Polyinfection in Fish Aeromoniasis: A Study of Co-Isolated Aeromonas Species in Aeromonas veronii Outbreaks
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Genomic Analysis
2.3. Phenotypic Characterization
2.4. In Vivo Virulence
3. Results and Discussion
3.1. Genomic Analysis
3.1.1. Genome Assemblies and Characteristics
3.1.2. Genome Similarity
3.1.3. Prediction of Genomic Islands, CRISPR Systems, Prophages, and Insertion Sequences
3.1.4. Antibiotic Resistance Genes
3.1.5. Virulence Factors
3.2. Phenotypic Characterization
3.2.1. Morphology and Biochemical Characteristics
3.2.2. Hemolytic Activity
3.2.3. Antibiotic Susceptibility
3.2.4. In Vivo Virulence
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Smyrli, M.; Prapas, A.; Rigos, G.; Kokkari, C.; Pavlidis, M.; Katharios, P. Aeromonas veronii Infection Associated with High Morbidity and Mortality in Farmed European Seabass Dicentrarchus labrax in the Aegean Sea, Greece. Fish Pathol. 2017, 52, 68–81. [Google Scholar] [CrossRef]
- Beaz-Hidalgo, R.; Alperi, A.; Buján, N.; Romalde, J.L.; Figueras, M.J. Comparison of phenotypical and genetic identification of Aeromonas strains isolated from diseased fish. Syst. Appl. Microbiol. 2010, 33, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Scarano, C.; Piras, F.; Virdis, S.; Ziino, G.; Nuvoloni, R.; Dalmasso, A.; De Santis, E.P.L.; Spanu, C. Antibiotic resistance of Aeromonas ssp. strains isolated from Sparus aurata reared in Italian mariculture farms. Int. J. Food Microbiol. 2018, 284, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Smyrli, M.; Triga, A.; Dourala, N.; Varvarigos, P.; Pavlidis, M.; Quoc, V.H.; Katharios, P. Comparative Study on A Novel Pathogen of European Seabass. Diversity of Aeromonas veronii in the Aegean Sea. Microorganisms 2019, 7, 504. [Google Scholar] [CrossRef]
- Amal, M.N.A.; Koh, C.B.; Nurliyana, M.; Suhaiba, M.; Nor-Amalina, Z.; Santha, S.; Diyana-Nadhirah, K.P.; Yusof, M.T.; Ina-Salwany, M.Y.; Zamri-Saad, M. A case of natural co-infection of Tilapia Lake Virus and Aeromonas veronii in a Malaysian red hybrid tilapia (Oreochromis niloticus × O. mossambicus) farm experiencing high mortality. Aquaculture 2018, 485, 12–16. [Google Scholar] [CrossRef]
- Chandrarathna, H.P.S.U.; Nikapitiya, C.; Dananjaya, S.H.S.; Wijerathne, C.U.B.; Wimalasena, S.H.M.P.; Kwun, H.J.; Heo, G.-J.; Lee, J.; De Zoysa, M. Outcome of co-infection with opportunistic and multidrug resistant Aeromonas hydrophila and A. veronii in zebrafish: Identification, characterization, pathogenicity and immune responses. Fish Shellfish. Immunol. 2018, 80, 573–581. [Google Scholar] [CrossRef]
- Fuentes-Valencia, M.A.; Osornio-Esquivel, J.L.; Martínez Palacios, C.A.; Contreras-Ávila, J.L.; Barriga-Tovar, E.; la Mora, G.I.; Arellano-Torres, A.; Baizabal-Aguirre, V.M.; Bravo-Patiño, A.; Cajero-Juárez, M.; et al. Bacterial and parasite co-infection in Mexican golden trout (Oncorhynchus chrysogaster) by Aeromonas bestiarum, Aeromonas sobria, Plesiomonas shigelloides and Ichthyobodo necator. BMC Vet. Res. 2022, 18, 137. [Google Scholar] [CrossRef]
- Proietti-Junior, A.A.; Lima, L.S.; Roges, E.M.; Rodrigues, Y.C.; Lima, K.V.B.; Rodrigues, D.P.; Tavares-Dias, M. Experimental co-infection by Aeromonas hydrophila and Aeromonas jandaei in pirarucu Arapaima gigas (Pisces: Arapaimidae). Aquac. Res. 2021, 52, 1688–1696. [Google Scholar] [CrossRef]
- Okon, E.M.; Okocha, R.C.; Taiwo, A.B.; Michael, F.B.; Bolanle, A.M. Dynamics of co-infection in fish: A review of pathogen-host interaction and clinical outcome. Fish Shellfish Immunol. Rep. 2023, 4, 100096. [Google Scholar] [CrossRef]
- Fernández-Bravo, A.; Kilgore, P.B.; Andersson, J.A.; Blears, E.; Figueras, M.J.; Hasan, N.A.; Colwell, R.R.; Sha, J.; Chopra, A.K. T6SS and ExoA of flesh-eating Aeromonas hydrophila in peritonitis and necrotizing fasciitis during mono- and polymicrobial infections. Proc. Natl. Acad. Sci. USA 2019, 116, 24084–24092. [Google Scholar] [CrossRef]
