Antimicrobial Resistance and Genetic Diversity of Pseudomonas aeruginosa Strains Isolated from Equine and Other Veterinary Samples
Abstract
1. Introduction
2. Materials and Methods
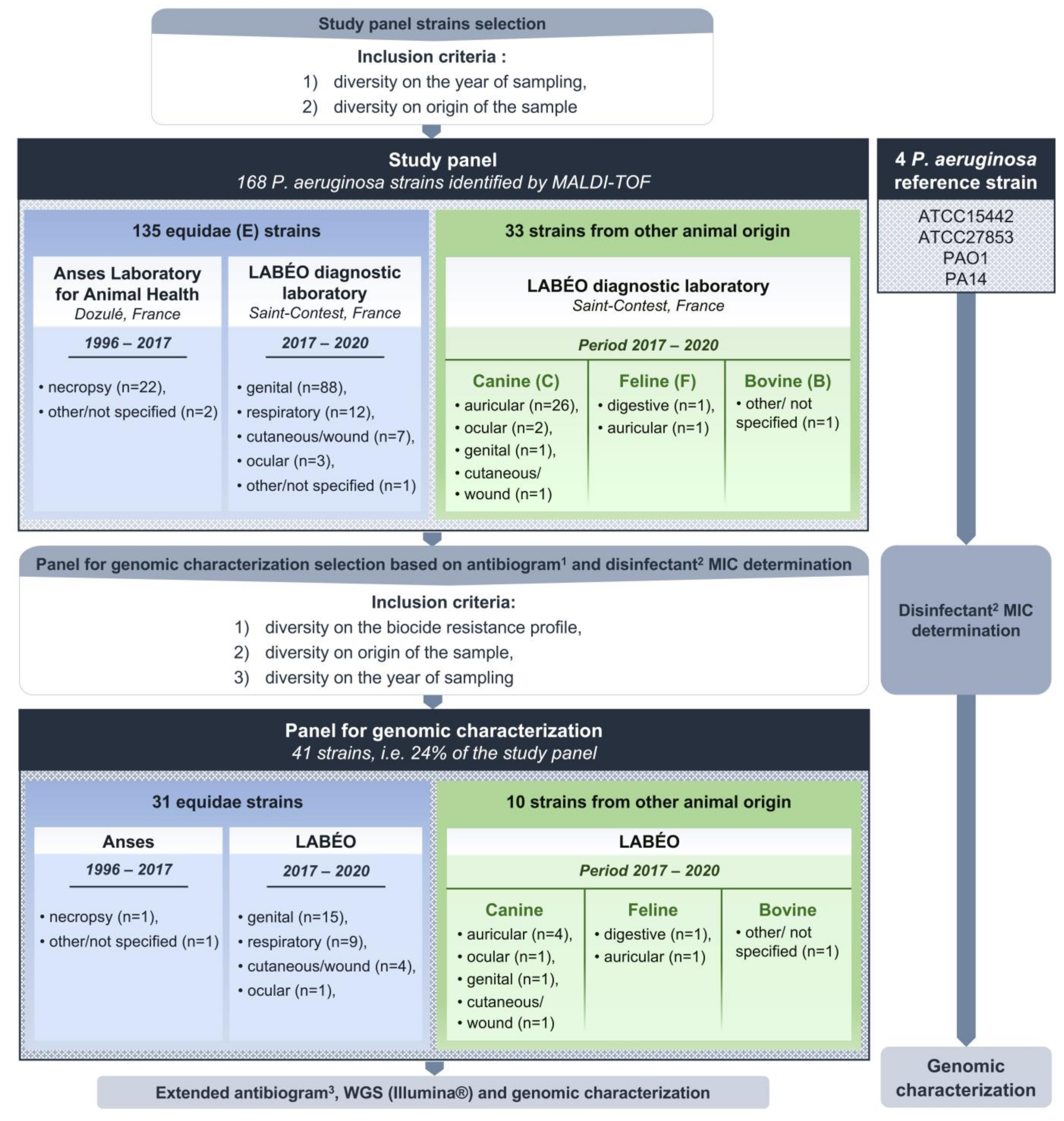
2.1. P. aeruginosa Bacterial Strains
2.2. Antimicrobial Susceptibility Testing
2.2.1. Antibiotic Susceptibility Testing
2.2.2. Quaternary Ammonium Compound Susceptibility Testing
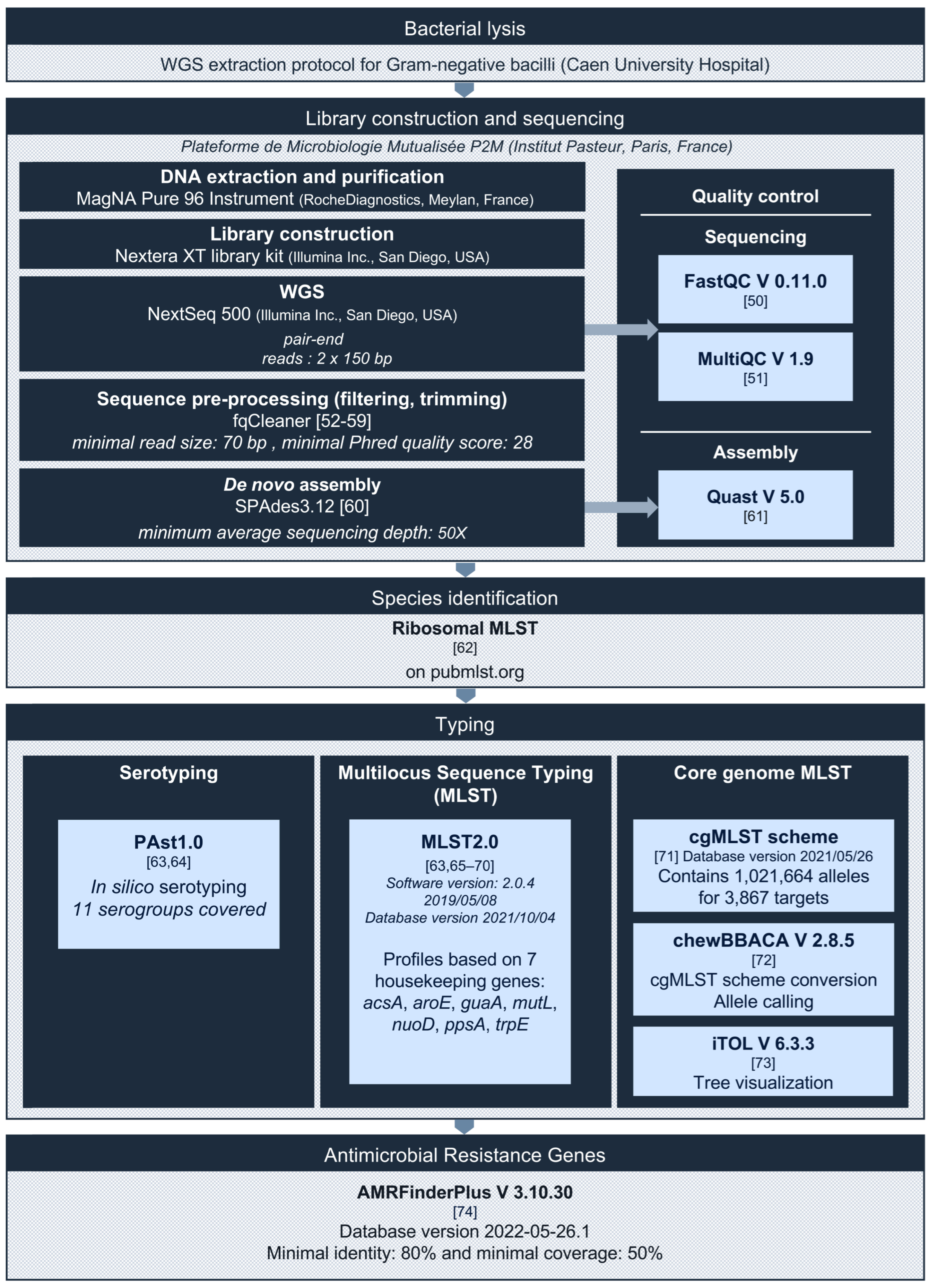
2.3. Whole Genome Sequencing and Bioinformatics Analysis
2.4. Statistical Analysis
3. Results
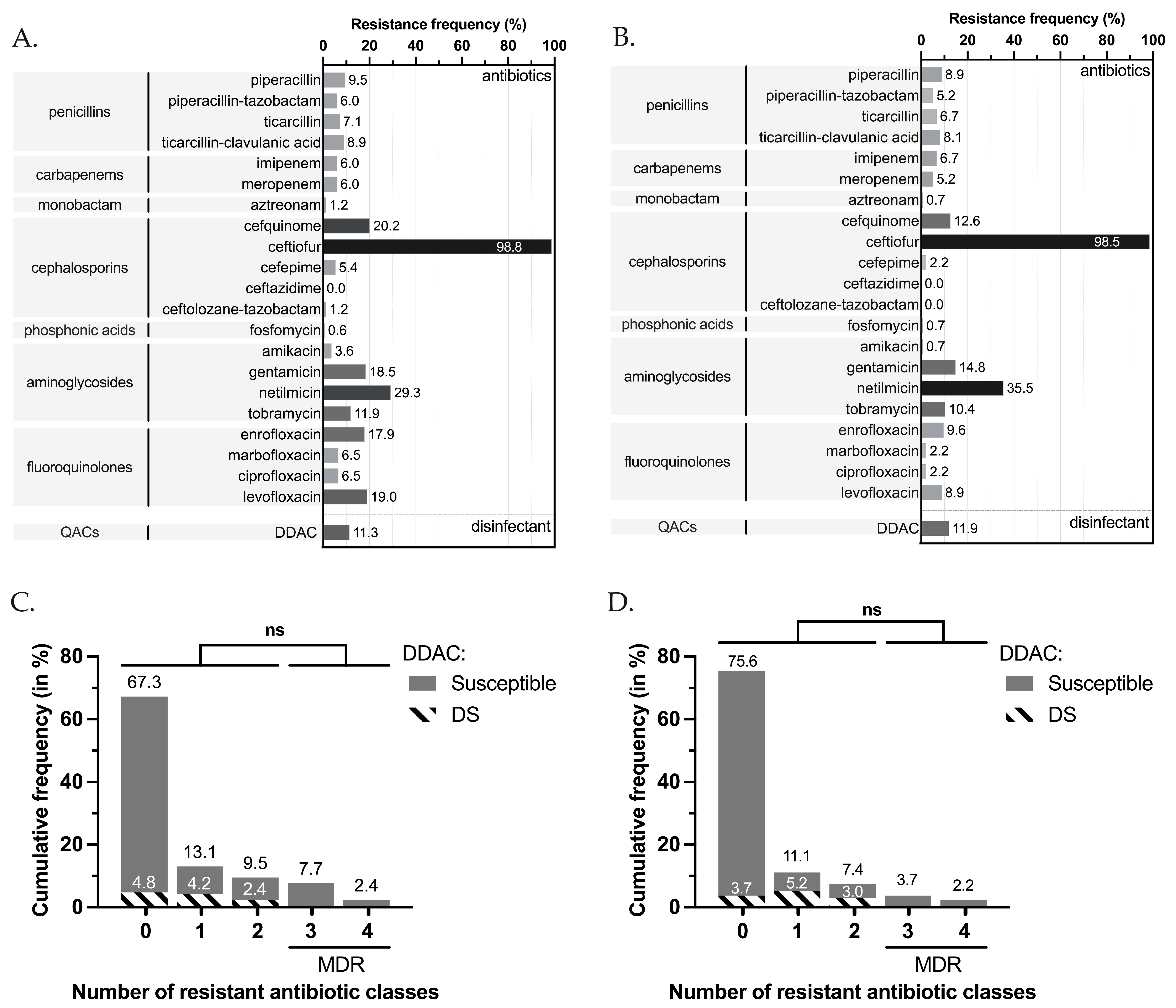
3.1. Antimicrobial and DDAC Resistance Phenotypes
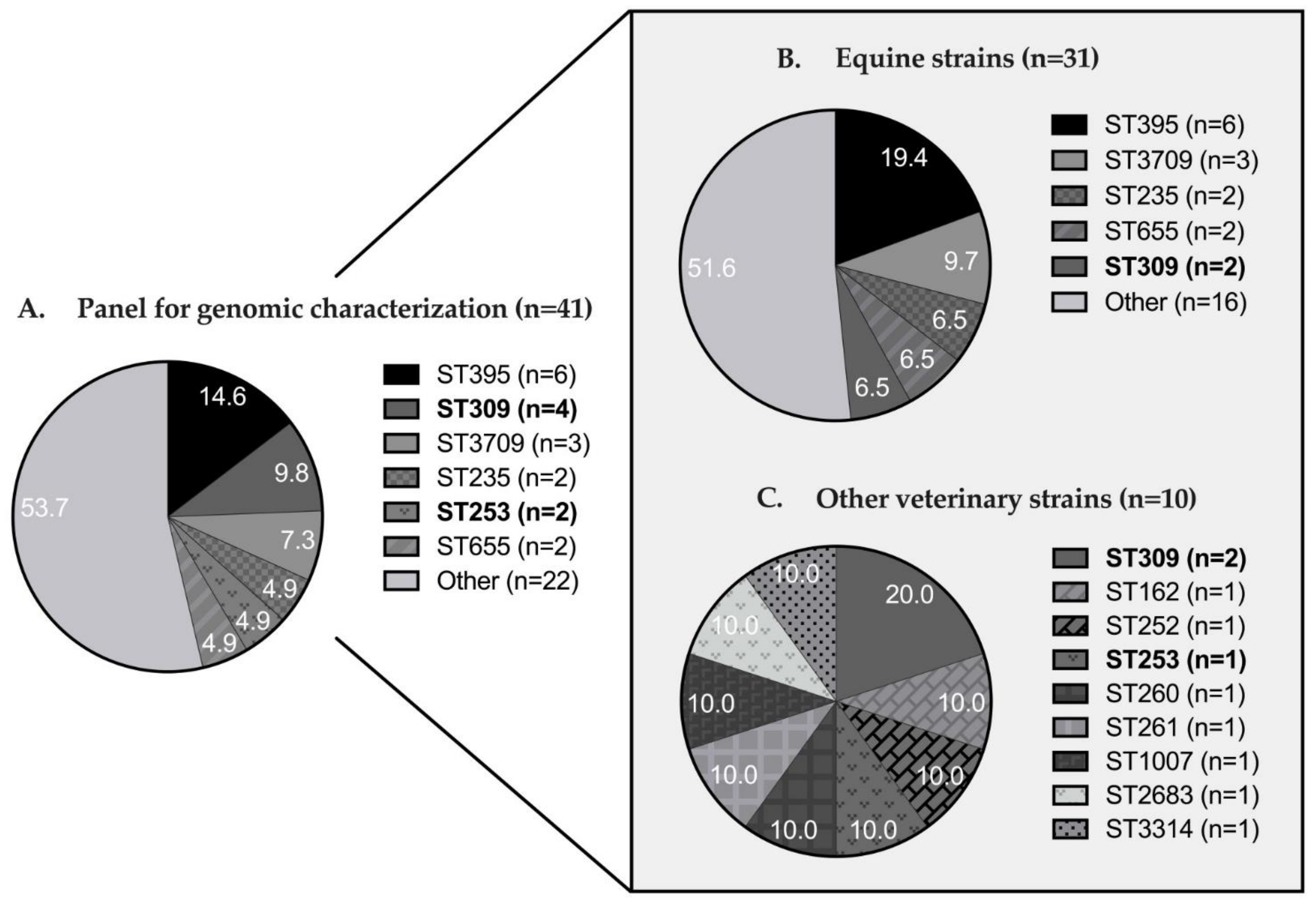
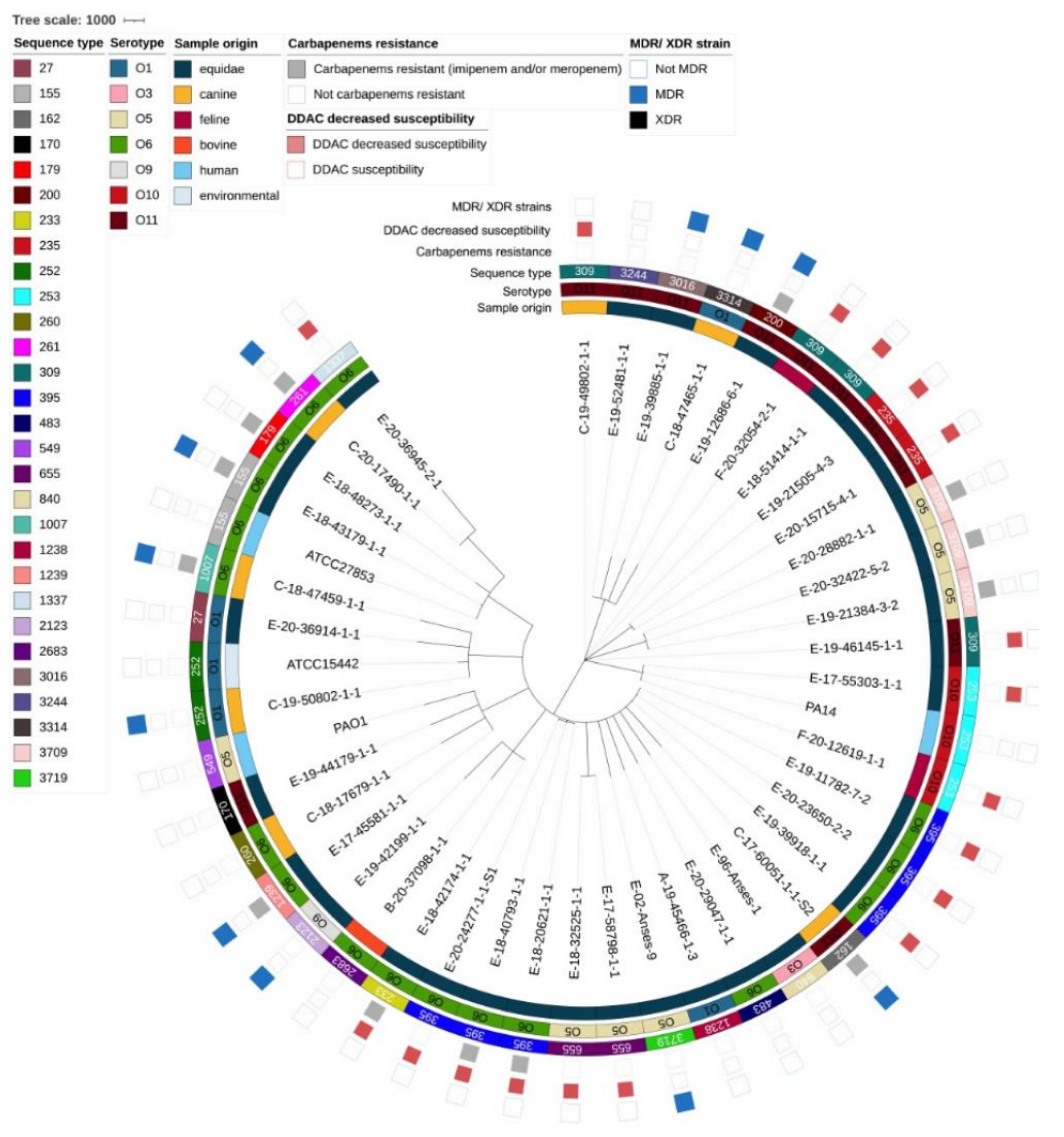
3.2. Genomic Diversity and Resistome Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Year of Sampling | DDAC Status | Total n | |
|---|---|---|---|
| Susceptible n | DS n | ||
| 1996 | 1 | - | 1 |
| 2002 | 1 | - | 1 |
| 2017 | 2 | 2 | 4 |
| 2018 | 5 | 5 | 10 |
| 2019 | 8 | 5 | 13 |
| 2020 | 6 | 6 | 12 |
| Total | 23 | 18 | 41 |
| Sample origin | DDAC status | Total n | |
| Susceptible n | DS n | ||
| Equine | 16 | 15 | 31 |
| Canine | 6 | 1 | 7 |
| Feline | - | 2 | 2 |
| Bovine | 1 | - | 1 |
| Total | 23 | 18 | 41 |
| Type of sample | DDAC status | Total n | |
| Susceptible n | DS n | ||
| Genital | 10 | 6 | 16 |
| Respiratory | 1 | 8 | 9 |
| Auricular | 4 | 1 | 5 |
| Cutaneous/wound | 4 | 1 | 5 |
| Other/not specified | 2 | - | 2 |
| Ocular | 1 | 1 | 2 |
| Digestive | - | 1 | 1 |
| Necropsy | 1 | - | 1 |
| Total | 23 | 18 | 41 |
Appendix B
| Antibiotics | Used for MDR/XDR Categorization | Source | Zone Diameter Breakpoints (mm) | |||
|---|---|---|---|---|---|---|
