Quorum Quenching with a Diffusible Signal Factor Analog in Stenotrophomonas maltophilia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains of Stenotrophomonas maltophilia
2.2. Trimethoprim-Sulfamethoxazole Susceptibility Test
2.3. Detection of Resistance Genes
2.4. Multilocus Sequence Typing
2.5. Biofilm Formation and Reduction
2.6. Motility and Motility Reduction Test
2.7. Viability Test
2.8. Statistical Analysis
3. Results
3.1. Trimethoprim–Sulfamethoxazole Susceptibility Test
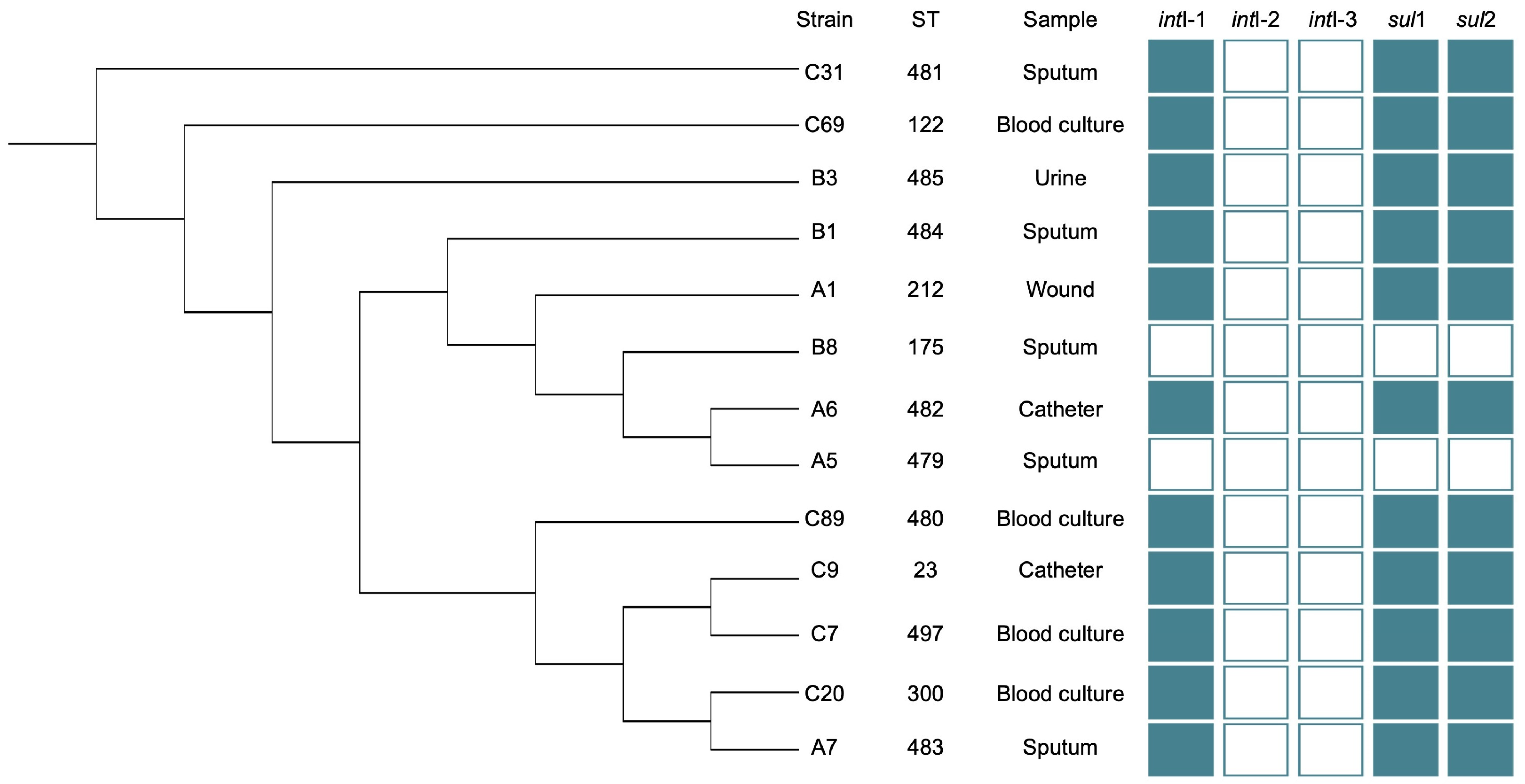
3.2. Detection of Resistance Genes
3.3. Multilocus Sequence Typing
3.4. Biofilm Formation and Reduction Assay
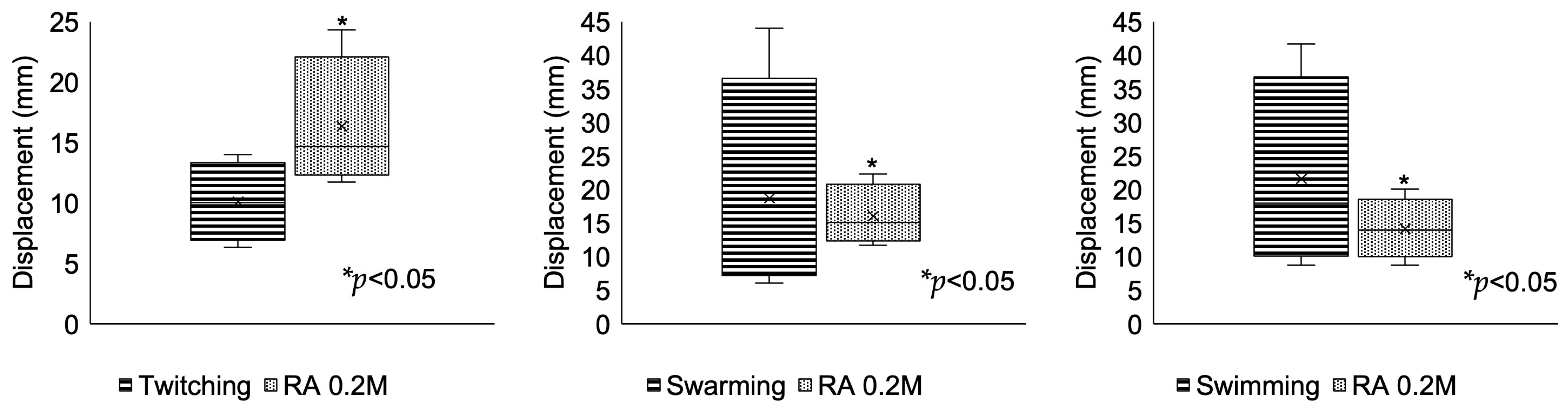
3.5. Motility and Interruption Assay
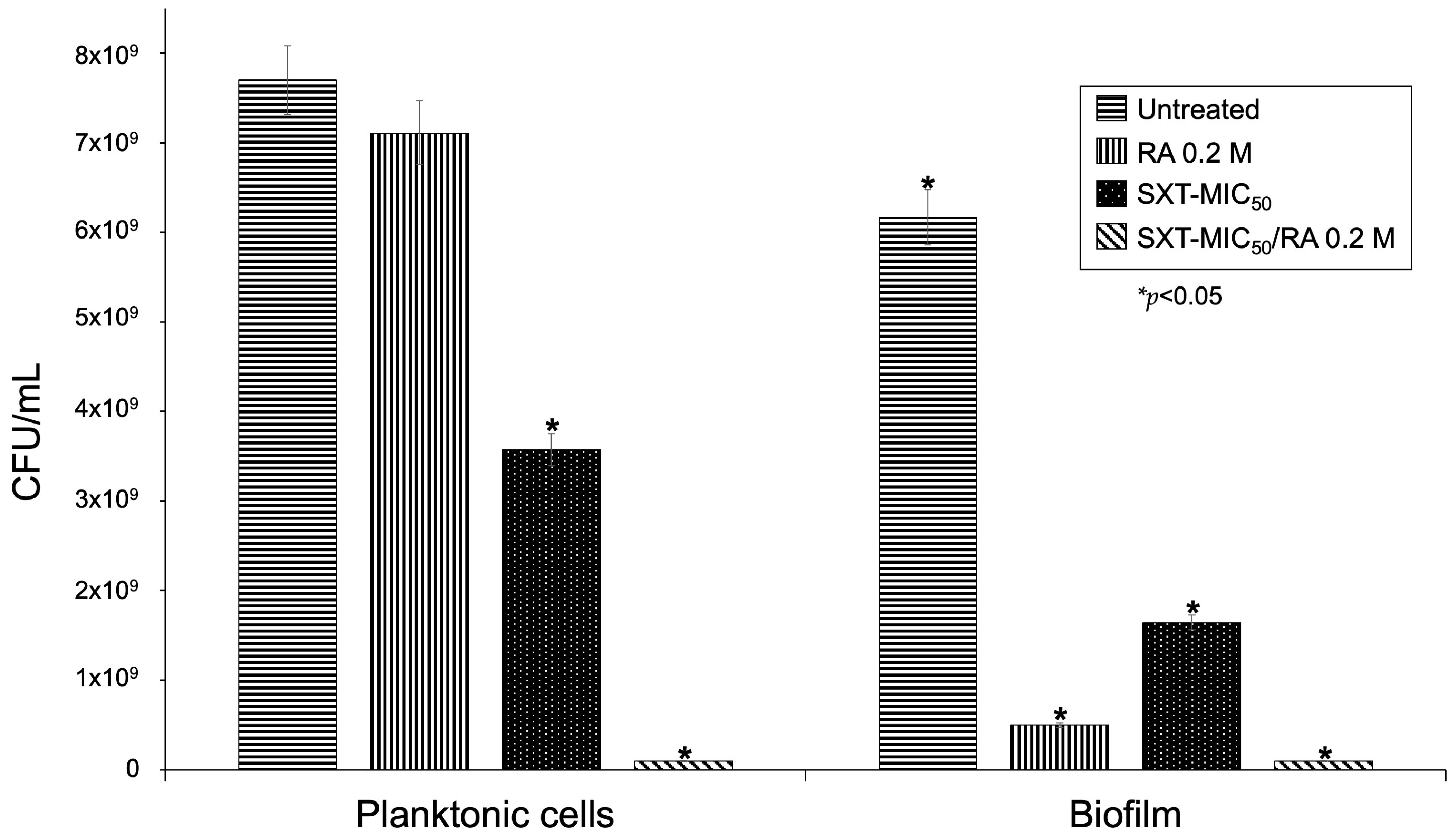
3.6. Viability Assay
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barbolla, R.; Catalano, M.; Orman, B.E.; Famiglietti, A.; Vay, C.; Smayevsky, J.; Centrón, D.; Pineiro, S.A. Class 1 integrons increase trimethoprim-sulfamethoxazole MICs against epidemiologically unrelated Stenotrophomonas maltophifilia isolates. Antimicrob. Agents Chemother. 2004, 48, 666–669. [Google Scholar] [CrossRef] [PubMed]
- Kaur, P.; Gautam, V.; Tewari, R. Distribution of class 1 integrons, sul1 and sul2 genes among clinical isolates of Stenotrophomonas maltophilia from a tertiary care hospital in North India. Microb. Drug Resist. 2015, 21, 380–385. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.F.; Chang, X.; Ye, Y.; Wang, Z.X.; Shao, Y.B.; Shi, W.; Li, X.; Li, J.B. Stenotrophomonas maltophilia resistance to trimethoprim/sulfamethoxazole mediated by acquisition of sul and dfrA genes in a plasmid-mediated class 1 integron. Int. J. Antimicrob. Agents 2011, 37, 230–234. [Google Scholar] [CrossRef] [PubMed]
- Hathroubi, S.; Mekni, M.A.; Domenico, P.; Nguyen, D.; Jacques, M. Biofilms: Microbial Shelters Against Antibiotics. Microb. Drug Resist. 2017, 23, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Magana, M.; Sereti, C.; Ioannidis, A.; Mitchell, C.A.; Ball, A.R.; Magiorkinis, E.; Chatzipanagiotou, S.; Hamblin, M.R.; Hadjifrangiskou, M.; Tegos, G.P. Options and Limitations in Clinical Investigation of Bacterial Biofilms. Clin. Microbiol. Rev. 2018, 31, e00084-16. [Google Scholar] [CrossRef] [PubMed]
