MexF | OprN | MexEF-OprN | ||
| MexG | MexH,I | OpmD | MexGHI-OpmD | ||
| MexJ | MexK | OpmH | MexJK-OpmH | ||
| MexM | MexN | OprM | MexMN-OprM | ||
| MexP | MexQ | - | |||
| MexV | MexW | OprM | MexVW-OprM | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baiomy, A.A.; Serry, F.E.; Kadry, A.A.; Yahya, G.; Doijad, S.; Mostafa, A.; Mraheil, M.A.; El-Ganiny, A.M. Genome Analysis of Pseudomonas aeruginosa Strains from Chronically Infected Patients with High Levels of Persister Formation. Pathogens 2023, 12, 426. https://doi.org/10.3390/pathogens12030426
Baiomy AA, Serry FE, Kadry AA, Yahya G, Doijad S, Mostafa A, Mraheil MA, El-Ganiny AM. Genome Analysis of Pseudomonas aeruginosa Strains from Chronically Infected Patients with High Levels of Persister Formation. Pathogens. 2023; 12(3):426. https://doi.org/10.3390/pathogens12030426
Chicago/Turabian StyleBaiomy, Amr A., Fathy E. Serry, Ashraf A. Kadry, Galal Yahya, Swapnil Doijad, Ahmed Mostafa, Mobarak Abu Mraheil, and Amira M. El-Ganiny. 2023. "Genome Analysis of Pseudomonas aeruginosa Strains from Chronically Infected Patients with High Levels of Persister Formation" Pathogens 12, no. 3: 426. https://doi.org/10.3390/pathogens12030426
APA StyleBaiomy, A. A., Serry, F. E., Kadry, A. A., Yahya, G., Doijad, S., Mostafa, A., Mraheil, M. A., & El-Ganiny, A. M. (2023). Genome Analysis of Pseudomonas aeruginosa Strains from Chronically Infected Patients with High Levels of Persister Formation. Pathogens, 12(3), 426. https://doi.org/10.3390/pathogens12030426