| S≥ | R< | ATU | ||||
| Penicillins | Piperacillin | yes | EUCAST 2021, Pseudomonas spp. [97] | 50 | 18 | 18–19 |
| Piperacillin-tazobactam | yes | EUCAST 2021, Pseudomonas spp. [97] | 50 | 18 | 18–19 | |
| Ticarcillin | yes | EUCAST 2021, Pseudomonas spp. [97] | 50 | 18 | ||
| Ticarcillin-clavulanic acid | yes | EUCAST 2021, Pseudomonas spp. [97] | 50 | 18 | ||
| Carbapenems | Imipenem | yes | EUCAST 2021, Pseudomonas spp. [97] | 50 | 20 | |
| Meropenem | yes | EUCAST 2021, Pseudomonas spp. [97] | 24 | 18 | ||
| Monobactam | Aztreonam | yes | EUCAST 2021, Pseudomonas spp. [97] | 50 | 18 | |
| Cephalosporins | Cefquinome | no | CASFM veterinary 2013, Enterobacteriaceae [98] | 22 | 19 | |
| Ceftiofur | no | CASFM veterinary 2013, Enterobacteriaceae [98] | 21 | 18 | ||
| Cefepime | yes | EUCAST 2021, Pseudomonas spp. [97] | 50 | 21 | ||
| Ceftazidime | yes | EUCAST 2021, Pseudomonas spp. [97] | 50 | 17 | ||
| Ceftolozane-tazobactam | yes | EUCAST 2021, Pseudomonas spp. [97] | 23 | 23 | ||
| Phosphonic acids | Fosfomycin | yes | EUCAST 2021, Pseudomonas spp. Based on clinical observations [97] | 12 | 7 | |
| Aminoglycosides | Amikacin | yes | EUCAST 2021, Pseudomonas spp. [97] | 15 | 15 | |
| Gentamicin | yes | EUCAST 2019, Pseudomonas spp. [99] | 15 | 15 | ||
| Netilmicin | yes | EUCAST 2019, Pseudomonas spp. [99] | 12 | 12 | ||
| Tobramycin | yes | EUCAST 2021, Pseudomonas spp. [97] | 18 | 18 | ||
| Fluoroquinolones | Enrofloxacin | no | CASFM veterinary 2013, Enterobacteriaceae [98] | 22 | 17 | |
| Marbofloxacin | no | CASFM veterinary 2013, Enterobacteriaceae [98] | 18 | 15 | ||
| Ciprofloxacin * | yes | EUCAST 2021, Pseudomonas spp. [97] | 50 | 26 | ||
