Amphiphysin AoRvs167-Mediated Membrane Curvature Facilitates Trap Formation, Endocytosis, and Stress Resistance in Arthrobotrys oligospora
Abstract
1. Introduction
2. Results
2.1. Sequence and Protein Structure Analysis of AoRvs167
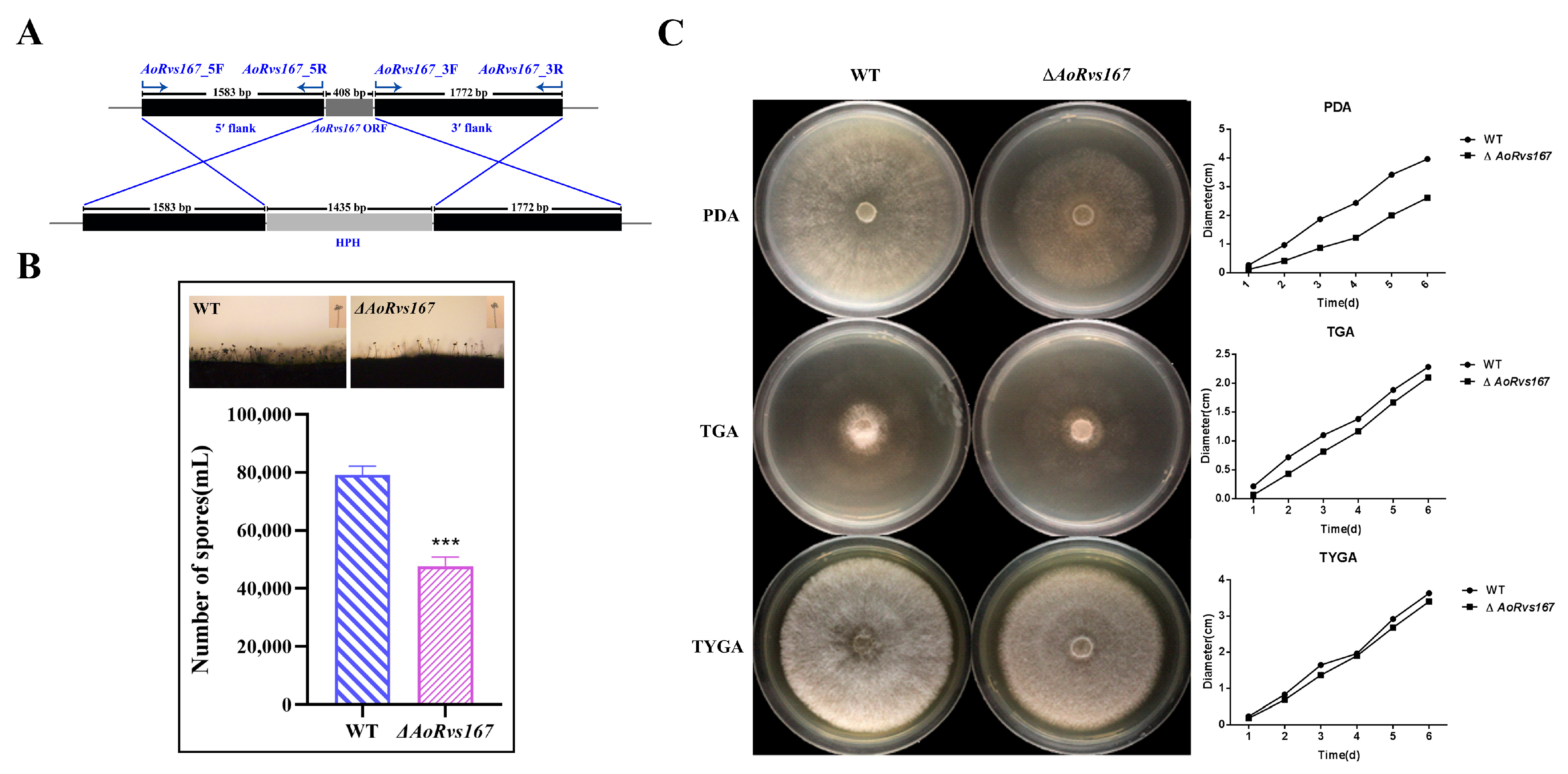
2.2. AoRvs167 Is Associated with the Colony Growth
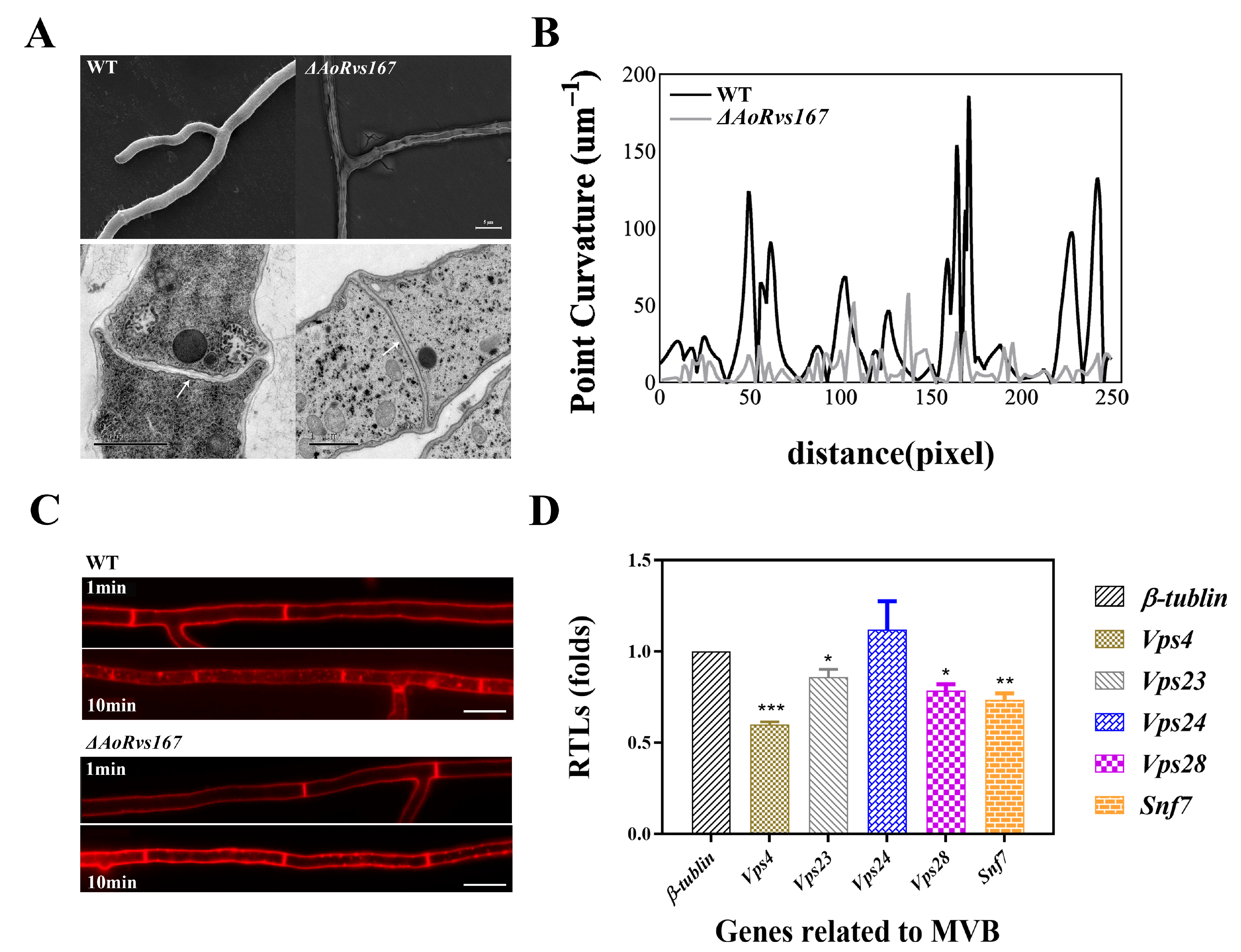
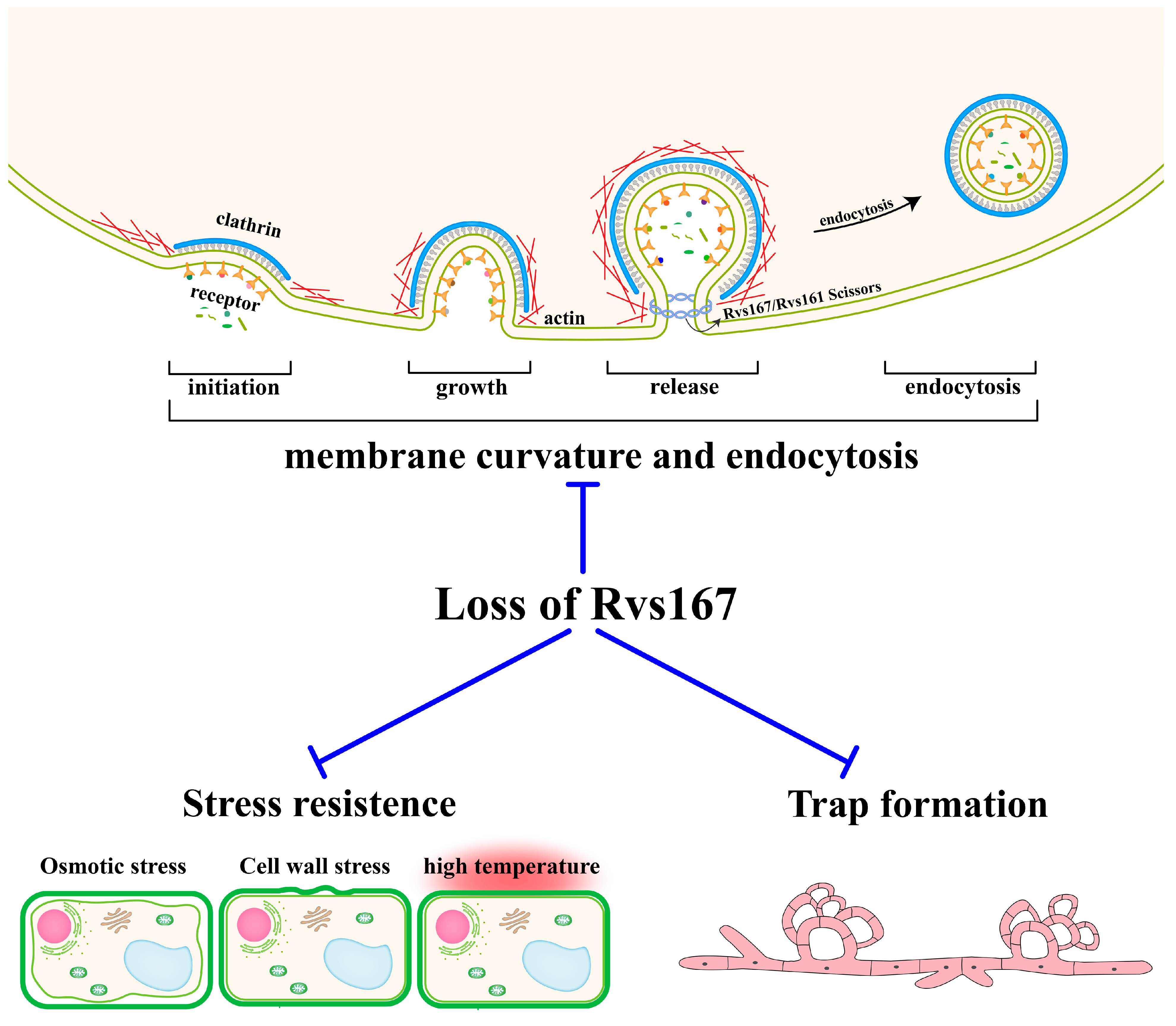
2.3. AoRvs167 Mediates Membrane Curvature and Endocytosis
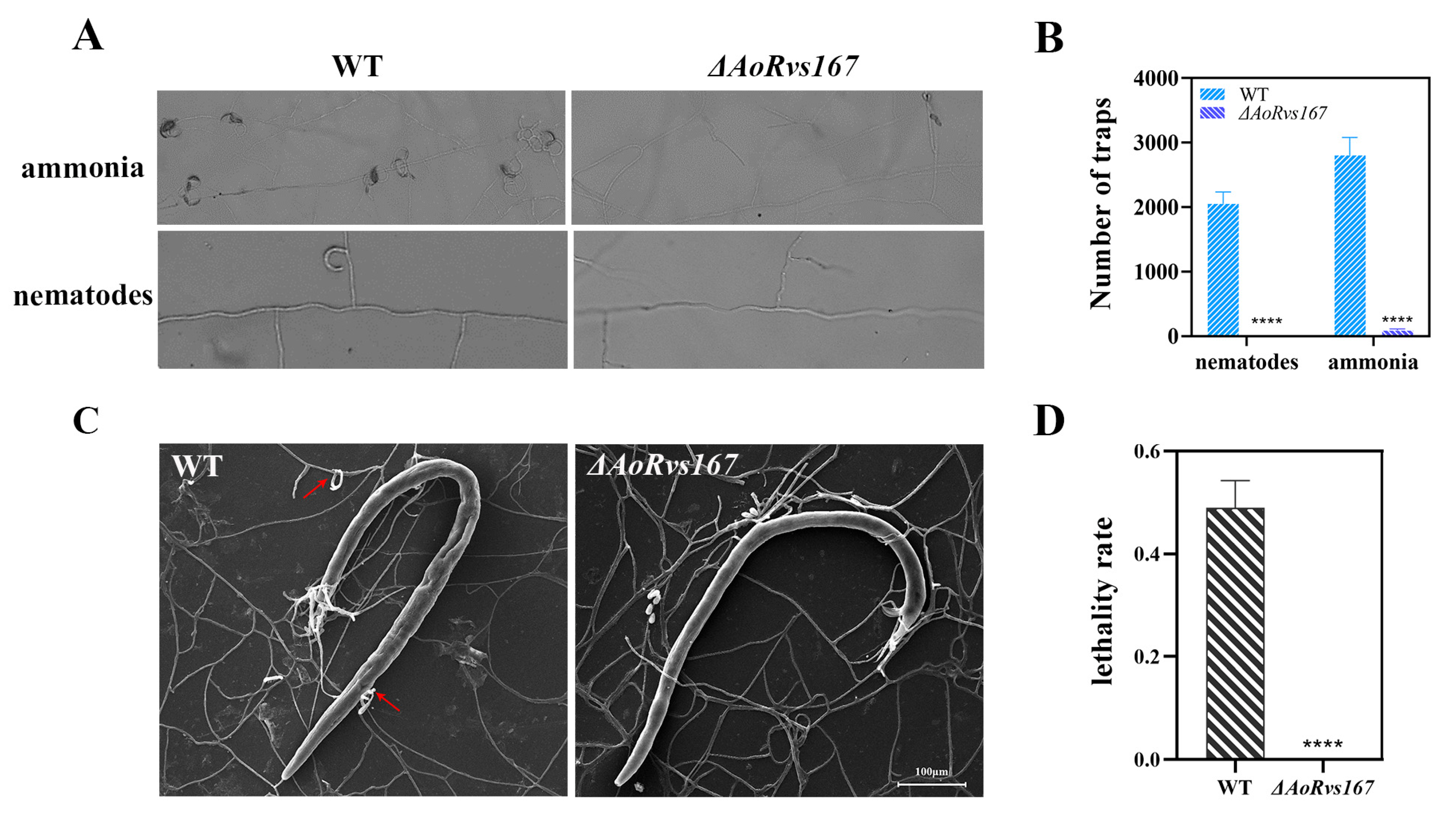
2.4. AoRvs167 Deletion Significantly Suppressed the Trap Generation
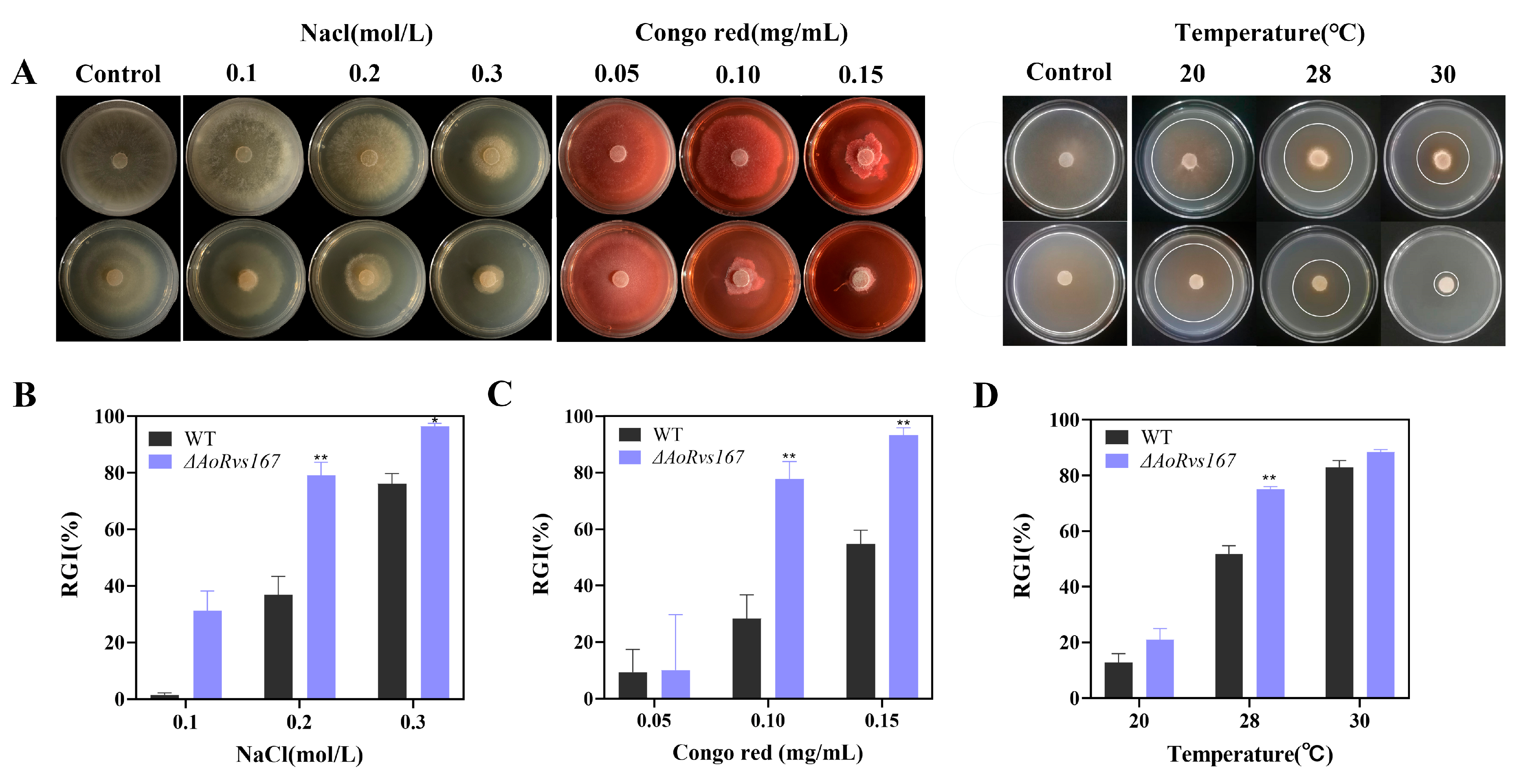
2.5. Rvs167 Gene Regulates Stress Resistance of A. oligospora
3. Discussion
4. Materials and Methods
4.1. Fungal Strains and Culture Conditions
4.2. Sequence and Phylogenetic Analyses of AoRvs167
4.3. Deletion of the AoRvs167
4.4. Comparison of Mycelial Growth, Sporulation and Hyphal Morphology
4.5. Endocytosis Analyses
4.6. TEM and SEM Sample Preparation
4.7. Trap Formation and Pathogenicity Assays
4.8. Stress Resistance
4.9. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- McMahon, H.T.; Boucrot, E. Membrane curvature at a glance. J. Cell Sci. 2015, 128, 1065–1070. [Google Scholar] [CrossRef] [PubMed]
- Bernardino de la Serna, J.; Schütz, G.J.; Eggeling, C.; Cebecauer, M. There is no simple model of the plasma membrane organization. Front. Cell Dev. Biol. 2016, 4, 106. [Google Scholar] [CrossRef] [PubMed]
- Harker-Kirschneck, L.; Hafner, A.E.; Yao, T.; Vanhille-Campos, C.; Jiang, X.; Pulschen, A.; Hurtig, F.; Hryniuk, D.; Culley, S.; Henriques, R. Physical mechanisms of ESCRT-III–driven cell division. Proc. Natl. Acad. Sci. USA 2022, 119, e2107763119. [Google Scholar] [CrossRef] [PubMed]
