Measurement of Autophagy Activity Reveals Time-Dependent, Bacteria-Specific Turnover during Mycobacterium tuberculosis Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Bacterial Culture Conditions
2.3. Infection Experiments
2.4. Colony-Forming Units (CFU) Enumeration
2.5. Western Blot Analysis
2.6. Immunofluorescence Sample Preparation and Image Acquisition (RAW 264.7 Cells)
2.7. Immunofluorescence (Confocal) Image Analysis
2.8. Statistical Analyses
3. Results
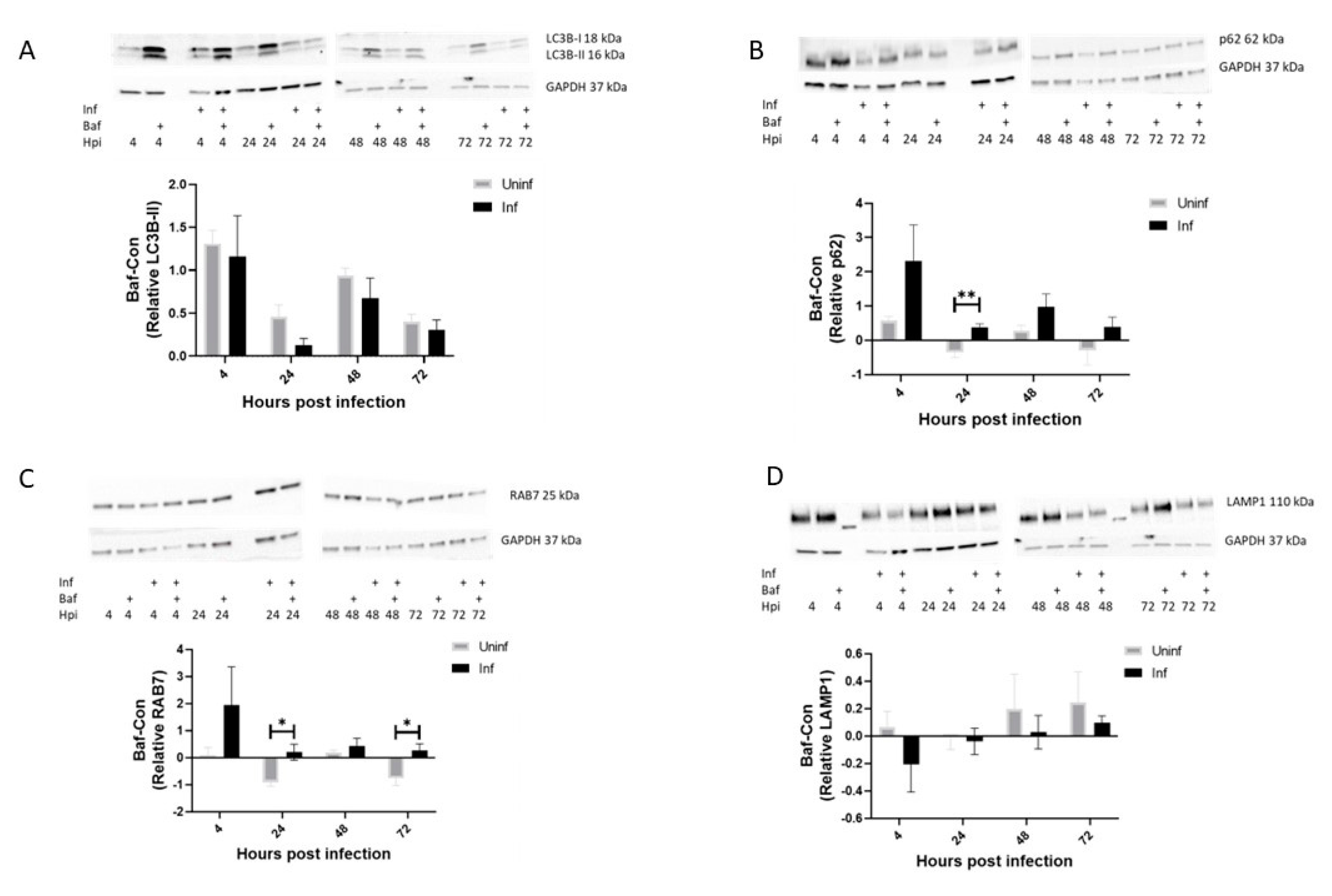
3.1. Autophagy Activity Is Higher in M. tb-Infected THP-1 Macrophages Compared to Uninfected Macrophages at 24 and 72 h Post-Infection
3.2. LC3B-II Induction in M. tb-Infected Macrophages Remains Stable over Time with Efficient Turnover at 4 and 48 h Post-Infection
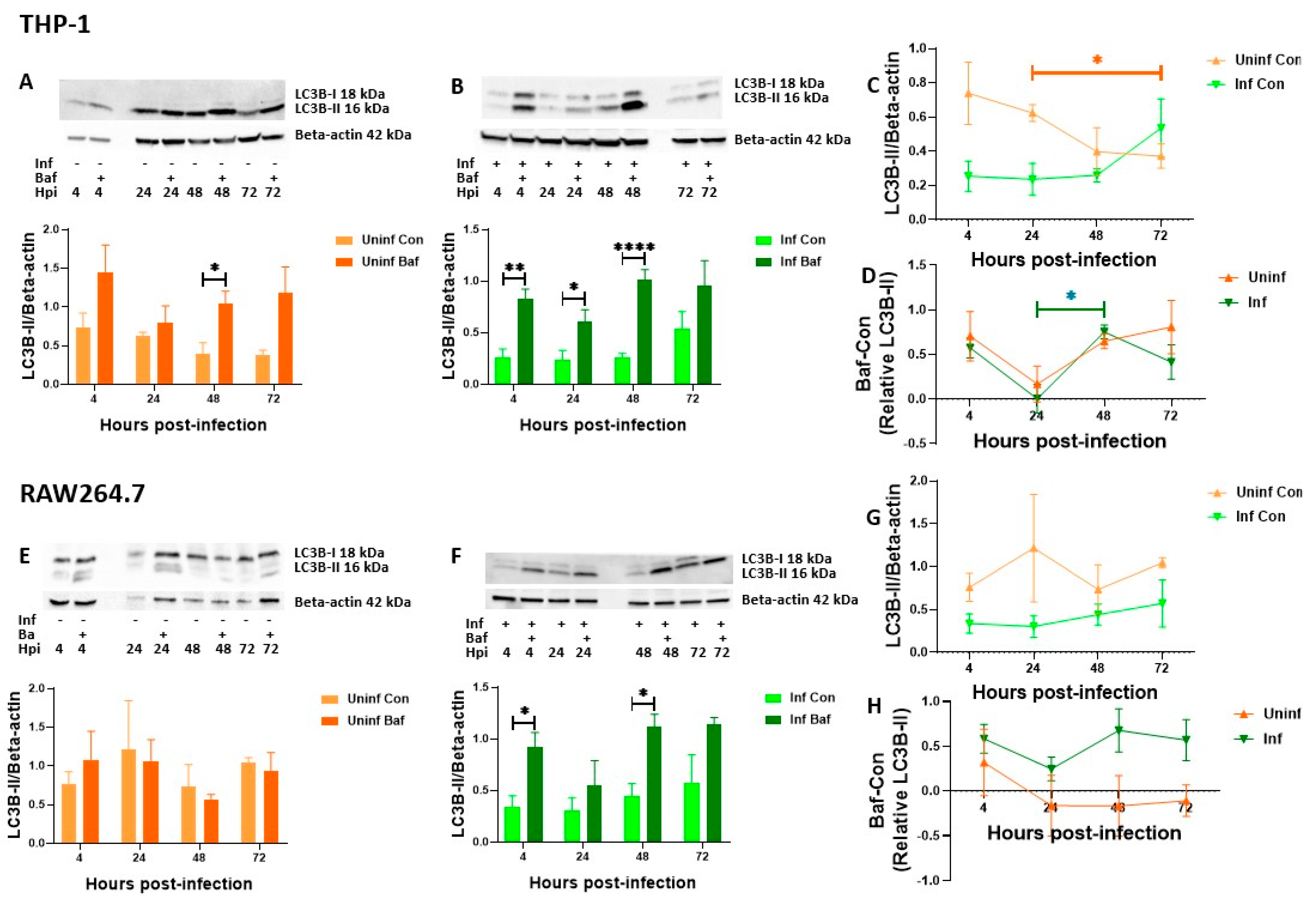
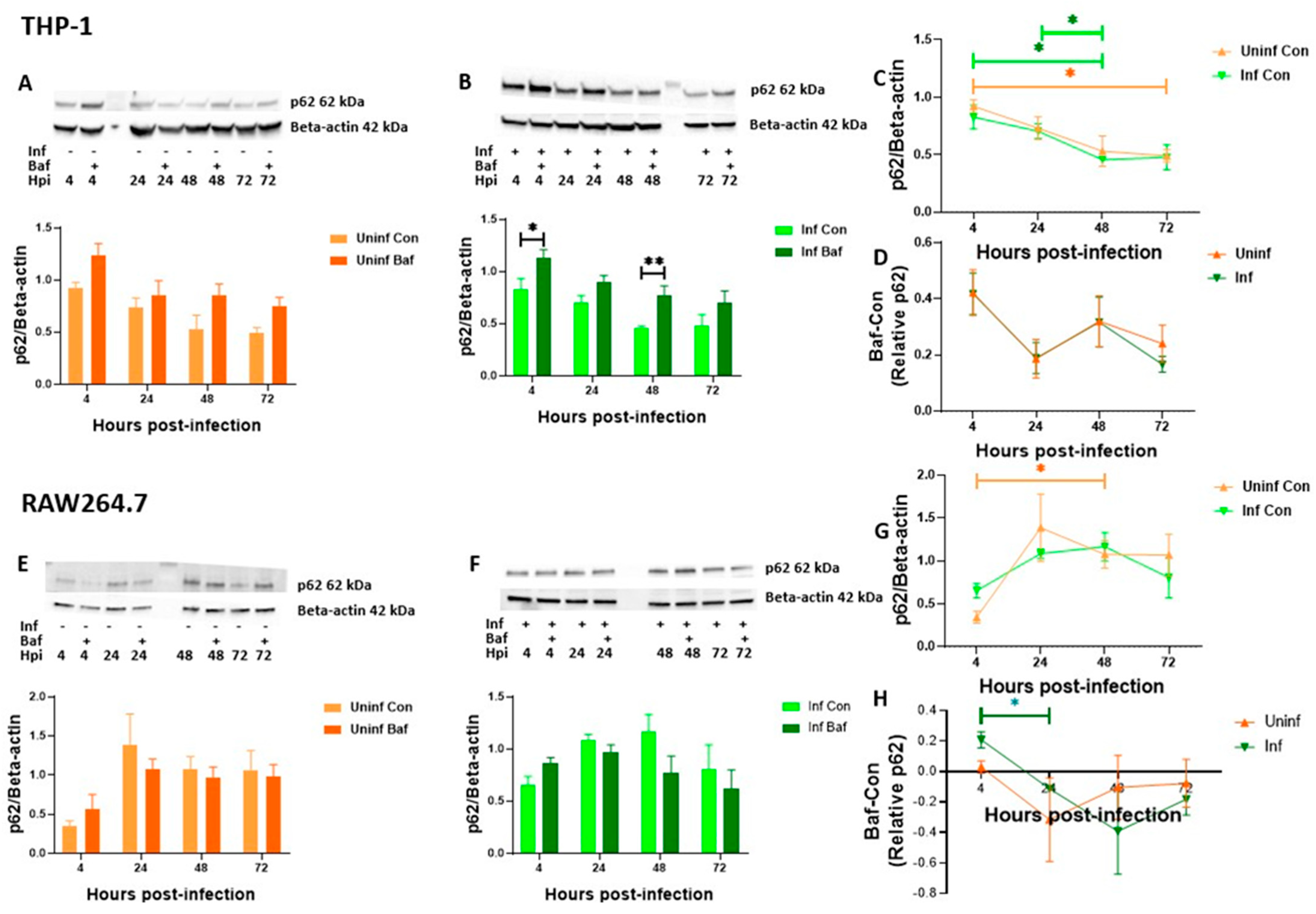