- Mosser, T.; Talagrand-Reboul, E.; Colston, S.M.; Graf, J.; Figueras, M.J.; Jumas-Bilak, E.; Lamy, B. Exposure to pairs of Aeromonas strains enhances virulence in the Caenorhabditis elegans infection model. Front. Microbiol. 2015, 6, 1218. [Google Scholar] [CrossRef]
- Saraceni, P.R.; Romero, A.; Figueras, A.; Novoa, B. Establishment of Infection Models in Zebrafish Larvae (Danio rerio) to Study the Pathogenesis of Aeromonas hydrophila. Front. Microbiol. 2016, 7, 1219. [Google Scholar] [CrossRef] [PubMed]
- Sibley, C.D.; Duan, K.; Fischer, C.; Parkins, M.D.; Storey, D.G.; Rabin, H.R.; Surette, M.G. Discerning the Complexity of Community Interactions Using a Drosophila Model of Polymicrobial Infections. PLoS Pathog. 2008, 4, e1000184. [Google Scholar] [CrossRef] [PubMed]
- Robert, A.; Talagrand-Reboul, E.; Figueras, M.-J.; Ruimy, R.; Boyer, L.; Lamy, B. Drosophila melanogaster Systemic Infection Model to Study Altered Virulence during Polymicrobial Infection by Aeromonas. Pathogens 2023, 12, 405. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yu, C.; Li, Z.; et al. SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. GigaScience 2018, 7, gix120. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed]
- Wattam, A.R.; Brettin, T.; Davis, J.J.; Gerdes, S.; Kenyon, R.; Machi, D.; Mao, C.; Olson, R.; Overbeek, R.; Pusch, G.D.; et al. Assembly, annotation, and comparative genomics in PATRIC, the all bacterial bioinformatics resource center. In Comparative Genomics: Methods and Protocols; Humana Press: New York, NY, USA, 2018; pp. 79–101. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Waterhouse, R.M.; Seppey, M.; Simão, F.A.; Manni, M.; Ioannidis, P.; Klioutchnikov, G.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO Applications from Quality Assessments to Gene Prediction and Phylogenomics. Mol. Biol. Evol. 2018, 35, 543–548. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef]
- Li, W.; O’Neill, K.R.; Haft, D.H.; DiCuccio, M.; Chetvernin, V.; Badretdin, A.; Coulouris, G.; Chitsaz, F.; Derbyshire, M.K.; Durkin, A.S.; et al. RefSeq: Expanding the Prokaryotic Genome Annotation Pipeline reach with protein family model curation. Nucleic Acids Res. 2021, 49, D1020–D1028. [Google Scholar] [CrossRef]
- Dhillon, B.K.; Laird, M.R.; Shay, J.A.; Winsor, G.L.; Lo, R.; Nizam, F.; Pereira, S.K.; Waglechner, N.; McArthur, A.G.; Langille, M.G.I.; et al. IslandViewer 3: More flexible, interactive genomic island discovery, visualization and analysis: Figure 1. Nucleic Acids Res. 2015, 43, W104–W108. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zheng, D.; Zhou, S.; Chen, L.; Yang, J. VFDB 2022: A general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022, 50, D912–D917. [Google Scholar] [CrossRef]
- Siguier, P.; Varani, A.; Perochon, J.; Chandler, M. Exploring bacterial insertion sequences with ISfinder: Objectives, uses, and future developments. In Mobile Genetic Elements. Methods in Molecular Biology; Humana Press: New York, NY, USA, 2012; pp. 91–103. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef]
- Lee, I.; Ouk Kim, Y.; Park, S.-C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Fernández-Álvarez, C.; Gijón, D.; Álvarez, M.; Santos, Y. First isolation of Aeromonas salmonicida subspecies salmonicida from diseased sea bass, Dicentrarchus labrax (L.), cultured in Spain. Aquac. Rep. 2016, 4, 36–41. [Google Scholar] [CrossRef]
- Hudzicki, J. Kirby-Bauer disk diffusion susceptibility test protocol. Am. Soc. Microbiol. 2009, 15, 55–63. [Google Scholar]
- Droubogiannis, S.; Katharios, P. Genomic and Biological Profile of a Novel Bacteriophage, Vibrio phage Virtus, Which Improves Survival of Sparus aurata Larvae Challenged with Vibrio harveyi. Pathogens 2022, 11, 630. [Google Scholar] [CrossRef] [PubMed]