- Mim, C.; Unger, V.M. Membrane curvature and its generation by BAR proteins. Trends Biochem. Sci. 2012, 37, 526–533. [Google Scholar] [CrossRef] [PubMed]
- Carman, P.J.; Dominguez, R. BAR domain proteins—A linkage between cellular membranes, signaling pathways, and the actin cytoskeleton. Biophys. Rev. 2018, 10, 1587–1604. [Google Scholar] [CrossRef] [PubMed]
- Youn, J.-Y.; Friesen, H.; Kishimoto, T.; Henne, W.M.; Kurat, C.F.; Ye, W.; Ceccarelli, D.F.; Sicheri, F.; Kohlwein, S.D.; McMahon, H.T. Dissecting BAR domain function in the yeast Amphiphysins Rvs161 and Rvs167 during endocytosis. Mol. Biol. Cell 2010, 21, 3054–3069. [Google Scholar] [CrossRef]
- Zhao, H.; Michelot, A.; Koskela, E.V.; Tkach, V.; Stamou, D.; Drubin, D.G.; Lappalainen, P. Membrane-sculpting BAR domains generate stable lipid microdomains. Cell Rep. 2013, 4, 1213–1223. [Google Scholar] [CrossRef]
- Feyder, S.; De Craene, J.-O.; Bär, S.; Bertazzi, D.L.; Friant, S. Membrane trafficking in the yeast Saccharomyces cerevisiae model. Int. J. Mol. Sci. 2015, 16, 1509–1525. [Google Scholar] [CrossRef]
- Prokic, I.; Cowling, B.S.; Laporte, J. Amphiphysin 2 (BIN1) in physiology and diseases. J. Mol. Med. 2014, 92, 453–463. [Google Scholar] [CrossRef]
- Ren, G.; Vajjhala, P.; Lee, J.S.; Winsor, B.; Munn, A.L. The BAR domain proteins: Molding membranes in fission, fusion, and phagy. Microbiol. Mol. Biol. Rev. 2006, 70, 37–120. [Google Scholar] [CrossRef]
- Herlo, R.; Lund, V.K.; Lycas, M.D.; Jansen, A.M.; Khelashvili, G.; Andersen, R.C.; Bhatia, V.; Pedersen, T.S.; Albornoz, P.B.; Johner, N. An amphipathic helix directs cellular membrane curvature sensing and function of the BAR domain protein PICK1. Cell Rep. 2018, 23, 2056–2069. [Google Scholar] [CrossRef] [PubMed]