| Levofloxacin | yes | EUCAST 2021, Pseudomonas spp. [97] | 50 | 22 | ||
Appendix C
References
- Walker, T.S.; Bais, H.P.; Déziel, E.; Schweizer, H.P.; Rahme, L.G.; Fall, R.; Vivanco, J.M. Pseudomonas aeruginosa-plant root interactions, pathogenicity, biofilm formation, and root exudation. Plant Physiol. 2004, 134, 320–331. [Google Scholar] [CrossRef] [PubMed]
- Lupo, A.; Haenni, M.; Madec, J.-Y. Antimicrobial resistance in Acinetobacter spp. and Pseudomonas spp. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- WHO Regional Office for Europe/European Centre for Disease Prevention and Control. Antimicrobial Resistance Surveillance in Europe: 2022–2020 Data; WHO Regional Office for Europe: Copenhagen, Denmark, 2022; p. 164. [Google Scholar]
- Résapath. Réseau d’Epidémiosurveillance de l’Antibiorésistance des Bactéries Pathogènes Animales, Bilan 2014; Agence Nationale de Sécurité Sanitaire de l’Alimentation, de l’Environnement et du Travail (Anses): Lyon, France; Ploufragan-Plouzané-Niort Laboratory: Ploufragan, France, 2015; p. 168. [Google Scholar]
- Hariharan, H.; Coles, M.; Poole, D.; Lund, L.; Page, R. Update on antimicrobial susceptibilities of bacterial isolates from canine and feline otitis externa. Can. Vet. J. 2006, 47, 3. [Google Scholar]
- Edwards, S.G.; Maggs, D.J.; Byrne, B.A.; Kass, P.H.; Lassaline, M.E. Effect of topical application of 0.5% proparacaine on corneal culture results from 33 dogs, 12 cats, and 19 horses with spontaneously arising ulcerative keratitis. Vet. Ophthalmol. 2019, 22, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Poonsuk, K.; Chuanchuen, R. Contribution of the MexXY multidrug efflux pump and other chromosomal mechanisms on aminoglycoside resistance in Pseudomonas aeruginosa isolates from canine and feline infections. J. Vet. Med. Sci. 2012, 74, 1575–1582. [Google Scholar] [CrossRef]