- Noon, K.R.; Guymon, R.; Crain, P.F.; McCloskey, J.A.; Thomm, M.; Lim, J.; Cavicchioli, R. Influence of Temperature on tRNA Modification in Archaea: Methanococcoides burtonii (Optimum Growth Temperature [Topt], 23 °C) and Stetteria hydrogenophila (Topt, 95 °C). J. Bacteriol. 2003, 185, 5483–5490. [Google Scholar] [CrossRef] [PubMed]
- Satapathy, S.S.; Dutta, M.; Ray, S.K. Higher tRNA diversity in thermophilic bacteria: A possible adaptation to growth at high temperature. Microbiol. Res. 2010, 165, 609–616. [Google Scholar] [CrossRef]
- Vincent, A.T.; Trudel, M.V.; Freschi, L.; Nagar, V.; Gagné-Thivierge, C.; Levesque, R.C.; Charette, S.J. Increasing genomic diversity and evidence of constrained lifestyle evolution due to insertion sequences in Aeromonas salmonicida. BMC Genom. 2016, 17, 44. [Google Scholar] [CrossRef]
- Vincent, A.T.; Charette, S.J. To Be or Not to Be Mesophilic, That Is the Question for Aeromonas salmonicida. Microorganisms 2022, 10, 240. [Google Scholar] [CrossRef]
- Triga, A.; Smyrli, M.; Katharios, P. Aeromoniasis. In Aquaculture Pathophysiology; Elsevier: Amsterdam, The Netherlands, 2022; pp. 317–327. [Google Scholar] [CrossRef]
- Vincent, A.T.; Rouleau, F.D.; Moineau, S.; Charette, S.J. Study of mesophilic Aeromonas salmonicida A527 strain sheds light on the species’ lifestyles and taxonomic dilemma. FEMS Microbiol. Lett. 2017, 364, fnx239. [Google Scholar] [CrossRef]
- Uhrynowski, W.; Decewicz, P.; Dziewit, L.; Radlinska, M.; Krawczyk, P.S.; Lipinski, L.; Adamska, D.; Drewniak, L. Analysis of the Genome and Mobilome of a Dissimilatory Arsenate Reducing Aeromonas sp. O23A Reveals Multiple Mechanisms for Heavy Metal Resistance and Metabolism. Front. Microbiol. 2017, 8, 936. [Google Scholar] [CrossRef]
- Drewniak, L.; Matlakowska, R.; Rewerski, B.; Sklodowska, A. Arsenic release from gold mine rocks mediated by the activity of indigenous bacteria. Hydrometallurgy 2010, 104, 437–442. [Google Scholar] [CrossRef]
- Lukasz, D.; Liwia, R.; Aleksandra, M.; Aleksandra, S. Dissolution of Arsenic Minerals Mediated by Dissimilatory Arsenate Reducing Bacteria: Estimation of the Physiological Potential for Arsenic Mobilization. BioMed Res. Int. 2014, 2014, 841892. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Tian, Z.; Gillings, M.R.; Zhang, Y.; Zhang, H.; Huyan, J.; Yang, M. Novel Transposon Tn 6433 Variants Accelerate the Dissemination of tet (E) in Aeromonas in an Aerobic Biofilm Reactor under Oxytetracycline Stresses. Environ. Sci. Technol. 2020, 54, 6781–6791. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Zhang, Y.; Wu, X.; Zhang, H.; Yang, M.; Tian, Z. Potential dissemination mechanism of the tetC gene in Aeromonas media from the aerobic biofilm reactor under oxytetracycline stresses. J. Environ. Sci. 2021, 105, 90–99. [Google Scholar] [CrossRef]
- Maslowska, K.H.; Makiela-Dzbenska, K.; Fijalkowska, I.J. The SOS system: A complex and tightly regulated response to DNA damage. Environ. Mol. Mutagen. 2019, 60, 368–384. [Google Scholar] [CrossRef] [PubMed]
- McDonnell, G.E.; McConnell, D.J. Overproduction, isolation, and DNA-binding characteristics of Xre, the repressor protein from the Bacillus subtilis defective prophage PBSX. J. Bacteriol. 1994, 176, 5831–5834. [Google Scholar] [CrossRef]
- Long, M.; Fan, H.; Gan, Z.; Jiang, Z.; Tang, S.; Xia, H.; Lu, Y. Comparative genomic analysis provides insights into taxonomy and temperature adaption of Aeromonas salmonicida. J. Fish Dis. 2023, 46, 545–561. [Google Scholar] [CrossRef]
- Siguier, P.; Gourbeyre, E.; Varani, A.; Ton-Hoang, B.; Chandler, M. Everyman’s Guide to Bacterial Insertion Sequences. Microbiol. Spectr. 2015, 3, 555–590. [Google Scholar] [CrossRef]
- Bello-López, J.M.; Cabrero-Martínez, O.A.; Ibáñez-Cervantes, G.; Hernández-Cortez, C.; Pelcastre-Rodríguez, L.I.; Gonzalez-Avila, L.U.; Castro-Escarpulli, G. Horizontal Gene Transfer and Its Association with Antibiotic Resistance in the Genus Aeromonas spp. Microorganisms 2019, 7, 363. [Google Scholar] [CrossRef]