- Sivadon, P.; Crouzet, M.; Aigle, M. Functional assessment of the yeast Rvs161 and Rvs167 protein domains. FEBS Lett. 1997, 417, 21–27. [Google Scholar] [CrossRef]
- Stamenova, S.D.; Dunn, R.; Adler, A.S.; Hicke, L. The Rsp5 ubiquitin ligase binds to and ubiquitinates members of the yeast CIN85-endophilin complex, Sla1-Rvs167. J. Biol. Chem. 2004, 279, 16017–16025. [Google Scholar] [CrossRef] [PubMed]
- Dagdas, Y.F.; Yoshino, K.; Dagdas, G.; Ryder, L.S.; Bielska, E.; Steinberg, G.; Talbot, N.J. Septin-mediated plant cell invasion by the rice blast fungus, Magnaporthe oryzae. Science 2012, 336, 1590–1595. [Google Scholar] [CrossRef] [PubMed]
- Douglas, L.M.; Martin, S.W.; Konopka, J.B. BAR domain proteins Rvs161 and Rvs167 contribute to Candida albicans endocytosis, morphogenesis, and virulence. Infect. Immun. 2009, 77, 4150–4160. [Google Scholar] [CrossRef] [PubMed]
- Gkourtsa, A.; van den Burg, J.; Strijbis, K.; Avula, T.; Bijvoets, S.; Timm, D.; Hochstenbach, F.; Distel, B. Identification and characterization of Rvs162/Rvs167-3, a novel N-BAR heterodimer in the human fungal pathogen Candida albicans. Eukaryot Cell 2015, 14, 182–193. [Google Scholar] [CrossRef]
- Kornitzer, D. Regulation of Candida albicans hyphal morphogenesis by endogenous signals. J. Fungi 2019, 5, 21. [Google Scholar] [CrossRef]
- Wang, P.; Shen, G. The endocytic adaptor proteins of pathogenic fungi: Charting new and familiar pathways. Med. Mycol. 2011, 49, 449–457. [Google Scholar] [CrossRef][Green Version]