- Sela, S.; Hammer-Muntz, O.; Krifucks, O.; Pinto, R.; Weisblit, L.; Leitner, G. Phenotypic and genotypic characterization of Pseudomonas aeruginosa strains isolated from mastitis outbreaks in dairy herds. J. Dairy Res. 2007, 74, 425–429. [Google Scholar] [CrossRef]
- Aleksandrov, M.; Petkov, A. Case of Pseudomonas aeruginosa infection in tropical snakes. Vet.-Meditsinski Nauk. 1985, 22, 53–61. [Google Scholar]
- Résapath. Réseau d’Epidémiosurveillance de l’Antibiorésistance des Bactéries Pathogènes Animales, Bilan 2021; Agence Nationale de Sécurité Sanitaire de l’Alimentation, de l’Environnement et du Travail (Anses): Lyon, France; Ploufragan-Plouzané-Niort Laboratory: Ploufragan, France, 2022; p. 48. [Google Scholar]
- Govan, J.R.; Sarasola, P.; Taylor, D.J.; Tatnell, P.J.; Russell, N.J.; Gacesa, P. Isolation of a mucoid alginate-producing Pseudomonas aeruginosa strain from the equine guttural pouch. J. Clin. Microbiol. 1992, 30, 595–599. [Google Scholar] [CrossRef]
- Moore, J.E.; Buckley, T.C.; Millar, B.C.; Gibson, P.; Cannon, G.; Egan, C.; Cosgrove, H.; Stanbridge, S.; Anzai, T.; Matsuda, M.; et al. Molecular surveillance of the incidence of Taylorella equigenitalis and Pseudomonas aeruginosa from horses in Ireland by sequence-specific PCR. Equine Vet. J. 2001, 33, 319–322. [Google Scholar] [CrossRef]
- Coman, D. Horserace Betting Levy Board: Codes of practice update. Vet Equine 2022, 6, 159–162. [Google Scholar] [CrossRef]
- Horserace Betting Levy Board Codes of Practice; HBLB: London, UK, 2004; p. 24.