3.3. THP-1 and RAW 264.7 Macrophages Exhibit Decreasing Levels of Autophagosomal Degradation over Time
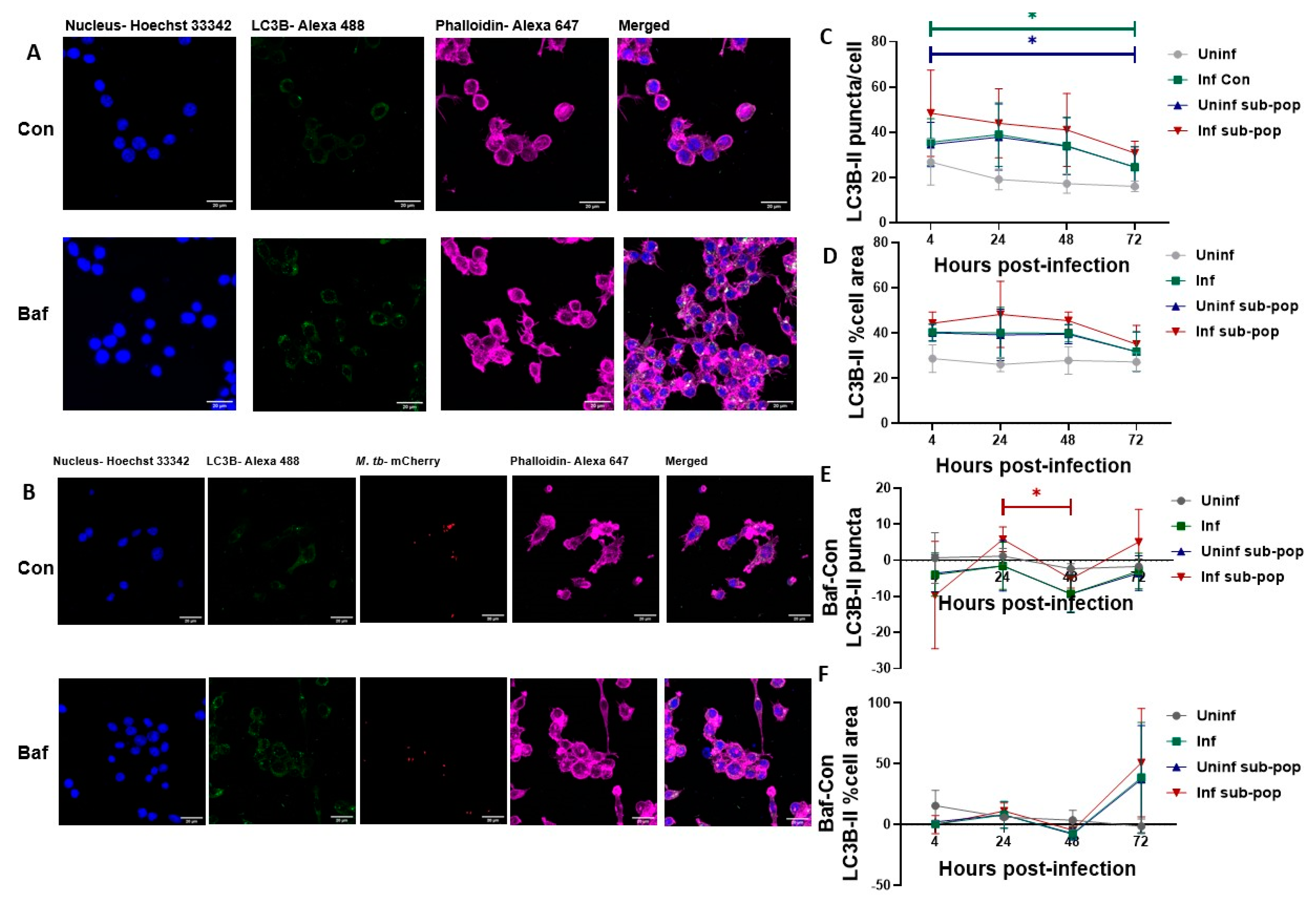
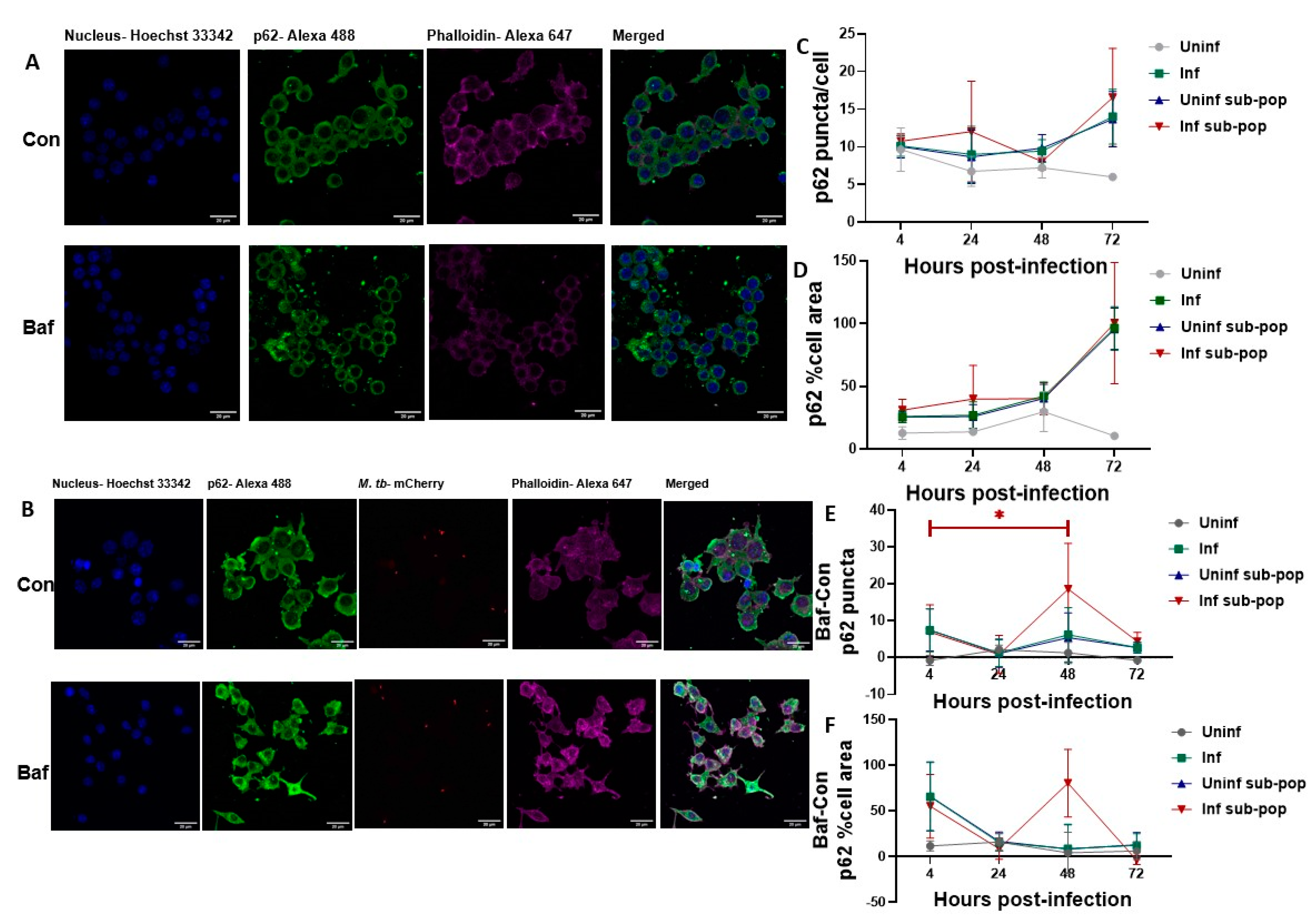
3.4. LC3B Puncta Count, and Area Are Increased in Bacteria-Containing RAW 264.7 Macrophages
3.5. P62 Puncta Turnover in the Infected Subpopulation of RAW 264.7 Macrophages Is Highest at 48 h
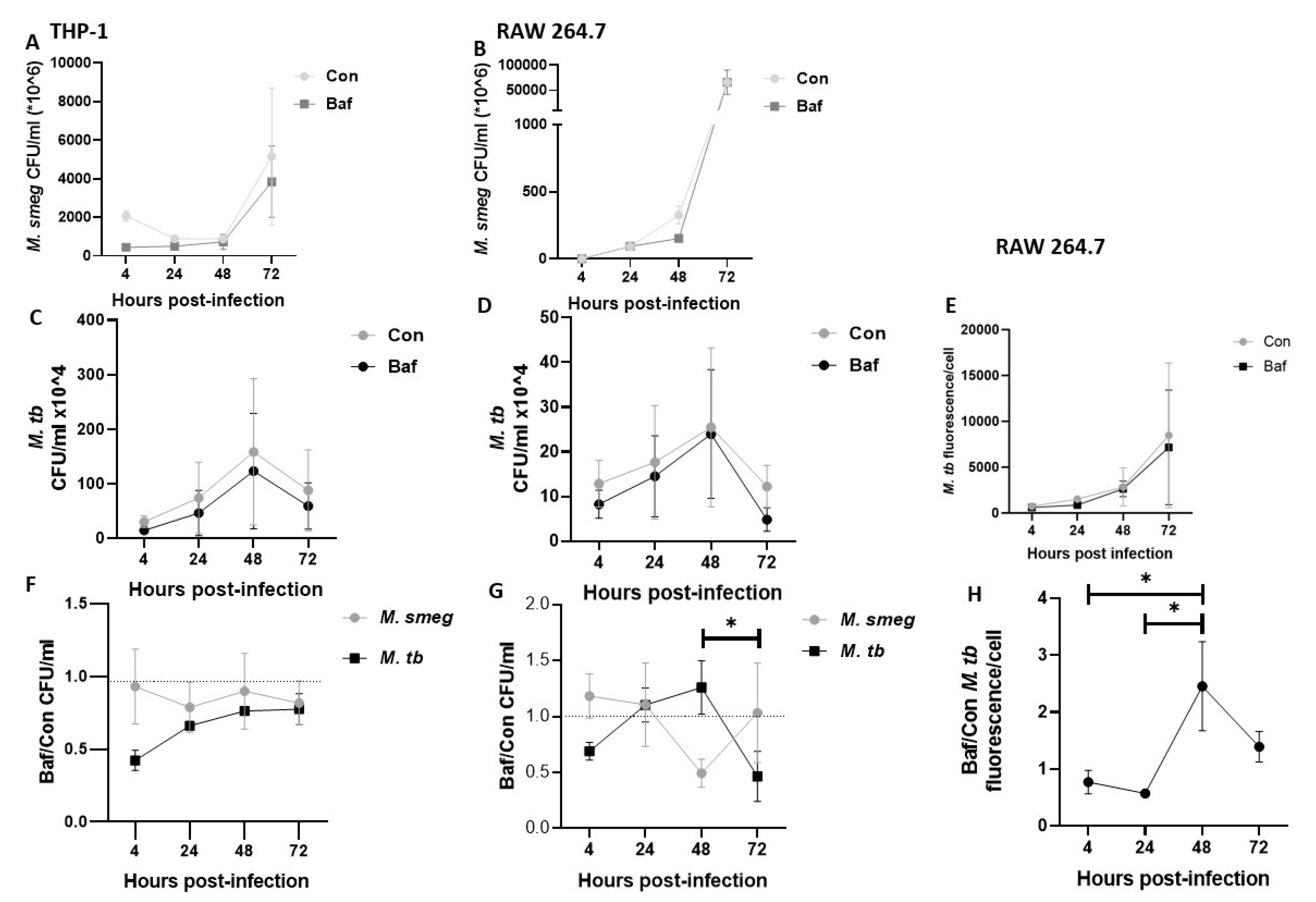