- López-Jácome, L.E.; Garza-Ramos, G.; Hernández-Durán, M.; Franco-Cendejas, R.; Loarca, D.; Romero-Martínez, D.; Nguyen, P.T.D.; Maeda, T.; González-Pedrajo, B.; Díaz-Guerrero, M.; et al. AiiM Lactonase Strongly Reduces Quorum Sensing Controlled Virulence Factors in Clinical Strains of Pseudomonas aeruginosa Isolated From Burned Patients. Front. Microbiol. 2019, 10, 2657. [Google Scholar] [CrossRef]
- AboZahra, R. Quorum sensing and interspecies interactions in Stenotrophomonas maltophilia. Microbiol. Res. J. Int. 2013, 3, 414–422. Available online: https://scholar.google.com.eg/citations?view_op=view_citation&hl=en&user=lqKKYwMAAAAJ&citation_for_view=lqKKYwMAAAAJ:d1gkVwhDpl0C (accessed on 11 September 2022). [CrossRef]
- Martínez, P.; Huedo, P.; Martinez-Servat, S.; Planell, R.; Ferrer-Navarro, M.; Daura, X.; Yero, D.; Gibert, I. Stenotrophomonas maltophilia responds to exogenous AHL signals through the LuxR solo SmoR (Smlt1839). Front. Cell. Infect. Microbiol. 2015, 5, 41. [Google Scholar] [CrossRef]
- Huedo, P.; Kumar, V.P.; Horgan, C.; Yero, D.; Daura, X.; Gibert, I.; O’Sullivan, T.P. Sulfonamide-based difusible signal factor analogs interfere with quorum sensing in Stenotrophomonas maltophilia and Burkholderia cepacian. Future Med. Chem. 2019, 11, 1565–1582. [Google Scholar] [CrossRef]
- Huedo, P.; Coves, X.; Daura, X.; Gibert, I.; Yero, D. Quorum Sensing Signaling and Quenching in the Multidrug-Resistant Pathogen Stenotrophomonas maltophilia. Front. Cell. Infect. Microbiol. 2018, 8, 122. [Google Scholar] [CrossRef]
- Paluch, E.; Rewak-Soroczyńska, J.; Jędrusik, I.; Mazurkiewicz, E.; Jermakow, K. Prevention of biofilm formation by quorum quenching. Appl. Microbiol. Biotechnol. 2020, 104, 1871–1881. [Google Scholar] [CrossRef]