- Whitlock, F.M.; Newton, J.R. A practitioner’s guide to understanding equine infectious disease diagnostics in the United Kingdom. Part 1: How to optimise sampling approaches and a guide to agent detection testing methods. Equine Vet. Educ. 2022, 34, 330–336. [Google Scholar] [CrossRef]
- RESPE. Saison de Monte 2021, Bilan Annuel; Réseau d’Epidémio-Surveillance en Pathologie Équine: Saint-Contest, France, 2021; p. 6. [Google Scholar]
- Allen, J.L.; Begg, A.P.; Browning, G.F. Outbreak of equine endometritis caused by a genotypically identical strain of Pseudomonas aeruginosa. J. Vet. Diagn. Investig. 2011, 23, 1236–1239. [Google Scholar] [CrossRef]
- Bruyas, J.-F.; Puyt, J.-D.; Hermange, T.; Betsch, J.-M.; Maillard, K.; Destrumelle, S. Thérapeutique anti-infectieuse raisonnée des métrites et endométrites de la jument. Prat. Vét. Equine 2013, 45, 7–16. [Google Scholar]
- Tiago, G.; Júlio, C.; António, R. Conception rate, uterine infection and embryo quality after artificial insemination and natural breeding with a stallion carrier of Pseudomonas aeruginosa: A Case Report. Acta Vet. Scand. 2012, 54, 20. [Google Scholar] [CrossRef]
- Omar, H.; Hambidge, M.; Firmanes, B.; Shabandri, A.M.; Wilsher, S. Bacteria isolated from equine uteri in the United Arab Emirates: A retrospective study. J. Equine Vet. Sci. 2022, 115, 104029. [Google Scholar] [CrossRef]
- Díaz-Bertrana, M.L.; Deleuze, S.; Pitti Rios, L.; Yeste, M.; Morales Fariña, I.; Rivera del Alamo, M.M. Microbial prevalence and antimicrobial sensitivity in equine endometritis in field conditions. Animals 2021, 11, 1476. [Google Scholar] [CrossRef]
- Mena, K.D.; Gerba, C.P. Risk assessment of Pseudomonas aeruginosa in water. In Reviews of Environmental Contamination and Toxicology; Whitacre, D.M., Ed.; Springer: Boston, MA, USA, 2009; Volume 201, pp. 71–115. ISBN 978-1-4419-0031-9. [Google Scholar]
- Stover, C.K.; Pham, X.Q.; Erwin, A.L.; Mizoguchi, S.D.; Warrener, P.; Hickey, M.J.; Brinkman, F.S.; Hufnagle, W.O.; Kowalik, D.J.; Lagrou, M.; et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 2000, 406, 959–964. [Google Scholar] [CrossRef]
- Gellatly, S.L.; Hancock, R.E.W. Pseudomonas aeruginosa: New insights into pathogenesis and host defenses. Pathog. Dis. 2013, 67, 159–173. [Google Scholar] [CrossRef]
- Azam, M.W.; Khan, A.U. Updates on the pathogenicity status of Pseudomonas aeruginosa. Drug Discov. Today 2019, 24, 350–359. [Google Scholar] [CrossRef]
- Botelho, J.; Grosso, F.; Peixe, L. Antibiotic resistance in Pseudomonas aeruginosa-mechanisms, epidemiology and evolution. Drug Resist. Updates 2019, 44, 100640. [Google Scholar] [CrossRef]
- Reig, S.; Le Gouellec, A.; Bleves, S. What is new in the anti–Pseudomonas aeruginosa clinical development pipeline since the 2017 WHO alert? Front. Cell. Infect. Microbiol. 2022, 12, 909731. [Google Scholar] [CrossRef] [PubMed]
- Adhimi, R.; Tayh, G.; Ghariani, S.; Chairat, S.; Chaouachi, A.; Boudabous, A.; Slama, K.B. Distribution, diversity and antibiotic resistance of Pseudomonas spp. isolated from the water dams in the north of Tunisia. Curr. Microbiol. 2022, 79, 188. [Google Scholar] [CrossRef] [PubMed]
- Fangous, M.-S.; Gosset, P.; Galakhoff, N.; Gouriou, S.; Guilloux, C.-A.; Payan, C.; Vallet, S.; Héry-Arnaud, G.; Le Berre, R. Priming with intranasal Lactobacilli prevents Pseudomonas aeruginosa acute pneumonia in mice. BMC Microbiol. 2021, 21, 195. [Google Scholar] [CrossRef]
- Oyardi, O.; Savage, P.B.; Guzel, C.B. Effects of ceragenins and antimicrobial peptides on the A549 cell line and an in vitro co-culture model of A549 cells and Pseudomonas aeruginosa. Pathogens 2022, 11, 1044. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Commitee for Medicinal Products for Veterinary. Categorisation of Antibiotics in the European Union; European Medicines Agency: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Russell, A. Biocide use and antibiotic resistance: The relevance of laboratory findings to clinical and environmental situations. Lancet Infect. Dis. 2003, 3, 794–803. [Google Scholar] [CrossRef]
- Yazdankhah, S.P.; Scheie, A.A.; Høiby, E.A.; Lunestad, B.-T.; Heir, E.; Fotland, T.Ø.; Naterstad, K.; Kruse, H. Triclosan and antimicrobial resistance in bacteria: An overview. Microb. Drug Resist. 2006, 12, 83–90. [Google Scholar] [CrossRef]
- Forbes, S.; Latimer, J.; Bazaid, A.; McBain, A.J. Altered competitive fitness, antimicrobial susceptibility, and cellular morphology in a triclosan-induced small-colony variant of Staphylococcus aureus. Antimicrob. Agents Chemother. 2015, 59, 4809–4816. [Google Scholar] [CrossRef]
- Wieland, N.; Boss, J.; Lettmann, S.; Fritz, B.; Schwaiger, K.; Bauer, J.; Hölzel, C.S. Susceptibility to disinfectants in antimicrobial-resistant and -susceptible isolates of Escherichia coli, Enterococcus faecalis and Enterococcus faecium from poultry-ESBL/AmpC-phenotype of E. coli is not associated with resistance to a quaternary ammonium compound, DDAC. J. Appl. Microbiol. 2017, 122, 1508–1517. [Google Scholar] [CrossRef]
- Rozman, U.; Pušnik, M.; Kmetec, S.; Duh, D.; Šostar Turk, S. Reduced susceptibility and increased resistance of bacteria against disinfectants: A systematic review. Microorganisms 2021, 9, 2550. [Google Scholar] [CrossRef]
- Nasr, A.M.; Mostafa, M.S.; Arnaout, H.H.; Elshimy, A.A.A. The effect of exposure to sub-inhibitory concentrations of hypochlorite and quaternary ammonium compounds on antimicrobial susceptibility of Pseudomonas aeruginosa. Am. J. Infect. Control 2018, 46, e57–e63. [Google Scholar] [CrossRef]
- Lloyd, D.H.; Page, S.W. Antimicrobial stewardship in veterinary medicine. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Yemiş, F.; Harmancı, N.Y. Classification, uses and environmental implications of disinfectants. Pak. J. Anal. Environ. Chem. 2020, 21, 179–192. [Google Scholar] [CrossRef]
- Kampf, G. Didecyldimethylammonium chloride. In Antiseptic Stewardship; Springer International Publishing: Cham, Switzerland, 2018; pp. 371–394. ISBN 978-3-319-98784-2. [Google Scholar]
- Samreen; Ahmad, I.; Malak, H.A.; Abulreesh, H.H. Environmental antimicrobial resistance and its drivers: A potential threat to public health. J. Glob. Antimicrob. Resist. 2021, 27, 101–111. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. PubChem Compound Summary for CID 23558, Didecyldimethylammonium Chloride. Available online: https://pubchem-ncbi-nlm-nih-gov.ezproxy.normandie-univ.fr/compound/Didecyldimethylammonium-chloride (accessed on 24 September 2022).