- Sørum, H.; L’Abée-Lund, T.M. Antibiotic resistance in food-related bacteria—A result of interfering with the global web of bacterial genetics. Int. J. Food Microbiol. 2002, 78, 43–56. [Google Scholar] [CrossRef]
- Piotrowska, M.; Popowska, M. Insight into the mobilome of Aeromonas strains. Front. Microbiol. 2015, 6, 494. [Google Scholar] [CrossRef]
- Govender, R.; Amoah, I.D.; Adegoke, A.A.; Singh, G.; Kumari, S.; Swalaha, F.M.; Bux, F.; Stenström, T.A. Identification, antibiotic resistance, and virulence profiling of Aeromonas and Pseudomonas species from wastewater and surface water. Environ. Monit. Assess. 2021, 193, 294. [Google Scholar] [CrossRef] [PubMed]
- Igbinosa, I.H.; Okoh, A.I. Antibiotic Susceptibility Profile of Aeromonas Species Isolated from Wastewater Treatment Plant. Sci. World J. 2012, 2012, 764563. [Google Scholar] [CrossRef] [PubMed]
- Palú, A.P.; Gomes, L.M.; Miguel, M.A.L.; Balassiano, I.T.; Queiroz, M.L.P.; Freitas-Almeida, A.C.; de Oliveira, S.S. Antimicrobial resistance in food and clinical Aeromonas isolates. Food Microbiol. 2006, 23, 504–509. [Google Scholar] [CrossRef] [PubMed]
- Skwor, T.; Stringer, S.; Haggerty, J.; Johnson, J.; Duhr, S.; Johnson, M.; Seckinger, M.; Stemme, M. Prevalence of Potentially Pathogenic Antibiotic-Resistant Aeromonas spp. in Treated Urban Wastewater Effluents versus Recipient Riverine Populations: A 3-Year Comparative Study. Appl. Environ. Microbiol. 2020, 86, e02053-19. [Google Scholar] [CrossRef]
- Nguyen, F.; Starosta, A.L.; Arenz, S.; Sohmen, D.; Dönhöfer, A.; Wilson, D.N. Tetracycline antibiotics and resistance mechanisms. Biol. Chem. 2014, 395, 559–575. [Google Scholar] [CrossRef]
- Hernández, A.; Sánchez, M.B.; Martínez, J.L. Quinolone Resistance: Much More than Predicted. Front. Microbiol. 2011, 2, 22. [Google Scholar] [CrossRef]
- Kim, J.H.; Hwang, S.Y.; Son, J.S.; Han, J.E.; Jun, J.W.; Shin, S.P.; Choresca, C.; Choi, Y.J.; Park, Y.H.; Park, S.C. Molecular characterization of tetracycline- and quinolone-resistant Aeromonas salmonicida isolated in Korea. J. Vet. Sci. 2011, 12, 41–48. [Google Scholar] [CrossRef][Green Version]
- Barnes, A.C.; Lewin, C.S.; Hastings, T.S.; Amyes, S.G.B. Cross resistance between oxytetracycline and oxolinic acid in Aeromonas salmonicida associated with alterations in outer membrane proteins. FEMS Microbiol. Lett. 1990, 72, 337–339. [Google Scholar] [CrossRef]
- Burr, S.E.; Stuber, K.; Wahli, T.; Frey, J. Evidence for a Type III Secretion System in Aeromonas salmonicida subsp. salmonicida. J. Bacteriol. 2002, 184, 5966–5970. [Google Scholar] [CrossRef]
- Dacanay, A.; Knickle, L.; Solanky, K.S.; Boyd, J.M.; Walter, J.A.; Brown, L.L.; Johnson, S.C.; Reith, M. Contribution of the type III secretion system (TTSS) to virulence of Aeromonas salmonicida subsp. salmonicida. Microbiology 2006, 152, 1847–1856. [Google Scholar] [CrossRef]
- Beaz-Hidalgo, R.; Figueras, M.J. Aeromonas spp. whole genomes and virulence factors implicated in fish disease. J. Fish Dis. 2013, 36, 371–388. [Google Scholar] [CrossRef] [PubMed]
- Dallaire-Dufresne, S.; Tanaka, K.H.; Trudel, M.V.; Lafaille, A.; Charette, S.J. Virulence, genomic features, and plasticity of Aeromonas salmonicida subsp. salmonicida, the causative agent of fish furunculosis. Vet. Microbiol. 2014, 169, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.H.; Vincent, A.T.; Emond-Rheault, J.-G.; Adamczuk, M.; Frenette, M.; Charette, S.J. Plasmid composition in Aeromonas salmonicida subsp. salmonicida 01-B526 unravels unsuspected type three secretion system loss patterns. BMC Genom. 2017, 18, 528. [Google Scholar] [CrossRef] [PubMed]
- Daher, R.K.; Filion, G.; Tan, S.G.E.; Dallaire-Dufresne, S.; Paquet, V.E.; Charette, S.J. Alteration of virulence factors and rearrangement of pAsa5 plasmid caused by the growth of Aeromonas salmonicida in stressful conditions. Vet. Microbiol. 2011, 152, 353–360. [Google Scholar] [CrossRef]