- De Rossi, P.; Nomura, T.; Andrew, R.J.; Masse, N.Y.; Sampathkumar, V.; Musial, T.F.; Sudwarts, A.; Recupero, A.J.; Le Metayer, T.; Hansen, M.T. Neuronal BIN1 regulates presynaptic neurotransmitter release and memory consolidation. Cell Rep. 2020, 30, 3520–3535.e7. [Google Scholar] [CrossRef]
- Guimas Almeida, C.; Sadat Mirfakhar, F.; Perdigão, C.; Burrinha, T. Impact of late-onset Alzheimer’s genetic risk factors on beta-amyloid endocytic production. Cell. Mol. Life Sci. 2018, 75, 2577–2589. [Google Scholar] [CrossRef]
- Su, H.; Zhao, Y.; Zhou, J.; Feng, H.; Jiang, D.; Zhang, K.Q.; Yang, J. Trapping devices of nematode-trapping fungi: Formation, evolution, and genomic perspectives. Biol. Rev. 2017, 92, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Dijksterhuis, J.; Veenhuis, M.; Harder, W.; Nordbring-Hertz, B. Nematophagous fungi: Physiological aspects and structure–function relationships. Adv. Microb. Physiol. 1994, 36, 111–143. [Google Scholar] [PubMed]
- Liu, X.; Zhang, K. Dactylella shizishanna sp. nov., from Shizi Mountain, China. Fungal Divers. 2003, 14, 103–107. [Google Scholar]
- Dhawan, S.; Narayana, R.; Babn, N.P. Influence of abiotic and biotic factors on growth of Paecilomyces lilacinus, Arthrobotrys oligospora and Pochonia chlamydosporia and parasitization of eggs/trapping of Meloidogyne incognita juveniles. Ann. Plant Prot. Sci. 2004, 12, 369–372. [Google Scholar]
- Wang, X.; Li, G.-H.; Zou, C.-G.; Ji, X.-L.; Liu, T.; Zhao, P.-J.; Liang, L.-M.; Xu, J.-P.; An, Z.-Q.; Zheng, X. Bacteria can mobilize nematode-trapping fungi to kill nematodes. Nat. Commun. 2014, 5, 5776. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.-M.; Zhang, K.-Q. Arthrobotrys oligospora: A model organism for understanding the interaction between fungi and nematodes. Mycology 2011, 2, 59–78. [Google Scholar] [CrossRef]