- Association Française de Normalisation. NF EN 13727+A2-Antiseptiques et Désinfectants Chimiques-Essai Quantitatif de Suspension pour l’Evaluation de l’Activité Bactéricide en Médecine; Association Française de Normalisation: Paris, France, 2015; p. 54. [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing. Antimicrobial Susceptibility Testing EUCAST Disk Diffusion Method; European Committee on Antimicrobial Susceptibility Testing: Växjö, Sweden, 2021; p. 22. [Google Scholar]
- Mikkelsen, H.; McMullan, R.; Filloux, A. The Pseudomonas aeruginosa reference strain PA14 displays increased virulence due to a mutation in ladS. PLoS ONE 2011, 6, e29113. [Google Scholar] [CrossRef]
- Association Française de Normalisation. NF U47-107-Méthodes D’analyse en Santé Animale-Guide de Réalisation des Antibiogrammes par la Méthode de Diffusion en Milieu Gélosé; Association Française de Normalisation: Paris, France, 2012; p. 13. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- ISO 20776-1:2019; Susceptibility Testing of Infectious Agents and Evaluation of Performance of Antimicrobial Susceptibility Test Devices—Part 1: Broth Micro-Dilution Reference Method for Testing the In Vitro Activity of Antimicrobial Agents against Rapidly Growing Aerobic Bacteria Involved in Infectious Diseases. International Organization for Standardization: Geneva, Switzerland, 2019; p. 19.
- Simon Andrews Babraham Bioinformatics-FastQC a Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 14 January 2022).
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Brown, C.T.; Howe, A.; Zhang, Q.; Pyrkosz, A.B.; Brom, T.H. A reference-free algorithm for computational normalization of shotgun sequencing data. arXiv 2012, arXiv:1203.4802. [Google Scholar]
- Criscuolo, A.; Brisse, S. AlienTrimmer: A tool to quickly and accurately trim off multiple short contaminant sequences from high-throughput sequencing reads. Genomics 2013, 102, 500–506. [Google Scholar] [CrossRef]
- Durai, D.A.; Schulz, M.H. Improving in silico normalization using read weights. Sci. Rep. 2019, 9, 5133. [Google Scholar] [CrossRef]
- Legrand, V.N.; Kergrohen, T.N.; Joly, N.N.; Criscuolo, A.N. ROCK: Digital normalization of whole genome sequencing data. J. Open Source Softw. 2022, 7, 3790. [Google Scholar] [CrossRef]
- Liu, Y.; Schröder, J.; Schmidt, B. Musket: A multistage k-mer spectrum-based error corrector for Illumina sequence data. Bioinformatics 2013, 29, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Roguski, Ł.; Deorowicz, S. DSRC 2—Industry-oriented compression of fastq files. Bioinformatics 2014, 30, 2213–2215. [Google Scholar] [CrossRef] [PubMed]
- Wedemeyer, A.; Kliemann, L.; Srivastav, A.; Schielke, C.; Reusch, T.B.; Rosenstiel, P. An improved filtering algorithm for big read datasets and its application to single-cell assembly. BMC Bioinform. 2017, 18, 324. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. J. Comput. Mol. Cell Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bliss, C.M.; Bennett, J.S.; Bratcher, H.B.; Brehony, C.; Colles, F.M.; Wimalarathna, H.; Harrison, O.B.; Sheppard, S.K.; Cody, A.J.; et al. Ribosomal multilocus sequence typing: Universal characterization of bacteria from domain to strain. Microbiol. Read. Engl. 2012, 158, 1005–1015. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Thrane, S.W.; Taylor, V.L.; Lund, O.; Lam, J.S.; Jelsbak, L. Application of whole-genome sequencing data for O-specific antigen analysis and in silico serotyping of Pseudomonas aeruginosa isolates. J. Clin. Microbiol. 2016, 54, 1782–1788. [Google Scholar] [CrossRef]
- Lemee, L.; Dhalluin, A.; Pestel-Caron, M.; Lemeland, J.-F.; Pons, J.-L. Multilocus sequence typing analysis of Human and animal Clostridium difficile isolates of various toxigenic types. J. Clin. Microbiol. 2004, 42, 2609–2617. [Google Scholar] [CrossRef]
- Bartual, S.G.; Seifert, H.; Hippler, C.; Luzon, M.A.D.; Wisplinghoff, H.; Rodríguez-Valera, F. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 2005, 43, 4382–4390. [Google Scholar] [CrossRef]
- Wirth, T.; Falush, D.; Lan, R.; Colles, F.; Mensa, P.; Wieler, L.H.; Karch, H.; Reeves, P.R.; Maiden, M.C.J.; Ochman, H.; et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 2006, 60, 1136–1151. [Google Scholar] [CrossRef]
- Jaureguy, F.; Landraud, L.; Passet, V.; Diancourt, L.; Frapy, E.; Guigon, G.; Carbonnelle, E.; Lortholary, O.; Clermont, O.; Denamur, E.; et al. Phylogenetic and genomic diversity of human bacteremic Escherichia coli strains. BMC Genom. 2008, 9, 560. [Google Scholar] [CrossRef]
- Griffiths, D.; Fawley, W.; Kachrimanidou, M.; Bowden, R.; Crook, D.W.; Fung, R.; Golubchik, T.; Harding, R.M.; Jeffery, K.J.M.; Jolley, K.A.; et al. Multilocus sequence typing of Clostridium difficile. J. Clin. Microbiol. 2010, 48, 770–778. [Google Scholar] [CrossRef]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef]
- Tönnies, H.; Prior, K.; Harmsen, D.; Mellmann, A. Establishment and evaluation of a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Pseudomonas aeruginosa. J. Clin. Microbiol. 2021, 59, e01987-20. [Google Scholar] [CrossRef]