- Tanaka, K.H.; Dallaire-Dufresne, S.; Daher, R.K.; Frenette, M.; Charette, S.J. An Insertion Sequence-Dependent Plasmid Rearrangement in Aeromonas salmonicida Causes the Loss of the Type Three Secretion System. PLoS ONE 2012, 7, e33725. [Google Scholar] [CrossRef]
- Citterio, B.; Biavasco, F. Aeromonas hydrophila virulence. Virulence 2015, 6, 417–418. [Google Scholar] [CrossRef]
- Reith, M.E.; Singh, R.K.; Curtis, B.; Boyd, J.M.; Bouevitch, A.; Kimball, J.; Munholland, J.; Murphy, C.; Sarty, D.; Williams, J.; et al. The genome of Aeromonas salmonicida subsp. salmonicida A449: Insights into the evolution of a fish pathogen. BMC Genom. 2008, 9, 427. [Google Scholar] [CrossRef]
- Tekedar, H.C.; Abdelhamed, H.; Kumru, S.; Blom, J.; Karsi, A.; Lawrence, M.L. Comparative Genomics of Aeromonas hydrophila Secretion Systems and Mutational Analysis of hcp1 and vgrG1 Genes From T6SS. Front. Microbiol. 2019, 9, 3216. [Google Scholar] [CrossRef]
- Merino, S.; Gavín, R.; Vilches, S.; Shaw, J.G.; Tomás, J.M. A Colonization Factor (Production of Lateral Flagella) of Mesophilic Aeromonas spp. Is Inactive in Aeromonas salmonicida Strains. Appl. Environ. Microbiol. 2003, 69, 663–667. [Google Scholar] [CrossRef]
- Canals, R.; Ramirez, S.; Vilches, S.; Horsburgh, G.; Shaw, J.G.; Tomás, J.M.; Merino, S. Polar Flagellum Biogenesis in Aeromonas hydrophila. J. Bacteriol. 2006, 188, 542–555. [Google Scholar] [CrossRef]
- McIntosh, D.; Austin, B. Atypical characteristics of the salmonid pathogen Aeromonas salmonicida. J. Gen. Microbiol. 1991, 137, 1341–1343. [Google Scholar] [CrossRef] [PubMed]
- Hadi, N.; Yang, Q.; Barnett, T.C.; Tabei, S.M.B.; Kirov, S.M.; Shaw, J.G. Bundle-Forming Pilus Locus of Aeromonas veronii bv. Sobria. Infect. Immun. 2012, 80, 1351–1360. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.L.; Arakawa, E.; Leung, K.Y. Novel Aeromonas hydrophila PPD134/91 Genes Involved in O-Antigen and Capsule Biosynthesis. Infect. Immun. 2002, 70, 2326–2335. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, Y.L.; Lau, Y.L.; Arakawa, E.; Leung, K.Y. Detection and genetic analysis of group II capsules in Aeromonas hydrophila. Microbiology 2003, 149, 1051–1060. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jimenez, N.; Lacasta, A.; Vilches, S.; Reyes, M.; Vazquez, J.; Aquillini, E.; Merino, S.; Regué, M.; Tomás, J.M. Genetics and Proteomics of Aeromonas salmonicida Lipopolysaccharide Core Biosynthesis. J. Bacteriol. 2009, 191, 2228–2236. [Google Scholar] [CrossRef]
- Radaev, S.; Dastidar, P.; Patel, M.; Woodard, R.W.; Gatti, D.L. Structure and Mechanism of 3-Deoxy-d-manno-octulosonate 8-Phosphate Synthase. J. Biol. Chem. 2000, 275, 9476–9484. [Google Scholar] [CrossRef]
- Taj, A.; Jia, L.; Sha, S.; Wang, C.; Ullah, H.; Haris, M.; Ma, X.; Ma, Y. Functional analysis and enzyme characterization of mannose-1-phosphate guanylyl transferase (ManB) from Mycobacterium tuberculosis. Res. Microbiol. 2022, 173, 103884. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, X.; Dacanay, A.; Harrison, B.A.; Fast, M.; Colquhoun, D.J.; Lund, V.; Brown, L.L.; Li, J.; Altman, E. Carbohydrate analysis and serological classification of typical and atypical isolates of Aeromonas salmonicida: A rationale for the lipopolysaccharide-based classification of A. salmonicida. Fish Shellfish Immunol. 2007, 23, 1095–1106. [Google Scholar] [CrossRef]
- Noonan, B.; Trust, T.J. The synthesis, secretion and role in virulence of the paracrystalline surface protein layers of Aeromonas salmonicida and A. hydrophila. FEMS Microbiol. Lett. 2006, 154, 1–7. [Google Scholar] [CrossRef]
- Garduño, R.A.; Phipps, B.M.; Baumeister, W.; Kay, W.W. Novel structural patterns in divalent cation-depleted surface layers of Aeromonas salmonicida. J. Struct. Biol. 1992, 109, 184–195. [Google Scholar] [CrossRef]