- Yang, X.; Ma, N.; Yang, L.; Zheng, Y.; Zhen, Z.; Li, Q.; Xie, M.; Li, J.; Zhang, K.-Q.; Yang, J. Two Rab GTPases play different roles in conidiation, trap formation, stress resistance, and virulence in the nematode-trapping fungus Arthrobotrys oligospora. Appl. Microbiol. Biotechnol. 2018, 102, 4601–4613. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Yang, X.; Xie, M.; Zhang, G.; Yang, L.; Bai, N.; Zhao, Y.; Li, D.; Zhang, K.-Q.; Yang, J. The Arf-GAP AoGlo3 regulates conidiation, endocytosis, and pathogenicity in the nematode-trapping fungus Arthrobotrys oligospora. Fungal Genet. Biol. 2020, 138, 103352. [Google Scholar] [CrossRef]
- Zhen, Z.; Xing, X.; Xie, M.; Yang, L.; Yang, X.; Zheng, Y.; Chen, Y.; Ma, N.; Li, Q.; Zhang, K.-Q. MAP kinase Slt2 orthologs play similar roles in conidiation, trap formation, and pathogenicity in two nematode-trapping fungi. Fungal Genet. Biol. 2018, 116, 42–50. [Google Scholar] [CrossRef]
- Zhen, Z.; Zhang, G.; Yang, L.; Ma, N.; Li, Q.; Ma, Y.; Niu, X.; Zhang, K.-Q.; Yang, J. Characterization and functional analysis of calcium/calmodulin-dependent protein kinases (CaMKs) in the nematode-trapping fungus Arthrobotrys oligospora. Appl. Microbiol. Biotechnol. 2019, 103, 819–832. [Google Scholar] [CrossRef]
- Zhou, D.; Zhu, Y.; Bai, N.; Yang, L.; Xie, M.; Yang, J.; Zhu, M.; Zhang, K.-Q.; Yang, J. AoATG5 plays pleiotropic roles in vegetative growth, cell nucleus development, conidiation, and virulence in the nematode-trapping fungus Arthrobotrys oligospora. Sci. China Life Sci. 2022, 65, 412–425. [Google Scholar] [CrossRef]