- Silva, M.; Machado, M.P.; Silva, D.N.; Rossi, M.; Moran-Gilad, J.; Santos, S.; Ramirez, M.; Carriço, J.A. ChewBBACA: A complete suite for gene-by-gene schema creation and strain identification. Microb. Genom. 2018, 4, e000166. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (ITOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef]
- Résapath. Réseau d’Epidémiosurveillance de l’Antibiorésistance des Bactéries Pathogènes Animales, Bilan 2020; Agence Nationale de Sécurité Sanitaire de l’Alimentation, de l’Environnement et du Travail (Anses): Lyon, France; Ploufragan-Plouzané-Niort Laboratory: Ploufragan, France, 2021; p. 42. [Google Scholar]
- Duchesne, R.; Castagnet, S.; Maillard, K.; Petry, S.; Cattoir, V.; Giard, J.-C.; Leon, A. In vitro antimicrobial susceptibility of equine clinical isolates from France, 2006–2016. J. Glob. Antimicrob. Resist. 2019, 19, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Léon, A.; Castagnet, S.; Maillard, K.; Paillot, R.; Giard, J.-C. Evolution of in vitro antimicrobial susceptibility of equine clinical isolates in France between 2016 and 2019. Anim. Open Access J. 2020, 10, 812. [Google Scholar] [CrossRef] [PubMed]
- Comité de l’Antibiogramme de la Société Française de Microbiologie (CA-SFM). Recommandations Vétérinaires 2021; Société Française de Microbiologie: Paris, France, 2021; p. 15. [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters; European Committee on Antimicrobial Susceptibility Testing: Växjö, Sweden, 2021; p. 116. [Google Scholar]
- Anses. Index des RCP (Résumé des Caractéristiques du Produit). Available online: http://www.ircp.anmv.anses.fr/ (accessed on 22 October 2022).
- Scott, A.; Pottenger, S.; Timofte, D.; Moore, M.; Wright, L.; Kukavica-Ibrulj, I.; Jeukens, J.; Levesque, R.C.; Freschi, L.; Pinchbeck, G.L.; et al. Reservoirs of resistance: Polymyxin resistance in veterinary-associated companion animal isolates of Pseudomonas aeruginosa. Vet. Rec. 2019, 185, 206. [Google Scholar] [CrossRef] [PubMed]
- Beier, R.C.; Foley, S.L.; Davidson, M.K.; White, D.G.; McDermott, P.F.; Bodeis-Jones, S.; Zhao, S.; Andrews, K.; Crippen, T.L.; Sheffield, C.L.; et al. Characterization of antibiotic and disinfectant susceptibility profiles among Pseudomonas aeruginosa veterinary isolates recovered during 1994–2003. J. Appl. Microbiol. 2015, 118, 326–342. [Google Scholar] [CrossRef] [PubMed]
- Rubin, J.; Walker, R.D.; Blickenstaff, K.; Bodeis-Jones, S.; Zhao, S. Antimicrobial resistance and genetic characterization of fluoroquinolone resistance of Pseudomonas aeruginosa isolated from canine infections. Vet. Microbiol. 2008, 131, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Van Spijk, J.N.; Schmitt, S.; Fürst, A.; Schoster, A. A retrospective analysis of antimicrobial resistance in bacterial pathogens in an equine hospital (2012–2015). Schweiz Arch. Tierheilkd 2016, 158, 433–442. [Google Scholar] [CrossRef]
- Pottier, M.; Gravey, F.; Castagnet, S.; Auzou, M.; Bénédicte, L.; Guérin, F.; Giard, J.-C.; Léon, A.; Hello, S.L. A ten-year microbiological study of Pseudomonas aeruginosa strains revealed diffusion of carbapenems and quaternary ammonium compounds resistant populations. medRxiv 2022. [Google Scholar] [CrossRef]
- Shen, J.; Pan, Y.; Fang, Y. Role of the outer membrane protein OprD2 in carbapenem-resistance mechanisms of Pseudomonas aeruginosa. PLoS ONE 2015, 10, e0139995. [Google Scholar] [CrossRef]
- Yoneyama, H.; Nakae, T. Mechanism of efficient elimination of protein D2 in outer membrane of imipenem-resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1993, 37, 2385–2390. [Google Scholar] [CrossRef]
- Yoon, E.-J.; Jeong, S.H. Mobile carbapenemase genes in Pseudomonas aeruginosa. Front. Microbiol. 2021, 12, 614058. [Google Scholar] [CrossRef]
- Aurilio, C.; Sansone, P.; Barbarisi, M.; Pota, V.; Giaccari, L.G.; Coppolino, F.; Barbarisi, A.; Passavanti, M.B.; Pace, M.C. Mechanisms of action of carbapenem resistance. Antibiotics 2022, 11, 421. [Google Scholar] [CrossRef]
- Haenni, M.; Bour, M.; Châtre, P.; Madec, J.-Y.; Plésiat, P.; Jeannot, K. Resistance of animal strains of Pseudomonas aeruginosa to carbapenems. Front. Microbiol. 2017, 8, 1847. [Google Scholar] [CrossRef]
- Sellera, F.P.; Da Silva, L.C.B.A.; Lincopan, N. Rapid Spread of critical priority carbapenemase-producing pathogens in companion animals: A One Health challenge for a post-pandemic world. J. Antimicrob. Chemother. 2021, 76, 2225–2229. [Google Scholar] [CrossRef]
- Al Bayssari, C.; Dabboussi, F.; Hamze, M.; Rolain, J.-M. Emergence of carbapenemase-producing Pseudomonas aeruginosa and Acinetobacter baumannii in livestock animals in Lebanon. J. Antimicrob. Chemother. 2015, 70, 950–951. [Google Scholar] [CrossRef]