- Paquet, V.E.; Durocher, A.F.; Charette, S.J. Aeromonas salmonicida intraspecies divergence revealed by the various strategies displayed when grazed by Tetrahymena pyriformis. FEMS Microbiol. Lett. 2022, 369, fnac067. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, E.E.; Kay, W.W.; Ainsworth, T.; Chamberlain, J.B.; Austen, R.A.; Buckley, J.T.; Trust, T.J. Loss of virulence during culture of Aeromonas salmonicida at high temperature. J. Bacteriol. 1981, 148, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Paquet, V.E.; Vincent, A.T.; Moineau, S.; Charette, S.J. Beyond the A-layer: Adsorption of lipopolysaccharides and characterization of bacteriophage-insensitive mutants of Aeromonas salmonicida subsp. salmonicida. Mol. Microbiol. 2019, 112, 667–677. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, J.; Cai, H.; Lin, M.; Zhang, Y.; Huang, L. Enhanced Hemolytic Activity of Mesophilic Aeromonas salmonicida SRW-OG1 Is Brought about by Elevated Temperatures. Microorganisms 2022, 10, 2033. [Google Scholar] [CrossRef] [PubMed]
- Suarez, G.; Khajanchi, B.K.; Sierra, J.C.; Erova, T.E.; Sha, J.; Chopra, A.K. Actin cross-linking domain of Aeromonas hydrophila repeat in toxin A (RtxA) induces host cell rounding and apoptosis. Gene 2012, 506, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Wick, M.J.; Frank, D.W.; Storey, D.G.; Iglewski, B.H. Structure, Function, And Regulation of Pseudomonas Aeruginosa Exotoxin A. Annu. Rev. Microbiol. 1990, 44, 335–363. [Google Scholar] [CrossRef]
- Park, S.Y.; Han, J.E.; Kwon, H.; Park, S.C.; Kim, J.H. Recent Insights into Aeromonas salmonicida and Its Bacteriophages in Aquaculture: A Comprehensive Review. J. Microbiol. Biotechnol. 2020, 30, 1443–1457. [Google Scholar] [CrossRef]
- Roesch, P.L.; Redford, P.; Batchelet, S.; Moritz, R.L.; Pellett, S.; Haugen, B.J.; Blattner, F.R.; Welch, R.A. Uropathogenic Escherichia coli use d-serine deaminase to modulate infection of the murine urinary tract. Mol. Microbiol. 2003, 49, 55–67. [Google Scholar] [CrossRef]
- Miller, R.A.; Walker, R.D.; Baya, A.; Clemens, K.; Coles, M.; Hawke, J.P.; Henricson, B.E.; Hsu, H.M.; Mathers, J.J.; Oaks, J.L.; et al. Antimicrobial Susceptibility Testing of Aquatic Bacteria: Quality Control Disk Diffusion Ranges for Escherichia coli ATCC 25922 and Aeromonas salmonicida subsp. salmonicida ATCC 33658 at 22 and 28 °C. J. Clin. Microbiol. 2003, 41, 4318–4323. [Google Scholar] [CrossRef]
- Yang, X.; Long, M.; Shen, X. Effector–Immunity Pairs Provide the T6SS Nanomachine its Offensive and Defensive Capabilities. Molecules 2018, 23, 1009. [Google Scholar] [CrossRef]
- Ikryannikova, L.N.; Kurbatov, L.K.; Gorokhovets, N.V.; Zamyatnin, A.A. Contact-Dependent Growth Inhibition in Bacteria: Do Not Get Too Close! Int. J. Mol. Sci. 2020, 21, 7990. [Google Scholar] [CrossRef] [PubMed]
- Cascarano, M.C.; Stavrakidis-Zachou, O.; Mladineo, I.; Thompson, K.D.; Papandroulakis, N.; Katharios, P. Mediterranean Aquaculture in a Changing Climate: Temperature Effects on Pathogens and Diseases of Three Farmed Fish Species. Pathogens 2021, 10, 1205. [Google Scholar] [CrossRef] [PubMed]
- Cain, A.K.; Barquist, L.; Goodman, A.L.; Paulsen, I.T.; Parkhill, J.; van Opijnen, T. A decade of advances in transposon-insertion sequencing. Nat. Rev. Genet. 2020, 21, 526–540. [Google Scholar] [CrossRef] [PubMed]

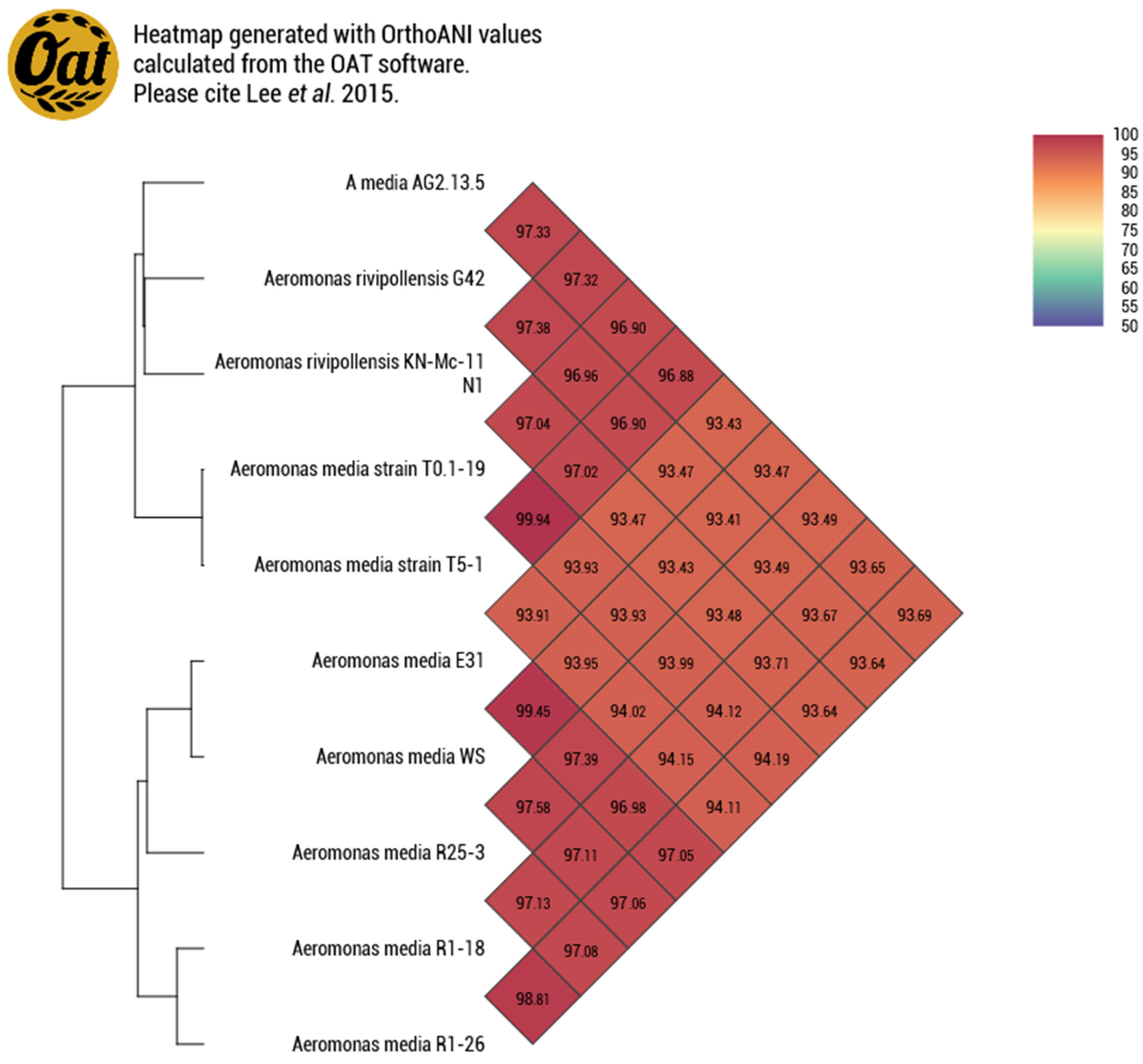
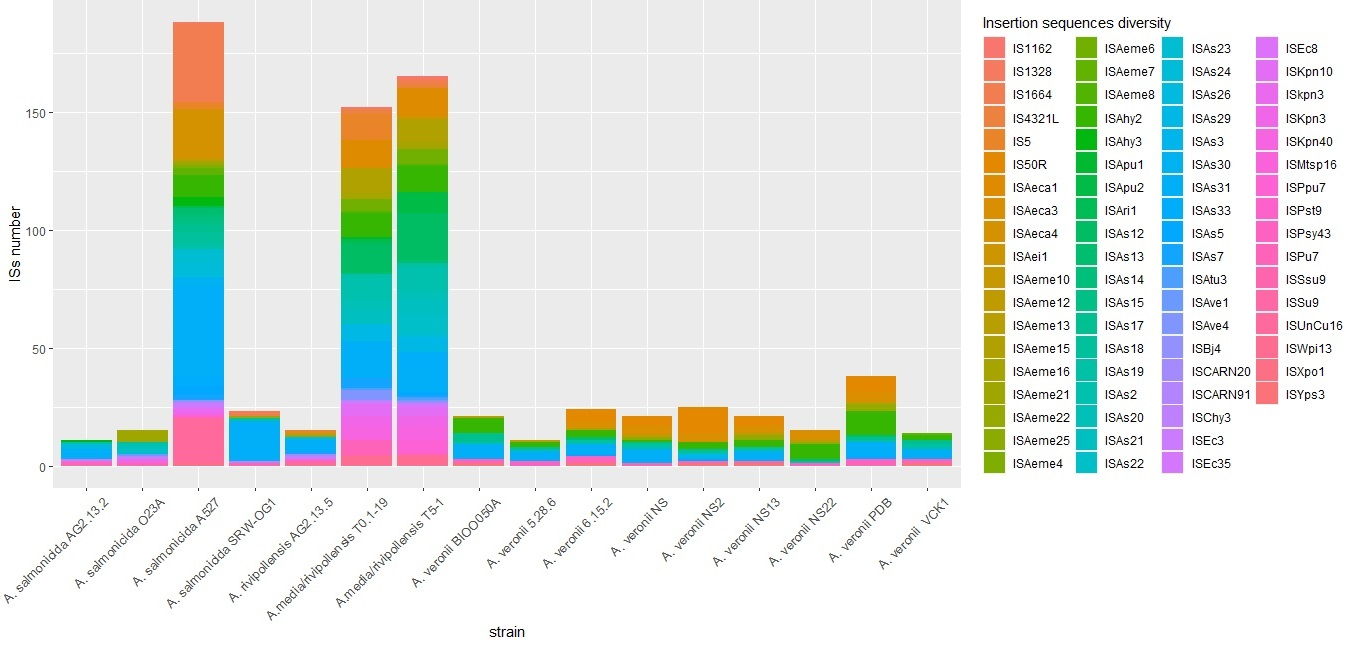
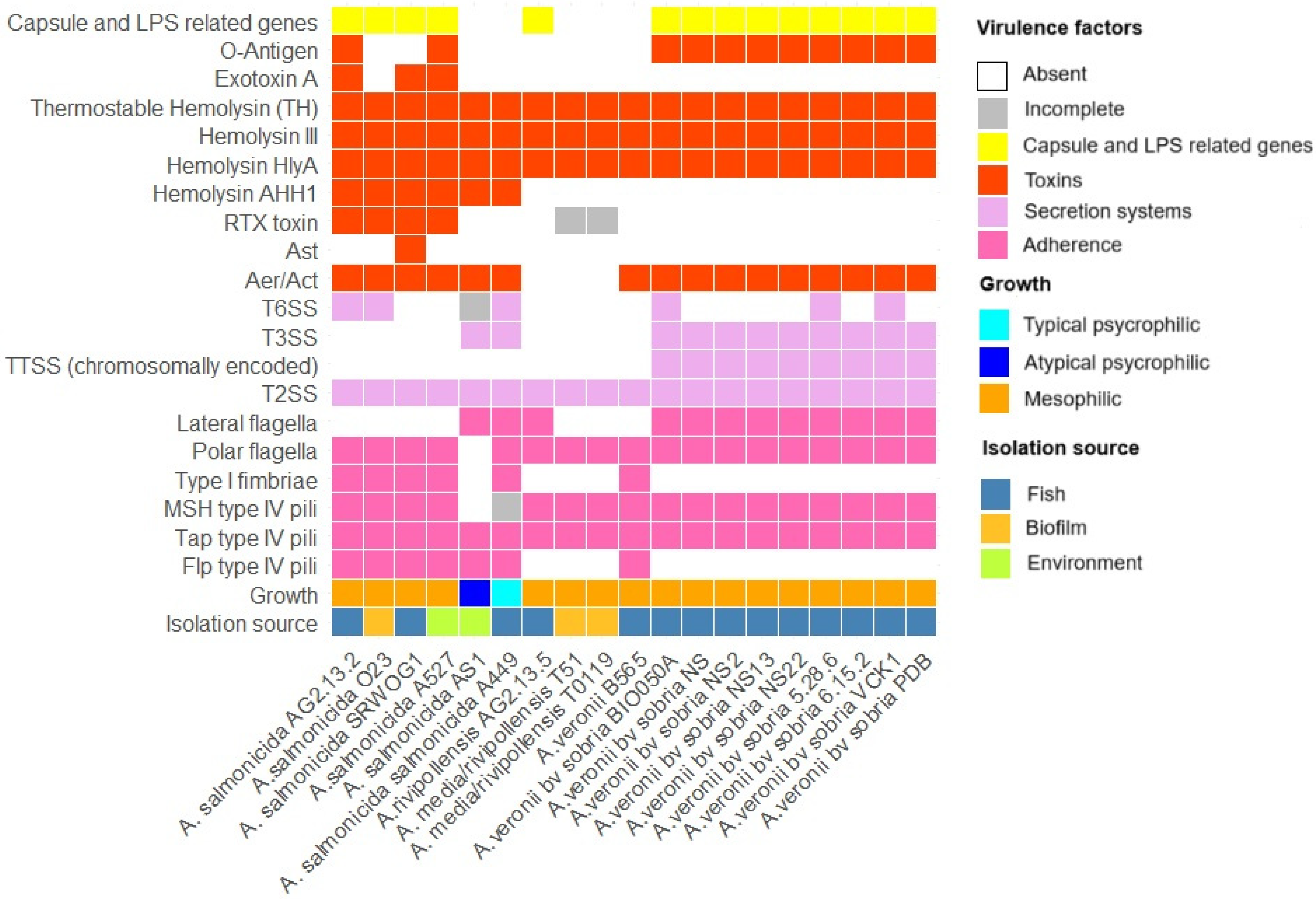


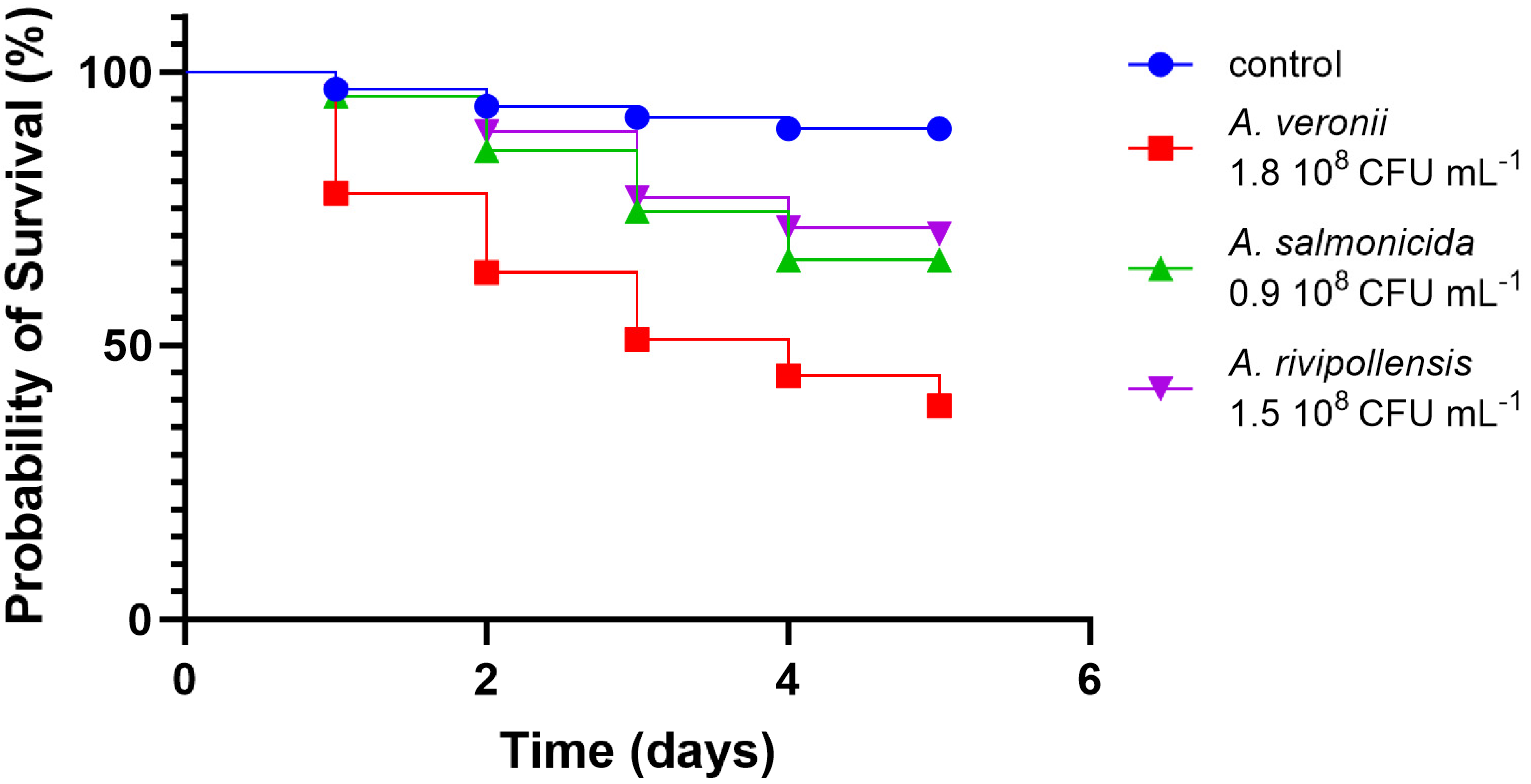
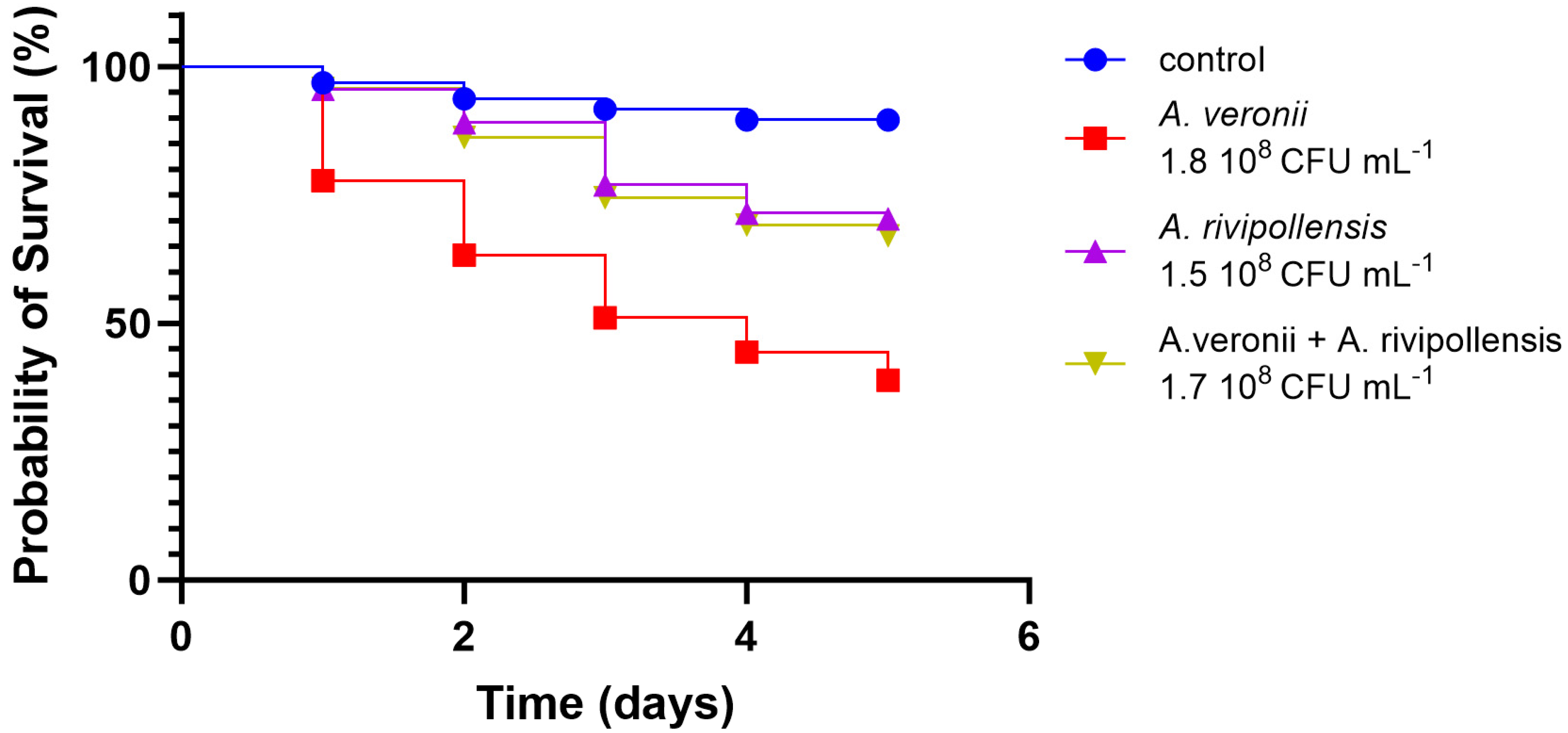
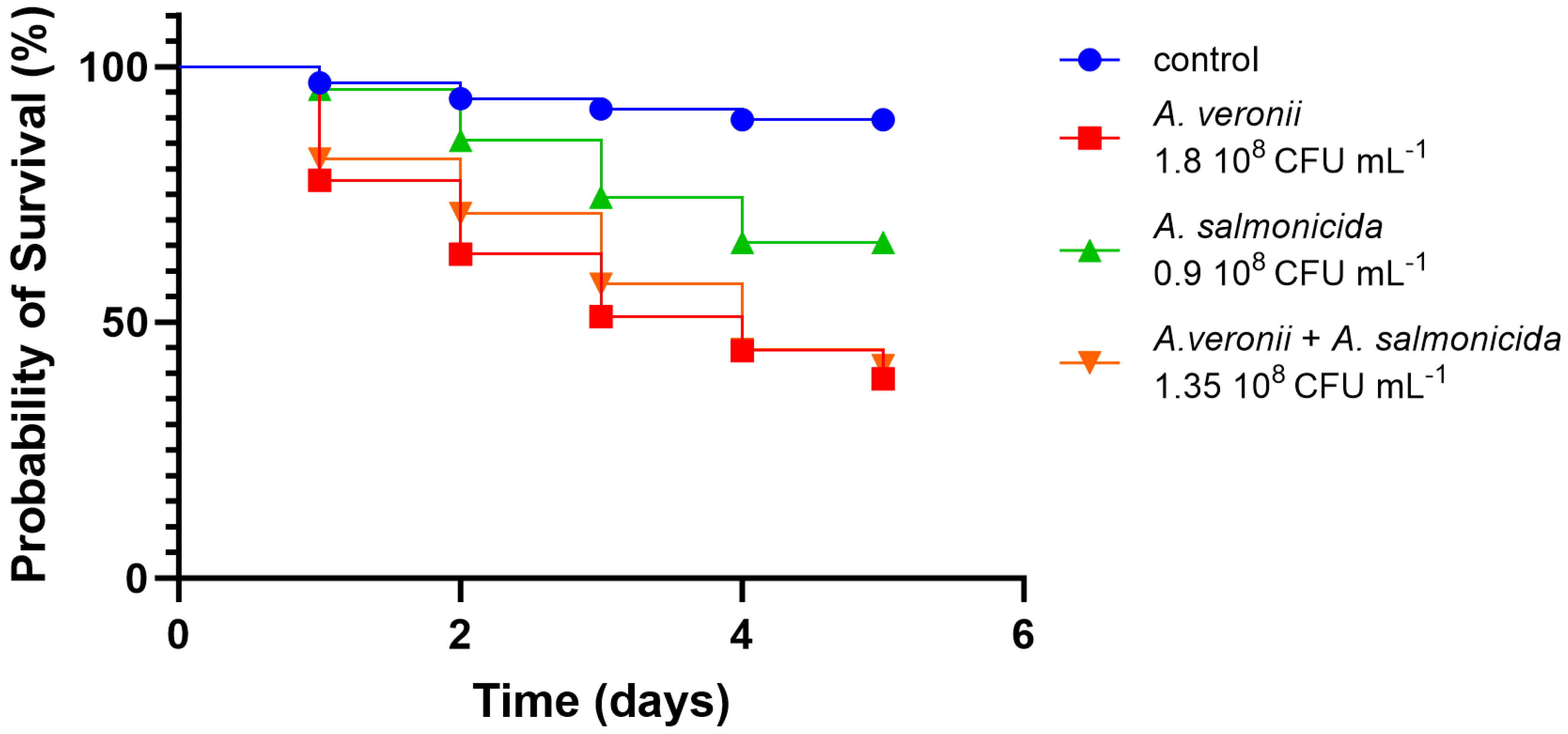
| A. salmonicida AG2.13.2 | A. salmonicida subsp. Salmonicida A449 | A. rivipollensis AG2.13.5 | Aeromonas rivipollensis KN-Mc-11N1 | |
|---|---|---|---|---|
| Length (bp) | 4,798,364 | 4,702,402 | 4,464,750 | 4,508,901 |
| Genes | 4437 | 4813 | 4128 | 4186 |