- Marques, C.N.; Davies, D.G.; Sauer, K. Control of Biofilms with the Fatty Acid Signaling Molecule cis-2-Decenoic Acid. Pharmaceuticals 2015, 8, 816–835. [Google Scholar] [CrossRef]
- Hamada, Y.; Kiso, Y. The application of bioisosteres in drug design for novel drug discovery: Focusing on acid protease inhibitors. Expert Opin. Drug Discov. 2012, 7, 903–922. [Google Scholar] [CrossRef]
- Tan, F.; She, P.; Zhou, L.; Liu, Y.; Chen, L.; Luo, Z.; Wu, Y. Bactericidal and Anti-biofilm Activity of the Retinoid Compound CD437 Against Enterococcus faecalis. Front. Microbiol. 2019, 10, 2301. [Google Scholar] [CrossRef]
- Kam, R.K.; Deng, Y.; Chen, Y.; Zhao, H. Retinoic acid synthesis and functions in early embryonic development. Cell Biosci. 2012, 2, 11. [Google Scholar] [CrossRef]
- Bayeva, N.; Coll, E.; Piskareva, O. Differentiating Neuroblastoma: A Systematic Review of the Retinoic Acid, Its Derivatives, and Synergistic Interactions. J. Pers. Med. 2021, 11, 211. [Google Scholar] [CrossRef]
- Campione, E.; Gaziano, R.; Marino, D.; Orlandi, A. Fungistatic activity of all-trans retinoic acid against Aspergillus fumigatus and Candida albicans. Drug Des. Dev. Ther. 2016, 10, 1551–1555. [Google Scholar] [CrossRef]
- Abdelhamid, L.; Luo, X.M. Retinoic Acid, Leaky Gut, and Autoimmune Diseases. Nutrients 2018, 10, 1016. [Google Scholar] [CrossRef]
- Bouriez, D.; Giraud, J.; Gronnier, C.; Varon, C. Efficiency of All-Trans Retinoic Acid on Gastric Cancer: A Narrative Literature Review. Int. J. Mol. Sci. 2018, 19, 3388. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, L.M.; Teixeira, F.M.E.; Sato, M.N. Impact of Retinoic Acid on Immune Cells and Inflammatory Diseases. Mediat. Inflamm. 2018, 2018, 3067126. [Google Scholar] [CrossRef] [PubMed]
- Pistoia, E.S.; Cosio, T.; Campione, E.; Pica, F.; Volpe, A.; Marino, D.; Di Francesco, P.; Monari, C.; Fontana, C.; Favaro, M.; et al. All-Trans Retinoic Acid Effect on Candida albicans Growth and Biofilm Formation. J. Fungi 2022, 8, 1049. [Google Scholar] [CrossRef]