- Abd El-Ghany, W.A. Pseudomonas aeruginosa infection of avian origin: Zoonosis and One Health implications. Vet. World 2021, 14, 2155–2159. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, X.; Schwarz, S.; Zhang, R.; Lei, L.; Liu, X.; Lin, D.; Shen, J. IMP-45-producing multidrug-resistant Pseudomonas aeruginosa of canine origin. J. Antimicrob. Chemother. 2014, 69, 2579–2581. [Google Scholar] [CrossRef]
- Hayashi, W.; Izumi, K.; Yoshida, S.; Takizawa, S.; Sakaguchi, K.; Iyori, K.; Minoshima, K.; Takano, S.; Kitagawa, M.; Nagano, Y.; et al. Antimicrobial resistance and Type III secretion system virulotypes of Pseudomonas aeruginosa isolates from dogs and cats in primary veterinary hospitals in Japan: Identification of the international high-risk clone Sequence Type 235. Microbiol. Spectr. 2021, 9, e00408-21. [Google Scholar] [CrossRef]
- Fernandes, M.R.; Sellera, F.P.; Moura, Q.; Carvalho, M.P.N.; Rosato, P.N.; Cerdeira, L.; Lincopan, N. Zooanthroponotic transmission of drug-resistant Pseudomonas aeruginosa, Brazil. Emerg. Infect. Dis. 2018, 24, 1160–1162. [Google Scholar] [CrossRef]
- Del Barrio-Tofiño, E.; López-Causapé, C.; Oliver, A. Pseudomonas aeruginosa epidemic high-risk clones and their association with horizontally-acquired β-lactamases: 2020 update. Int. J. Antimicrob. Agents 2020, 56, 106196. [Google Scholar] [CrossRef]
- Comité de l’Antibiogramme de la Société Française de Microbiologie (CA-SFM). Recommandations Vétérinaires 2013; Société Française de Microbiologie: Paris, France, 2013; p. 13. [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters; European Committee on Antimicrobial Susceptibility Testing: Växjö, Sweden, 2019; p. 100. [Google Scholar]

| Year of Sampling | DDAC Status | ||
|---|---|---|---|
| Susceptible | DS | ||
| n (%) | |||
| 1996 | 1 (100.0) | - | |
| 1997 | 1 (100.0) | - | |
| 1998 | 2 (100.0) | - | |
| 1999 | 2 (100.0) | - | |
| 2002 | 2 (100.0) | - | |
| 2003 | 2 (100.0) | - | |
| 2004 | 3 (100.0) | - | |
| 2005 | 3 (100.0) | - | |
| 2007 | 1 (100.0) | - | |
| 2008 | 1 (100.0) | - | |
| 2010 | 3 (100.0) | - | |
| 2011 | 1 (100.0) | - | |
| 2015 | 1 (100.0) | - | |
| 2017 | 7 (77.8) | 2 (22.2) | |
| 2018 | 14 (73.7) | 5 (26.3) | |
| 2019 | 61 (92.4) | 5 (7.6) | |
| 2020 | 44 (86.3) | 7 (13.7) | |
| Total | 149 (88.7) | 19 (11.3) | |
| Sample Origin | DDAC Status | p-values | |
| Susceptible | DS | ||
| n (%) | |||
| Equine | 119 (88.1) | 16 (11.9) | >0.9999 |
| Canine | 29 (96.7) | 1 (3.3) | 0.2023 |
| Feline | - | 2 (100) | - |
| Bovine | 1 (100) | - | - |
| Total | 149 (88.7) | 19 (11.3) | - |
| Type of Sample | DDAC status | p-values | |
| Susceptible | DS | ||
| n (%) | |||
| Genital | 82 (92.1) | 7 (7.9) | 0.1504 |
| Other animal species | 1 (100) | - | - |
| Equine | 81 (92) | 7 (8) | - |
| Auricular | 26 (96.3) | 1 (3.7) | 0.3164 |
| Other animal species | 26 (96.3) | 1 (3.7) | - |
| Necropsy | 22 (100) | - | 0.139 |
| Equine | 22 (100) | - | - |
| Respiratory | 4 (33.3) | 8 (66.7) | <0.0001 |
| Equine | 4 (33.3) | 8 (66.7) | - |
| Cutaneous/wound | 7 (87.5) | 1 (12.5) | >0.9999 |
| Other animal species | 1 (100) | - | - |
| Equine | 6 (85.7) | 1 (14.3) | - |
| Ocular | 4 (80) | 1 (20) | 0.4555 |
| Other animal species | 1 (50) | 1 (50) | - |
| Equine | 3 (100) | - | - |
| Other/not specified | 4 (100) | - | - |
| Other animal species | 1 (100) | - | - |
| Equine | 3 (100) | - | - |
| Digestive | - | 1 (100) | - |
| Other animal species | - | 1 (100) | - |
| Total | 149 (88.7) | 19 (11.3) | - |
| Strain Origin | Serotype | |||||||
|---|---|---|---|---|---|---|---|---|
| O1 | O3 | O5 | O6 | O9 | O10 | O11 | Total | |
| Equine | 2 | 1 | 6 | 12 | 1 | 1 | 8 | 31 |
| Canine | 2 | 3 | 2 | 7 | ||||
| Feline | 1 | 1 | 2 | |||||
| Bovine | 1 | 1 | ||||||
| Total n (%) | 4 (9.8) | 1 (2.4) | 6 (14.6) | 16 (39.0) | 1 (2.4) | 2 (4.9) | 11 (26.8) | 41 (100.0) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pottier, M.; Castagnet, S.; Gravey, F.; Leduc, G.; Sévin, C.; Petry, S.; Giard, J.-C.; Le Hello, S.; Léon, A. Antimicrobial Resistance and Genetic Diversity of Pseudomonas aeruginosa Strains Isolated from Equine and Other Veterinary Samples. Pathogens 2023, 12, 64. https://doi.org/10.3390/pathogens12010064
Pottier M, Castagnet S, Gravey F, Leduc G, Sévin C, Petry S, Giard J-C, Le Hello S, Léon A. Antimicrobial Resistance and Genetic Diversity of Pseudomonas aeruginosa Strains Isolated from Equine and Other Veterinary Samples. Pathogens. 2023; 12(1):64. https://doi.org/10.3390/pathogens12010064
Chicago/Turabian StylePottier, Marine, Sophie Castagnet, François Gravey, Guillaume Leduc, Corinne Sévin, Sandrine Petry, Jean-Christophe Giard, Simon Le Hello, and Albertine Léon. 2023. "Antimicrobial Resistance and Genetic Diversity of Pseudomonas aeruginosa Strains Isolated from Equine and Other Veterinary Samples" Pathogens 12, no. 1: 64. https://doi.org/10.3390/pathogens12010064
APA StylePottier, M., Castagnet, S., Gravey, F., Leduc, G., Sévin, C., Petry, S., Giard, J.-C., Le Hello, S., & Léon, A. (2023). Antimicrobial Resistance and Genetic Diversity of Pseudomonas aeruginosa Strains Isolated from Equine and Other Veterinary Samples. Pathogens, 12(1), 64. https://doi.org/10.3390/pathogens12010064