| CDS | 4341 | 4673 | 4035 | 4025 |
| Pseudogenes | 48 | 238 | 59 | 140 |
| rRNAs | 1, 1, 1 (5S, 16S, 23S) | 10, 9, 9 (5S, 16S, 23S) | 1, 1 (16S, 23S) | 11, 10, 10 (5S, 16S, 23S) |
| tRNA | 88 | 108 | 84 | 124 |
| ncRNAs | 5 | 9 | 7 | 6 |
| Strain | Resistance Gene | Locus Tag | Resistance Profile |
|---|---|---|---|
| A. salmonicida AG2.13.2 | cphA5 | LCG94_02875 | Subclass B2 β-lactamase |
| tetE, tetR | LCG94_08210, LCG94_08205 | Tetracycline efflux pump | |
| uhpT | LCG94_00425 | Fosfomycin | |
| OXA-12 | LCG94_03955 | Class D β-lactamase | |
| ampC | LCG94_12450 | Class C beta-lactamase | |
| tuf | LCG94_09150 | Elfamycin resistance gene | |
| macB-tolC | LCG94_11330, LCG94_00440 | Macrolide efflux pump | |
| qnrB | LCG94_06945 | Quinolone resistance pentapeptide | |
| mdtL | LCG94_10650, LCG94_03795 | Multidrug efflux pump | |
| uhpT | LCG94_00425 | Related to fosfomycin resistance | |
| pgsA | LCG94_15430 | Protein-altering, cell wall charge-conferring antibiotic resistance | |
| katG | LCG94_17960 | Isoniazid drug resistance | |
| oxyR | LCG94_12130 | Hydrogen peroxide-inducible gene activator | |
| A. rivipollensis AG2.13.5 | macB-tolC | RXZ09_11995, RXZ09_15840 | Macrolide efflux pump |
| qnrB | RXZ09_19125 | Quinolone resistance pentapeptide | |
| mdtL | RXZ09_01745 RXZ09_18315 | Multidrug efflux pump | |
| ampC | RXZ09_03805 | Class C beta-lactamase | |
| uhpT | RXZ09_12110 | Related to fosfomycin resistance | |
| OXA-504 | RXZ09_18590 | Class D β-lactamase | |
| tuf | RXZ09_17225 | Elfamycin resistance gene | |
| katG | RXZ09_09250 | Isoniazid drug resistance | |
| oxyR | RXZ09_03480 | Hydrogen peroxide-inducible gene activator |
| A. salmonicida subsp. salmonicida ATCC33658 | A. salmonicida subsp. achromogenes ATCC33659 | A. salmonicida subsp. masoucida ATCC27013 | A. salmonicida subsp. smithia ATCC49393 | A. salmonicida AG2.13.2 |
|---|---|---|---|---|
| 2028.35 | 2028.35 | 2028.35 | 2028.35 | 2027.3204 |
| 2127.46 | 2105.56 | 2127.46 | 2461.93 | 2128.6416 |
| 3240.09 | 2127.46 | 3011.77 | 2920.31 | 2524.7395 |
| 3435.62 | 3112.82 | 3240.09 | 3082.85 | 3042.0710 |
| 3595.69 | 3293.22 | 3372.66 | 3240.09 | 3151.8762 |
| 4260.10 | 3400.97 | 3435.62 | 3435.62 | 3597.1121 |
| 4371.13 | 3595.69 | 3595.69 | 3739.54 | 3842.3545 |
| 4595.81 | 4117.59 | 3762.02 | 4260.10 | 4171.2969 |
| 5156.84 | 5105.56 | 4260.10 | 4371.13 | 4258.7832 |
| 5534.26 | 6086.98 | 4371.13 | 4595.74 | 4347.0786 |
| 6086.98 | 6478.28 | 4595.81 | 5156.84 | 4468.8955 |
| 6478.28 | 6804.55 | 5156.84 | 5534.56 | 4592.4971 |
| 6850.39 | 6850.39 | 5534.26 | 5930.05 | 4655.8027 |
| 7333.91 | 7195.51 | 6086.76 | 6850.39 | 4700.5498 |
| 8072.35 | 7333.91 | 6477.70 | 7283.06 | 4976.9204 |
| 8936.98 | 8072.35 | 6747.92 | 8072.35 | 5051.1475 |
| 8884.05 | 7195.51 | 5089.3784 | ||
| 8936.98 | 7333.89 | 5156.0093 | ||
| 8072.35 | 6086.2578 | |||
| 8936.98 | 6306.0918 | |||
| 9014.04 | 6481.3032 | |||
| 7195.9482 | ||||
| 7333.4922 | ||||
| 7659.6675 | ||||
| 8343.4707 | ||||
| 8938.5283 | ||||
| 9185.4803 | ||||
| 9401.5156 |
| Substrate | A. salmonicida AG2.13.2 | A. rivipollensis AG2.13.5 | Substrate | A. salmonicida AG2.13.2 | A. rivipollensis AG2.13.5 |
|---|---|---|---|---|---|
| Negative control | − | − | Glycyl-L-proline | + | + |
| Dextrin | + | + | L-alanine | + | + |
| D-maltose | + | + | L-arginine | + | + |
| D-trehalose | + | + | L-aspartic acid | + | + |
| D-cellobiose | + | + | L-glutamic acid | + | + |
| Gentiobiose | − | − | L-histidine | + | + |
| Sucrose | + | + | L-pyroglutamic | − | − |
| D-turanose | − | − | L-serine | + | + |
| Stachyose | − | − | Lincomycin | + | + |
| Positive control | + | + | Guanidine HCL | + | + |
| pH 6 | + | + | Niaproof 4 | + | + |
| pH 5 | − | − | Pectin | intermediate | − |
| D-raffinose | − | − | D-galacturonic | intermediate | − |
| α-d-lactose | − | − | L-galactonic acid lactone | intermediate | − |