- ElBaradei, A.; Yakout, M.A. Stenotrophomonas maltophilia: Genotypic Characterization of Virulence Genes and The Effect of Ascorbic Acid on Biofilm Formation. Curr. Microbiol. 2022, 79, 180. [Google Scholar] [CrossRef]
- García, G.; Girón, J.; Yañez, J. Stenotrophomonas maltophilia an Its Ability to Form Biofilms. Microbiol. Res. 2023, 14, 1. [Google Scholar] [CrossRef]
- CLSI Supplement M100; Performance Standards for Antimicrobial Susceptibility Testing. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. Available online: https://scholar.google.com/scholar_lookup?title=CLSI+Supplement+M100&publication_year=2020& (accessed on 21 July 2020).
- White, P.A.; McIver, C.J.; Deng, Y.; Rawlinson, W.D. Characterisation of two new gene cassettes, aadA5 and dfrA17. FEMS Microbiol. Lett. 2000, 182, 265–269. [Google Scholar] [CrossRef]
- Machado, E.; Cantón, R.; Baquero, F.; Galán, J.C.; Rollán, A.; Peixe, L.; Coque, T.M. Integron contento f exdended-spectrum-beta-lactamase-producing Escherichia coli strains over 12 years in a single hospital in Madrid, Spain. Antimicrob. Agents Chemoter. 2005, 49, 1823–1829. [Google Scholar] [CrossRef]
- Goldstein, C.; Lee, M.D.; Sanchez, S.; Hudson, C.; Phillips, B.; Register, B.; Grady, M.; Liebert, C.; Summers, A.O.; White, D.G.; et al. Incidence of class 1 and 2 integrases in clinical and commensal bacteria from livestock, companion animals, and exotics. Antimicrob. Agents Chemother. 2000, 45, 723–726. [Google Scholar] [CrossRef]
- Toleman, M.A.; Bennett, P.M.; Walsh, T.R. Common regions e.g. orf513 and antibiotic resistance: IS91-like elements evolving complex class 1 integrons. J. Antimicrob. Chemother. 2006, 58, 1–6. [Google Scholar] [CrossRef]
- Pitcher, D.G.; Saunders, N.A.; Owen, R.J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Appl. Microbiol. 1989, 8, 151–156. [Google Scholar] [CrossRef]
- Kaiser, S.; Biehler, K.; Jonas, D.A. Stenotrophomonas maltophilia multilocus sequence typing scheme for inferring population structure. J. Bacteriol. 2009, 191, 2934–2943. [Google Scholar] [CrossRef]