3.6. Fold Change of M. tb Bacilli Is Highest at 48 h Post-Infection
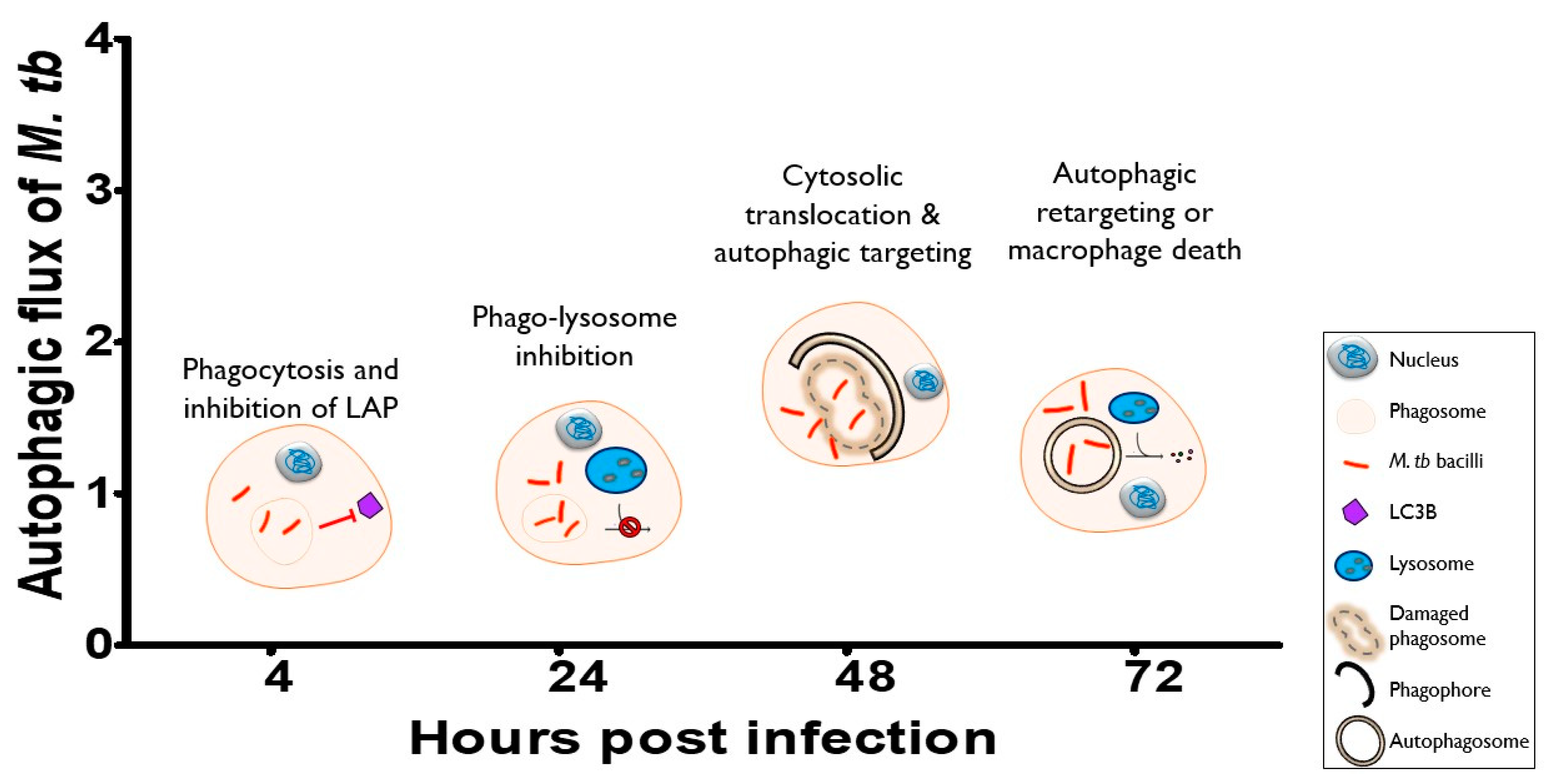
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sakula, A. Robert Koch: Centenary of the discovery of the tubercle bacillus, 1882. Thorax 1982, 37, 246–251. [Google Scholar] [CrossRef]
- WHO Global Tuberculosis Report 2022. Available online: https://www.who.int/publications-detail-redirect/9789240061729 (accessed on 14 December 2022).