- Zhou, L.; Li, M.; Cui, P.; Tian, M.; Xu, Y.; Zheng, X.; Zhang, K.; Li, G.; Wang, X. Arrestin-Coding Genes Regulate Endocytosis, Sporulation, Pathogenicity, and Stress Resistance in Arthrobotrys oligospora. Front. Cell. Infect. Microbiol. 2022, 12, 754333. [Google Scholar] [CrossRef] [PubMed]
- Seiler, N. Ammonia and Alzheimer’s disease. Neurochem. Int. 2002, 41, 189–207. [Google Scholar] [CrossRef]
- Vlanti, A.; Diallinas, G. The Aspergillus nidulans FcyB cytosine-purine scavenger is highly expressed during germination and in reproductive compartments and is downregulated by endocytosis. Mol. Microbiol. 2008, 68, 959–977. [Google Scholar] [CrossRef] [PubMed]
- Myers, M.D.; Ryazantsev, S.; Hicke, L.; Payne, G.S. Calmodulin promotes N-BAR domain-mediated membrane constriction and endocytosis. Dev. Cell 2016, 37, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Tanahashi, R.; Afiah, T.S.N.; Nishimura, A.; Watanabe, D.; Takagi, H. The C2 domain of the ubiquitin ligase Rsp5 is required for ubiquitination of the endocytic protein Rvs167 upon change of nitrogen source. FEMS Yeast Res. 2020, 20, foaa058. [Google Scholar] [CrossRef] [PubMed]
- Toume, M.; Tani, M. Yeast lacking the amphiphysin family protein Rvs167 is sensitive to disruptions in sphingolipid levels. FEBS J. 2016, 283, 2911–2928. [Google Scholar] [CrossRef]
- Breton, A.M.; Schaeffer, J.; Aigle, M. The yeast Rvs161 and Rvs167 proteins are involved in secretory vesicles targeting the plasma membrane and in cell integrity. Yeast 2001, 18, 1053–1068. [Google Scholar] [CrossRef]