- Francisco, A.P.; Vaz, C.; Monteiro, P.T.; Melo-Cristino, J.; Ramirez, M.; Carriço, J.A. PHYLOViZ: Phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinform. 2012, 13, 87. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Stepanović, S.; Vuković, D.; Hola, V.; Bonaventura, G.D.; Djukić, S.; Ćirković, I.; Ruzicka, F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 2007, 115, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Di Bonaventura, G.; Spedicato, I.; D’Antonio, D.; Robuffo, I.; Piccolomini, R. Biofilm formation by Stenotrophomonas maltophilia: Modulation by quinolones, trimethoprim-sulfamethoxazole, and ceftazidime. Antimicrob. Agents Chemoter. 2004, 48, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.H.; Kornberg, A. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2000, 97, 4885–4890. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Live (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Flores-Treviño, S.; Bocanegra-Ibarias, P.; Camacho-Ortiz, A.; Morfín-Otero, R.; Salazar-Sesatty, H.A.; Garza-González, E. Stenotrophomonas maltophilia biofilm: Its role in infection diseases. Expert Rev. Anti Infect. Ther. 2019, 17, 877–893. [Google Scholar] [CrossRef]
- Cruz-Córdova, A.; Mancilla-Rojano, J.; Luna-Pineda, V.M.; Escalona-Venegas, G.; Cázares-Domínguez, V.; Ormsby, C.; Franco-Hernández, I.; Zavala-Vega, S.; Hernández, M.A.; Medina-Pelcastre, M.; et al. Molecular Epidemiology, Antibiotic Resistance, and Virulence Traits of Stenotrophomonas maltophilia Strains Associated With an Outbreak in a Mexican Tertiary Care Hospital. Front. Cell. Infect. Microbiol. 2020, 10, 50. [Google Scholar] [CrossRef]
- Usta, E.; Eroğlu, C.; Yanık, K.; Karadağ, A.; Güney, A.K.; Günaydın, M. Klinik Stenotrophomonas maltophilia izolatlarında sınıf 1, 2, 3 integron varlığının ve antibiyotik direnci ile ilişkilerinin araştırılması. Mikrobiyol. Bul. 2015, 49, 35–46. [Google Scholar] [CrossRef]
- Kaushik, M.; Kumar, S.; Kapoor, R.K.; Virdi, J.S.; Gulati, P. Integrons in Enterobacteriaceae: Diversity, distribution and epidemiology. Int. J. Antimicrob. Agents 2018, 51, 167–176. [Google Scholar] [CrossRef]