| D-melibiose | − | − | D-gluconic acid | + | + |
| Β-Methyl-D-glucoside | + | + | D-glucuronic acid | − | − |
| D-salicin | − | − | Glucuronamide | intermediate | intermediate |
| N-acetyl-D-glucosamine | + | + | Mucic acid | − | − |
| N-acetyl-β-D-mannosamine | − | − | Quininic acid | − | − |
| N-acetyl-D-galactosamine | − | − | D-saccharic acid | − | − |
| N-acetyl neuraminic acid | − | − | Vancomycin | + | + |
| 1% NaCl | + | + | Tetrazolium violet | + | + |
| 4% NaCl | + | + | Tetrazolium blue | + | + |
| 8% NaCl | − | − | P-Hydroxy-phenylacetic acid | − | − |
| A-D-glucose | + | + | Methyl pyruvate | + | + |
| D-mannose | + | + | D-lactic acid methyl ester | − | − |
| D-Fructose | + | + | L-lactic acid | − | − |
| D-galactose | + | + | Citric acid | + | − |
| 3-methyl glucose | − | − | A-keto-glutaric acid | − | − |
| D-fucose | − | intermediate | D-malic acid | − | − |
| L-fucose | − | − | |||
| L-rhamnose | − | − | L-malic acid | + | + |
| Inosine | + | + | Bromo-succinic acid | intermediate | − |
| 1% sodium lactate | + | + | Nalidixic acid | + | − |
| Fusidic acid | − | − | Lithium chloride | + | − |
| D-Serine | + | − | Potassium tellurite | − | − |
| D-sorbitol | + | − | Tween 40 | + | + |
| D-mannitol | + | + | γ-Amino-butyric acid | − | − |
| D-arabitol | intermediate | − | α- Hydroxy-butyric acid | − | − |
| Myo-inositol | − | − | β- Hydroxy-D, L-butyric acid | − | intermediate |
| Glycerol | + | + | A-keto-butyric acid | intermediate | − |
| D-glucose 6-PO4 | + | + | Acetoacetic acid | + | + |
| D-Fructose 6-PO4 | + | + | Propionic acid | intermediate | − |
| D-aspartic acid | − | − | Acetic acid | + | + |
| D-serine | + | + | Formic acid | + | + |
| Troleandomycin | + | + | Aztreonam | + | intermediate |
| Rifamycin SV | + | + | Sodium butyrate | intermediate | + |
| Minocycline | intermediate | − | Sodium bromates | − | − |
| Gelatin | + | − |
| Strain | Drug | Content (μg) | IZD (mm) 24 h | Sensitivity |
|---|---|---|---|---|
| A. salmonicida salmonicida LMG3780 | Oxytetracycline | 30 | 36 | S |
| Tetracycline | 30 | 35 | S | |
| Piperacillin | 100 | 30 | S | |
| Sulfamethoxazole | 25 | 34.5 | S | |
| Florfenicol | 30 | 38 | S | |
| Ampicillin | 10 | 23 | S | |
| Flumequine | 30 | 38 | S | |
| Oxolinic acid | 2 | 35 | S | |
| A. salmonicida AG2.13.2 | Oxytetracycline | 30 | 10 | R |
| Tetracycline | 30 | 15 | I | |
| Piperacillin | 100 | 29.5 | S | |
| Sulfamethoxazole | 25 | 30 | S | |
| Florfenicol | 30 | 27 | I | |
| Ampicillin | 10 | 0 | R | |
| Flumequine | 30 | 24 | S | |
| Oxolinic acid | 2 | 11 | R | |
| A. rivipollensis AG2.13.5 | Oxytetracycline | 30 | 33 | S |
| Tetracycline | 30 | 34.5 | S | |
| Piperacillin | 100 | 34 | S | |
| Sulfamethoxazole | 25 | 32 | S | |
| Florfenicol | 30 | 30 | S | |
| Ampicillin | 10 | 0 | R | |
| Flumequine | 30 | 40 | S | |
| Oxolinic acid | 2 | 34 | S |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cantillo Villa, Y.; Triga, A.; Katharios, P. Polyinfection in Fish Aeromoniasis: A Study of Co-Isolated Aeromonas Species in Aeromonas veronii Outbreaks. Pathogens 2023, 12, 1337. https://doi.org/10.3390/pathogens12111337
Cantillo Villa Y, Triga A, Katharios P. Polyinfection in Fish Aeromoniasis: A Study of Co-Isolated Aeromonas Species in Aeromonas veronii Outbreaks. Pathogens. 2023; 12(11):1337. https://doi.org/10.3390/pathogens12111337
Chicago/Turabian StyleCantillo Villa, Yanelys, Adriana Triga, and Pantelis Katharios. 2023. "Polyinfection in Fish Aeromoniasis: A Study of Co-Isolated Aeromonas Species in Aeromonas veronii Outbreaks" Pathogens 12, no. 11: 1337. https://doi.org/10.3390/pathogens12111337
APA StyleCantillo Villa, Y., Triga, A., & Katharios, P. (2023). Polyinfection in Fish Aeromoniasis: A Study of Co-Isolated Aeromonas Species in Aeromonas veronii Outbreaks. Pathogens, 12(11), 1337. https://doi.org/10.3390/pathogens12111337