- Rekha, R.S.; Mily, A.; Sultana, T.; Haq, A.; Ahmed, S.; Mostafa Kamal, S.M.; van Schadewijk, A.; Hiemstra, P.S.; Gudmundsson, G.H.; Agerberth, B.; et al. Immune responses in the treatment of drug-sensitive pulmonary tuberculosis with phenylbutyrate and vitamin D3 as host directed therapy. BMC Infect. Dis. 2018, 18, 303. [Google Scholar] [CrossRef]
- Young, C.; Walzl, G.; Du Plessis, N. Therapeutic host-directed strategies to improve outcome in tuberculosis. Mucosal Immunol. 2020, 13, 190–204. [Google Scholar] [CrossRef]
- Kiran, D.; Podell, B.K.; Chambers, M.; Basaraba, R.J. Host-directed therapy targeting the Mycobacterium tuberculosis granuloma: A review. Semin. Immunopathol. 2016, 38, 167–183. [Google Scholar] [CrossRef]
- Kolloli, A.; Subbian, S. Host-Directed Therapeutic Strategies for Tuberculosis. Front. Med. 2017, 4, 171. [Google Scholar] [CrossRef]
- Palucci, I.; Delogu, G. Host Directed Therapies for Tuberculosis: Futures Strategies for an Ancient Disease. CHE 2018, 63, 172–180. [Google Scholar] [CrossRef]
- Ray, K.; Bobard, A.; Danckaert, A.; Paz-Haftel, I.; Clair, C.; Ehsani, S.; Tang, C.; Sansonetti, P.; Van Nhieu, G.T.; Enninga, J. Tracking the dynamic interplay between bacterial and host factors during pathogen-induced vacuole rupture in real time. Cell. Microbiol. 2010, 12, 545–556. [Google Scholar] [CrossRef]
- Gujral, H.; Kushwaha, A.K.; Khurana, S. Utilization of Time Series Tools in Life-sciences and Neuroscience. J. Exp. Neurosci. 2020, 15, 2633105520963045. [Google Scholar] [CrossRef]
- Bussi, C.; Gutierrez, M.G. Mycobacterium tuberculosis infection of host cells in space and time. FEMS Microbiol. Rev. 2019, 43, 341–361. [Google Scholar] [CrossRef]
- Gordon, K.; Smith, A.F.M. Modeling and Monitoring Biomedical Times Series. J. Am. Stat. Assoc. 1990, 85, 328–337. [Google Scholar]
- Jamwal, S.V.; Mehrotra, P.; Singh, A.; Siddiqui, Z.; Basu, A.; Rao, K.V.S. Mycobacterial escape from macrophage phagosomes to the cytoplasm represents an alternate adaptation mechanism. Sci. Rep. 2016, 6, 23089. [Google Scholar] [CrossRef]
- Leake, E.S.; Myrvik, Q.N.; Wright, M.J. Phagosomal membranes of Mycobacterium bovis BCG-immune alveolar macrophages are resistant to disruption by Mycobacterium tuberculosis H37Rv. Infect. Immun. 1984, 45, 443–446. [Google Scholar] [CrossRef]
- Myrvik, Q.N.; Leake, E.S.; Wright, M.J. Disruption of phagosomal membranes of normal alveolar macrophages by the H37Rv strain of Mycobacterium tuberculosis. A correlate of virulence. Am. Rev. Respir. Dis. 1984, 129, 322–328. [Google Scholar]
- Simeone, R.; Sayes, F.; Song, O.; Gröschel, M.I.; Brodin, P.; Brosch, R.; Majlessi, L. Cytosolic Access of Mycobacterium tuberculosis: Critical Impact of Phagosomal Acidification Control and Demonstration of Occurrence In Vivo. PLoS Pathog. 2015, 11, e1004650. [Google Scholar] [CrossRef] [PubMed]
- Simeone, R.; Bobard, A.; Lippmann, J.; Bitter, W.; Majlessi, L.; Brosch, R.; Enninga, J. Phagosomal Rupture by Mycobacterium tuberculosis Results in Toxicity and Host Cell Death. PLoS Pathog. 2012, 8, e1002507. [Google Scholar] [CrossRef]
- van der Wel, N.; Hava, D.; Houben, D.; Fluitsma, D.; van Zon, M.; Pierson, J.; Brenner, M.; Peters, P.J. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell 2007, 129, 1287–1298. [Google Scholar] [CrossRef]
- Manzanillo, P.S.; Ayres, J.S.; Watson, R.O.; Collins, A.C.; Souza, G.; Rae, C.S.; Schneider, D.S.; Nakamura, K.; Shiloh, M.U.; Cox, J.S. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature 2013, 501, 512–516. [Google Scholar] [CrossRef]
- Watson, R.O.; Manzanillo, P.S.; Cox, J.S. Extracellular, M. tuberculosis DNA Targets Bacteria for Autophagy by Activating the Host DNA-Sensing Pathway. Cell 2012, 150, 803–815. [Google Scholar] [CrossRef]
- Zullo, A.J.; Lee, S. Mycobacterial Induction of Autophagy Varies by Species and Occurs Independently of Mammalian Target of Rapamycin Inhibition. J. Biol. Chem. 2012, 287, 12668–12678. [Google Scholar] [CrossRef]
- Deretic, V. Autophagy in Immunity and Cell-Autonomous Defense Against Intracellular Microbes. Immunol. Rev. 2011, 240, 92–104. [Google Scholar] [CrossRef]
- Gong, L.; Devenish, R.J.; Prescott, M. Autophagy as a macrophage response to bacterial infection. IUBMB Life 2012, 64, 740–747. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, M.G.; Master, S.S.; Singh, S.B.; Taylor, G.A.; Colombo, M.I.; Deretic, V. Autophagy Is a Defense Mechanism Inhibiting BCG and Mycobacterium tuberculosis Survival in Infected Macrophages. Cell 2004, 119, 753–766. [Google Scholar] [CrossRef] [PubMed]
- Lerner, T.R.; Queval, C.J.; Lai, R.P.; Russell, M.R.G.; Fearns, A.; Greenwood, D.J.; Collinson, L.; Wilkinson, R.J.; Gutierrez, M.G. Mycobacterium tuberculosis Cords in the Cytosol of Live Lymphatic Endothelial Cells to Evade Host Immune Surveillance. Available online: https://insight.jci.org/articles/view/136937/pdf (accessed on 9 May 2020).