- Fundación Instituto de Inmunología de Colombia. Available online: http://www.saludcapital.gov.co/DSP/Infecciones%20Asociadas%20a%20Atencin%20en%20Salud/Comites/2017/Agosto/Tipificaci%C3%B3n_Molecular.pdf (accessed on 3 August 2019).
- Esposito, A.; Pompilio, A.; Bettua, C.; Crocetta, V.; Giacobazzi, E.; Fiscarelli, E.; Jousson, O.; Di Bonaventura, G. Evolution of Stenotrophomonas maltophilia in Cystic Fibrosis Lung over Chronic Infection: A Genomic and Phenotypic Population Study. Front. Microbiol. 2017, 8, 1590. [Google Scholar] [CrossRef]
- Landová, B.; Šilhán, J. Conformational changes of DNA repair glycosylase MutM triggered by DNA binding. FEBS Lett. 2020, 594, 3032–3044. [Google Scholar] [CrossRef] [PubMed]
- Ryan, R.P. Cyclic di-GMP signalling and the regulation of bacterial virulence. Microbiology 2013, 159 Pt 7, 1286–1297. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Song, S.; Yang, C.; Sun, X.; Huang, Y.; Li, K.; Zhao, S.; Zhang, Y.; Deng, Y. Disruption of Quorum Sensing and Virulence in Burkholderia cenocepacia by a Structural Analogue of the cis-2-Dodecenoic Acid Signal. Appl. Environ. Microbiol. 2019, 85, e00105-19. [Google Scholar] [CrossRef] [PubMed]
- An, S.Q.; Murtagh, J.; Twomey, K.B.; Gupta, M.K.; O’Sullivan, T.P.; Ingram, R.; Valvano, M.A.; Tang, J.L. Modulation of antibiotic sensitivity and biofilm formation in Pseudomonas aeruginosa by interspecies signal analogues. Nat. Commun. 2019, 10, 2334. [Google Scholar] [CrossRef]
- Kim, H.R.; Lee, D.; Eom, Y.B. Anti-biofilm and Anti-Virulence Efficacy of Celastrol Against Stenotrophomonas maltophilia. Int. J. Med. Sci. 2018, 15, 617–627. [Google Scholar] [CrossRef]
- Montoya-Hinojosa, E.; Bocanegra-Ibarias, P.; Garza-González, E.; Alonso-Ambriz, Ó.M.; Salazar-Mata, G.A.; Villarreal-Treviño, L.; Pérez-Alba, E.; Camacho-Ortiz, A.; Morfín-Otero, R.; Rodríguez-Noriega, E.; et al. Discrimination of biofilm-producing Stenotrophomonas maltophilia clinical strains by matrix-assisted laser desorption ionization-time of flight. PLoS ONE 2020, 15, e0244751. [Google Scholar] [CrossRef]