- Omura, F.; Takagi, M.; Kodama, Y. Compromised chitin synthesis in lager yeast affects its Congo red resistance and release of mannoproteins from the cells. FEMS Microbiol. Lett. 2020, 367, fnaa181. [Google Scholar] [CrossRef]
- Navarro, P.; Durrens, P.; Aigle, M. Protein–protein interaction between the RVS161 and RVS167 gene products of Saccharomyces cerevisiae. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1997, 1343, 187–192. [Google Scholar] [CrossRef]
- Colot, H.V.; Park, G.; Turner, G.E.; Ringelberg, C.; Crew, C.M.; Litvinkova, L.; Weiss, R.L.; Borkovich, K.A.; Dunlap, J.C. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 2006, 103, 10352–10357. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Cramer, P. AlphaFold2 and the future of structural biology. Nat. Struct. Mol. Biol. 2021, 28, 704–705. [Google Scholar] [CrossRef] [PubMed]
- Desta, I.T.; Porter, K.A.; Xia, B.; Kozakov, D.; Vajda, S. Performance and Its Limits in Rigid Body Protein-Protein Docking. Structure 2020, 28, 1071–1081.e3. [Google Scholar] [CrossRef]
- Kozakov, D.; Beglov, D.; Bohnuud, T.; Mottarella, S.E.; Xia, B.; Hall, D.R.; Vajda, S. How good is automated protein docking? Proteins 2013, 81, 2159–2166. [Google Scholar] [CrossRef]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef] [PubMed]
- Vajda, S.; Yueh, C.; Beglov, D.; Bohnuud, T.; Mottarella, S.E.; Xia, B.; Hall, D.R.; Kozakov, D. New additions to the ClusPro server motivated by CAPRI. Proteins 2017, 85, 435–444. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- DeLano, W.L. Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Cryst. 2002, 40, 82–92. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Le, S.Q.; Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef] [PubMed]