- Adikesavalu, H.; Gopalaswamy, R.; Kumar, A.; Ranganathan, U.D.; Shanmugam, S. Autophagy Induction as a Host-Directed Therapeutic Strategy against Mycobacterium tuberculosis Infection. Medicina 2021, 57, 522. [Google Scholar] [CrossRef] [PubMed]
- Paik, S.; Kim, J.K.; Chung, C.; Jo, E.-K. Autophagy: A new strategy for host-directed therapy of tuberculosis. Virulence 2018, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Strong, E.J.; Lee, S. Targeting Autophagy as a Strategy for Developing New Vaccines and Host-Directed Therapeutics Against Mycobacteria. Front. Microbiol. 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Strong, E.J.; Jurcic Smith, K.L.; Saini, N.K.; Ng, T.W.; Porcelli, S.A.; Lee, S. Identification of Autophagy-Inhibiting Factors of Mycobacterium tuberculosis by High-Throughput Loss-of-Function Screening. Infect. Immun. 2020, 88. [Google Scholar] [CrossRef]
- Houben, D.; Demangel, C.; van Ingen, J.; Perez, J.; Baldeón, L.; Abdallah, A.M.; Caleechurn, L.; Bottai, D.; van Zon, M.; de Punder, K.; et al. ESX-1-mediated translocation to the cytosol controls virulence of mycobacteria. Cell. Microbiol. 2012, 14, 1287–1298. [Google Scholar] [CrossRef]
- Augenstreich, J.; Arbues, A.; Simeone, R.; Haanappel, E.; Wegener, A.; Sayes, F.; Chevalier, F.L.; Chalut, C.; Malaga, W.; Guilhot, C.; et al. ESX-1 and phthiocerol dimycocerosates of Mycobacterium tuberculosis act in concert to cause phagosomal rupture and host cell apoptosis. Cell. Microbiol. 2017, 19, e12726. [Google Scholar] [CrossRef]
- de Wet, S.; Du Toit, A.; Loos, B. Spermidine and Rapamycin Reveal Distinct Autophagy Flux Response and Cargo Receptor Clearance Profile. Cells 2021, 10, 95. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Acevedo Arozena, A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 1–222. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdalla, F.C.; Abeliovich, H.; Abraham, R.T.; Acevedo-Arozena, A.; Adeli, K.; Agholme, L.; Agnello, M.; Agostinis, P.; Aguirre-Ghiso, J.A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012, 8, 445–544. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)1. Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef] [PubMed]
- Sahani, M.H.; Itakura, E.; Mizushima, N. Expression of the autophagy substrate SQSTM1/p62 is restored during prolonged starvation depending on transcriptional upregulation and autophagy-derived amino acids. Autophagy 2014, 10, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Saqib, M.; Gupta, A.; Kumar, P.; Bhaskar, S. Autophagy induction by Mycobacterium indicus pranii promotes Mycobacterium tuberculosis clearance from RAW 264.7 macrophages. PLoS ONE 2017, 12, e0189606. [Google Scholar] [CrossRef]
- Kumar, R.; Sahu, S.K.; Kumar, M.; Jana, K.; Gupta, P.; Gupta, U.D.; Kundu, M.; Basu, J. MicroRNA 17-5p regulates autophagy in Mycobacterium tuberculosis-infected macrophages by targeting Mcl-1 and STAT3. Cell. Microbiol. 2016, 18, 679–691. [Google Scholar] [CrossRef]
- Sahu, S.K.; Kumar, M.; Chakraborty, S.; Banerjee, S.K.; Kumar, R.; Gupta, P.; Jana, K.; Gupta, U.D.; Ghosh, Z.; Kundu, M.; et al. MicroRNA 26a (miR-26a)/KLF4 and CREB-C/EBPβ regulate innate immune signaling, the polarization of macrophages and the trafficking of Mycobacterium tuberculosis to lysosomes during infection. PLoS Pathog. 2017, 13, e1006410. [Google Scholar] [CrossRef]
- Pahari, S.; Khan, N.; Aqdas, M.; Negi, S.; Kaur, J.; Agrewala, J.N. Infergen Stimulated Macrophages Restrict Mycobacterium tuberculosis Growth by Autophagy and Release of Nitric Oxide. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Sanjurjo, L.; Amézaga, N.; Vilaplana, C.; Cáceres, N.; Marzo, E.; Valeri, M.; Cardona, P.-J.; Sarrias, M.-R. The Scavenger Protein Apoptosis Inhibitor of Macrophages (AIM) Potentiates the Antimicrobial Response against Mycobacterium tuberculosis by Enhancing Autophagy. PLoS ONE 2013, 8, e79670. [Google Scholar] [CrossRef]
- Liu, J.; Ming, S.; Song, W.; Meng, X.; Xiao, Q.; Wu, M.; Wu, Y.; Xie, H.; Zhou, J.; Zhong, H.; et al. B and T lymphocyte attenuator regulates autophagy in mycobacterial infection via the AKT/mTOR signal pathway. Int. Immunopharmacol. 2021, 91, 107215. [Google Scholar] [CrossRef]
- Bongiovanni, B.; Mata-Espinosa, D.; D’Attilio, L.; Leon-Contreras, J.C.; Marquez-Velasco, R.; Bottasso, O.; Hernandez-Pando, R.; Bay, M.L. Effect of cortisol and/or DHEA on THP1-derived macrophages infected with Mycobacterium tuberculosis. Tuberculosis 2015, 95, 562–569. [Google Scholar] [CrossRef]
- Zulauf, K.E.; Sullivan, J.T.; Braunstein, M. The SecA2 pathway of Mycobacterium tuberculosis exports effectors that work in concert to arrest phagosome and autophagosome maturation. PLoS Pathog. 2018, 14, e1007011. [Google Scholar] [CrossRef] [PubMed]
- Karim, A.F.; Chandra, P.; Chopra, A.; Siddiqui, Z.; Bhaskar, A.; Singh, A.; Kumar, D. Express Path Analysis Identifies a Tyrosine Kinase Src-centric Network Regulating Divergent Host Responses to Mycobacterium tuberculosis Infection. J. Biol. Chem. 2011, 286, 40307–40319. [Google Scholar] [CrossRef]