| Gene | Primer 1 | Sequence 5′–3′ | Molecular Size (pb) | Reference |
|---|---|---|---|---|
| intI-1 | intI1-F intI1-R | TCATGGCTTGTTATGACTGT GTAGGGCTTATTATGCACGC | 580 | [25] |
| intI-2 | intI2-F int2-R | CACGGATATGCCACAAAAAGGT GTAGCAAACGAGTGACGAAATG | 240 | [26] |
| intI-3 | intI3-F int3-R | AGTGGGTGGCGAATGAGTG TGTTCTTGTATCGGCAGGTG | 758 | [27] |
| sul1 | sul1-F sul1-R | ATGGTGACGGTGTTCGGCATTCTGA CTAGGCATGATCTAACCCTCGGTCT | 900 | [28] |
| sul2 | sul2-F | GCGCTCAAGGCAGATGGCATT | 293 | [2] |
| sul2-R | GCGTTTGATACCGGCACCCGT |
| Strains | Trimethoprim/Sulfamethoxazole MIC (µg/mL) | Susceptibility |
|---|---|---|
| A1 | 16/304 | R |
| A5 | 2/38 | S |
| A6 | 16/304 | R |
| A7 | 16/304 | R |
| B1 | 16/304 | R |
| B3 | 8/152 | R |
| B8 | 0.25/4.75 | S |
| C7 | 32/608 | R |
| C9 | 32/608 | R |
| C20 | 16/304 | R |
| C31 | 16/304 | R |
| C69 | 16/304 | R |
| C89 | 16/304 | R |
| S. maltophilia ATCC 17666 | 16/304 | R |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guillén-Navarro, D.; González-Vázquez, R.; León-Ávila, G.; Giono-Cerezo, S. Quorum Quenching with a Diffusible Signal Factor Analog in Stenotrophomonas maltophilia. Pathogens 2023, 12, 1448. https://doi.org/10.3390/pathogens12121448
Guillén-Navarro D, González-Vázquez R, León-Ávila G, Giono-Cerezo S. Quorum Quenching with a Diffusible Signal Factor Analog in Stenotrophomonas maltophilia. Pathogens. 2023; 12(12):1448. https://doi.org/10.3390/pathogens12121448
Chicago/Turabian StyleGuillén-Navarro, Dafne, Rosa González-Vázquez, Gloria León-Ávila, and Silvia Giono-Cerezo. 2023. "Quorum Quenching with a Diffusible Signal Factor Analog in Stenotrophomonas maltophilia" Pathogens 12, no. 12: 1448. https://doi.org/10.3390/pathogens12121448
APA StyleGuillén-Navarro, D., González-Vázquez, R., León-Ávila, G., & Giono-Cerezo, S. (2023). Quorum Quenching with a Diffusible Signal Factor Analog in Stenotrophomonas maltophilia. Pathogens, 12(12), 1448. https://doi.org/10.3390/pathogens12121448