- Tunlid, A.; Åhman, J.; Oliver, R. Transformation of the nematode-trapping fungus Arthrobotrys oligospora. FEMS Microbiol. Lett. 1999, 173, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Dai, Z.; Zhang, K.; Li, G.; Zhao, P. Pathogenicity and metabolites of endoparasitic nematophagous fungus Drechmeria coniospora YMF1. 01759 against nematodes. Microorganisms 2021, 9, 1735. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, P.; Tian, M.; Huang, J.; Zheng, X.; Guo, Y.; Li, G.; Wang, X. Amphiphysin AoRvs167-Mediated Membrane Curvature Facilitates Trap Formation, Endocytosis, and Stress Resistance in Arthrobotrys oligospora. Pathogens 2022, 11, 997. https://doi.org/10.3390/pathogens11090997
Cui P, Tian M, Huang J, Zheng X, Guo Y, Li G, Wang X. Amphiphysin AoRvs167-Mediated Membrane Curvature Facilitates Trap Formation, Endocytosis, and Stress Resistance in Arthrobotrys oligospora. Pathogens. 2022; 11(9):997. https://doi.org/10.3390/pathogens11090997
Chicago/Turabian StyleCui, Peijie, Mengqing Tian, Jinrong Huang, Xi Zheng, Yingqi Guo, Guohong Li, and Xin Wang. 2022. "Amphiphysin AoRvs167-Mediated Membrane Curvature Facilitates Trap Formation, Endocytosis, and Stress Resistance in Arthrobotrys oligospora" Pathogens 11, no. 9: 997. https://doi.org/10.3390/pathogens11090997
APA StyleCui, P., Tian, M., Huang, J., Zheng, X., Guo, Y., Li, G., & Wang, X. (2022). Amphiphysin AoRvs167-Mediated Membrane Curvature Facilitates Trap Formation, Endocytosis, and Stress Resistance in Arthrobotrys oligospora. Pathogens, 11(9), 997. https://doi.org/10.3390/pathogens11090997