- Chandra, V.; Bhagyaraj, E.; Nanduri, R.; Ahuja, N.; Gupta, P. NR1D1 ameliorates Mycobacterium tuberculosis clearance through regulation of autophagy. Autophagy 2015, 11, 1987–1997. [Google Scholar] [CrossRef]
- Gu, X.; Gao, Y.; Mu, D.-G.; Fu, E.-Q. MiR-23a-5p modulates mycobacterial survival and autophagy during Mycobacterium tuberculosis infection through TLR2/MyD88/NF-κB pathway by targeting TLR2. Exp. Cell Res. 2017, 354, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Köster, S.; Upadhyay, S.; Chandra, P.; Papavinasasundaram, K.; Yang, G.; Hassan, A.; Grigsby, S.J.; Mittal, E.; Park, H.S.; Jones, V.; et al. Mycobacterium tuberculosis is protected from NADPH oxidase and LC3-associated phagocytosis by the LCP protein CpsA. Proc. Natl. Acad. Sci. USA 2017, 114, E8711–E8720. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J. Detection of LC3-Associated Phagocytosis (LAP). Curr. Protoc. Cell Biol. 2020, 87, e104. [Google Scholar] [CrossRef]
- Duan, Z.; Chen, Q.; Du, L.; Tong, J.; Xu, S.; Zeng, R.; Ma, Y.; Chen, X.; Li, M. Phagocytosis of Candida albicans Inhibits Autophagic Flux in Macrophages. Oxid Med. Cell Longev. 2018, 2018, 4938649. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of Cells and Tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef] [PubMed]
- Gump, J.M.; Staskiewicz, L.; Morgan, M.J.; Bamberg, A.; Riches, D.W.H.; Thorburn, A. Autophagy variation within a cell population determines cell fate via selective degradation of Fap-1. Nat. Cell Biol. 2014, 16, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Nazarova, E.V.; Russell, D.G. Growing and handling of Mycobacterium tuberculosis for macrophage infection assays. Methods Mol. Biol. 2017, 1519, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Jaisinghani, N.; Dawa, S.; Singh, K.; Nandy, A.; Menon, D.; Bhandari, P.D.; Khare, G.; Tyagi, A.; Gandotra, S. Necrosis Driven Triglyceride Synthesis Primes Macrophages for Inflammation During Mycobacterium tuberculosis Infection. Front. Immunol. 2018, 9, 1490. [Google Scholar] [CrossRef] [PubMed]
- Mahamed, D.; Boulle, M.; Ganga, Y.; Mc Arthur, C.; Skroch, S.; Oom, L.; Catinas, O.; Pillay, K.; Naicker, M.; Rampersad, S.; et al. Intracellular growth of Mycobacterium tuberculosis after macrophage cell death leads to serial killing of host cells. eLife 2017, 6, e22028. [Google Scholar] [CrossRef] [PubMed]
- Beckwith, K.S.; Beckwith, M.S.; Ullmann, S.; Sætra, R.S.; Kim, H.; Marstad, A.; Åsberg, S.E.; Strand, T.A.; Haug, M.; Niederweis, M.; et al. Plasma membrane damage causes NLRP3 activation and pyroptosis during Mycobacterium tuberculosis infection. Nat. Commun. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.-W.; Jacobs, W.R. Critical role for NLRP3 in necrotic death triggered by Mycobacterium tuberculosis. Cell Microbiol. 2011, 13, 1371–1384. [Google Scholar] [CrossRef]
- Bohsali, A.; Abdalla, H.; Velmurugan, K.; Briken, V. The non-pathogenic mycobacteria M. smegmatis and M. fortuitum induce rapid host cell apoptosis via a caspase-3 and TNF dependent pathway. BMC Microbiol. 2010, 10, 237. [Google Scholar] [CrossRef]
- Leisching, G.; Pietersen, R.D.; Wiid, I.; Baker, B. Virulence, biochemistry, morphology and host-interacting properties of detergent-free cultured mycobacteria: An update. Tuberculosis. 2016, 100, 53–60. [Google Scholar] [CrossRef]
- Leisching, G.; Pietersen, R.D.; Mpongoshe, V.; van Heerden, C.; van Helden, P.; Wiid, I.; Baker, B. The Host Response to a Clinical MDR Mycobacterial Strain Cultured in a Detergent-Free Environment: A Global Transcriptomics Approach. PLoS ONE 2016, 11, e0153079. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4824497 (accessed on 13 April 2020). [CrossRef]
- du Toit, A.; Hofmeyr, J.-H.S.; Gniadek, T.J.; Loos, B. Measuring autophagosome flux. Autophagy 2018, 14, 1060–1071. [Google Scholar] [CrossRef]
- Sumpter, R.; Levine, B. Autophagy and Innate Immunity: Triggering, Targeting and Tuning. Semin. Cell Dev. Biol. 2010, 21, 699–711. [Google Scholar] [CrossRef]
- Assanga, I. Cell growth curves for different cell lines and their relationship with biological activities. Int. J. Biotechnol. Mol. Biol. Res. 2013, 4, 60–70. [Google Scholar] [CrossRef]
- Li, Z.; Ji, X.; Wang, D.; Liu, J.; Zhang, X. Autophagic flux is highly active in early mitosis and differentially regulated throughout the cell cycle. Oncotarget 2016, 7, 39705–39718. [Google Scholar] [CrossRef]
- Romao, S.; Gasser, N.; Becker, A.C.; Guhl, B.; Bajagic, M.; Vanoaica, D.; Ziegler, U.; Roesler, J.; Dengjel, J.; Reichenbach, J.; et al. Autophagy proteins stabilize pathogen-containing phagosomes for prolonged MHC II antigen processing. J. Cell Biol. 2013, 203, 757–766. [Google Scholar] [CrossRef]
- Romao, S.; Münz, C. LC3-associated phagocytosis. Autophagy 2014, 10, 526–528. [Google Scholar] [CrossRef]
- Lerner, T.R.; Borel, S.; Greenwood, D.J.; Repnik, U.; Russell, M.R.G.; Herbst, S.; Jones, M.L.; Collinson, L.M.; Griffiths, G.; Gutierrez, M.G. Mycobacterium tuberculosis replicates within necrotic human macrophages. J. Cell Biol. 2017, 216, 583–594. [Google Scholar] [CrossRef]
- Bernard, E.M.; Fearns, A.; Bussi, C.; Santucci, P.; Peddie, C.J.; Lai, R.J.; Collinson, L.M.; Gutierrez, M.G.M. tuberculosis infection of human iPSC-derived macrophages reveals complex membrane dynamics during xenophagy evasion. J. Cell Sci. 2020, 134. [Google Scholar] [CrossRef]
- Lerner, T.R.; Queval, C.J.; Fearns, A.; Repnik, U.; Griffiths, G.; Gutierrez, M.G. Phthiocerol dimycocerosates promote access to the cytosol and intracellular burden of Mycobacterium tuberculosis in lymphatic endothelial cells. BMC Biol. 2018, 16. [Google Scholar] [CrossRef]
- Quigley, J.; Hughitt, V.K.; Velikovsky, C.A.; Mariuzza, R.A.; El-Sayed, N.M.; Briken, V. The Cell Wall Lipid PDIM Contributes to Phagosomal Escape and Host Cell Exit of Mycobacterium tuberculosis. mBio 2017, 8, e00148-17. [Google Scholar] [CrossRef]
- Ufimtseva, E.; Eremeeva, N.; Bayborodin, S.; Umpeleva, T.; Vakhrusheva, D.; Skornyakov, S. Mycobacterium tuberculosis with different virulence reside within intact phagosomes and inhibit phagolysosomal biogenesis in alveolar macrophages of patients with pulmonary tuberculosis. Tuberculosis 2019, 114, 77–90. [Google Scholar] [CrossRef]
- Dallenga, T.; Repnik, U.; Corleis, B.; Eich, J.; Reimer, R.; Griffiths, G.W.; Schaible, U.E. M. tuberculosis-Induced Necrosis of Infected Neutrophils Promotes Bacterial Growth following Phagocytosis by Macrophages. Cell Host Microbe 2017, 22, 519–530. [Google Scholar] [CrossRef]
- Li, F.; Gao, B.; Xu, W.; Chen, L.; Xiong, S. The Defect in Autophagy Induction by Clinical Isolates of Mycobacterium tuberculosis Is Correlated with Poor Tuberculosis Outcomes. PLoS ONE 2016, 11, e0147810. [Google Scholar] [CrossRef]
- Cao, Q.-H.; Liu, F.; Yang, Z.-L.; Fu, X.-H.; Yang, Z.-H.; Liu, Q.; Wang, L.; Wan, X.-B.; Fan, X.-J. Prognostic value of autophagy related proteins ULK1, Beclin 1, ATG3, ATG5, ATG7, ATG9, ATG10, ATG12, LC3B and p62/SQSTM1 in gastric cancer. Am. J. Transl. Res. 2016, 8, 3831–3847. [Google Scholar] [PubMed]
- Tang, D.Y.L.; Ellis, R.A.; Lovat, P.E. Prognostic Impact of Autophagy Biomarkers for Cutaneous Melanoma. Front. Oncol. 2016, 6, 236. [Google Scholar] [CrossRef]
- Masuda, G.; Yashiro, M.; Kitayama, K.; Miki, Y.; Kasashima, H.; Kinoshita, H.; Morisaki, T.; Fukuoka, T.; Hasegawa, T.; Sakurai, K.; et al. Clinicopathological Correlations of Autophagy-related Proteins LC3, Beclin 1 and p62 in Gastric Cancer. Anticancer. Res. 2016, 36, 129–136. [Google Scholar] [PubMed]
- Schmitz, K.J.; Ademi, C.; Bertram, S.; Schmid, K.W.; Baba, H.A. Prognostic relevance of autophagy-related markers LC3, p62/sequestosome 1, Beclin-1 and ULK1 in colorectal cancer patients with respect to KRAS mutational status. World J. Surg Onc. 2016, 14, 189. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Sun, C.; Tian, D.; Li, Y.; Gao, X.; He, S.; Li, T. Expression and clinical significances of Beclin1, LC3 and mTOR in colorectal cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 3882–3891. [Google Scholar]
- Choi, J.; Jung, W.; Koo, J.S. Expression of autophagy-related markers beclin-1, light chain 3A, light chain 3B and p62 according to the molecular subtype of breast cancer. Histopathology 2013, 62, 275–286. [Google Scholar] [CrossRef]
- Bortnik, S.; Gorski, S.M. Clinical Applications of Autophagy Proteins in Cancer: From Potential Targets to Biomarkers. Int. J. Mol. Sci. 2017, 18, 1496. [Google Scholar] [CrossRef]
- Zhang, C.; Jiang, J.; Wang, L.; Zheng, L.; Xu, J.; Qi, X.; Huang, H.; Lu, J.; Li, K.; Wang, H. Identification of Autophagy-Associated Biomarkers and Corresponding Regulatory Factors in the Progression of Colorectal Cancer. Front. Genet. 2020, 11, 245. [Google Scholar] [CrossRef]
- Bensalem, J.; Hattersley, K.J.; Hein, L.K.; Teong, X.T.; Carosi, J.M.; Hassiotis, S.; Grose, R.H.; Fourrier, C.; Heilbronn, L.K.; Sargeant, T.J. Measurement of autophagic flux in humans: An optimized method for blood samples. Autophagy 2020, 10, 1–18. [Google Scholar] [CrossRef]
- Sargeant, T.J.; Bensalem, J. Human autophagy measurement: An underappreciated barrier to translation. Trends Mol. Med. 2021, 27, 1091–1094. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Okugbeni, N.; du Toit, A.; Cole-Holman, V.; Johnson, G.; Loos, B.; Kinnear, C. Measurement of Autophagy Activity Reveals Time-Dependent, Bacteria-Specific Turnover during Mycobacterium tuberculosis Infection. Pathogens 2023, 12, 24. https://doi.org/10.3390/pathogens12010024
Okugbeni N, du Toit A, Cole-Holman V, Johnson G, Loos B, Kinnear C. Measurement of Autophagy Activity Reveals Time-Dependent, Bacteria-Specific Turnover during Mycobacterium tuberculosis Infection. Pathogens. 2023; 12(1):24. https://doi.org/10.3390/pathogens12010024
Chicago/Turabian StyleOkugbeni, Naomi, André du Toit, Victoria Cole-Holman, Glynis Johnson, Ben Loos, and Craig Kinnear. 2023. "Measurement of Autophagy Activity Reveals Time-Dependent, Bacteria-Specific Turnover during Mycobacterium tuberculosis Infection" Pathogens 12, no. 1: 24. https://doi.org/10.3390/pathogens12010024
APA StyleOkugbeni, N., du Toit, A., Cole-Holman, V., Johnson, G., Loos, B., & Kinnear, C. (2023). Measurement of Autophagy Activity Reveals Time-Dependent, Bacteria-Specific Turnover during Mycobacterium tuberculosis Infection. Pathogens, 12(1), 24. https://doi.org/10.3390/pathogens12010024






