Virulence Mechanisms of Common Uropathogens and Their Intracellular Localisation within Urothelial Cells
Abstract
:1. Introduction
2. Results
2.1. Part 1: Visualisation of Uropathogens In Vivo
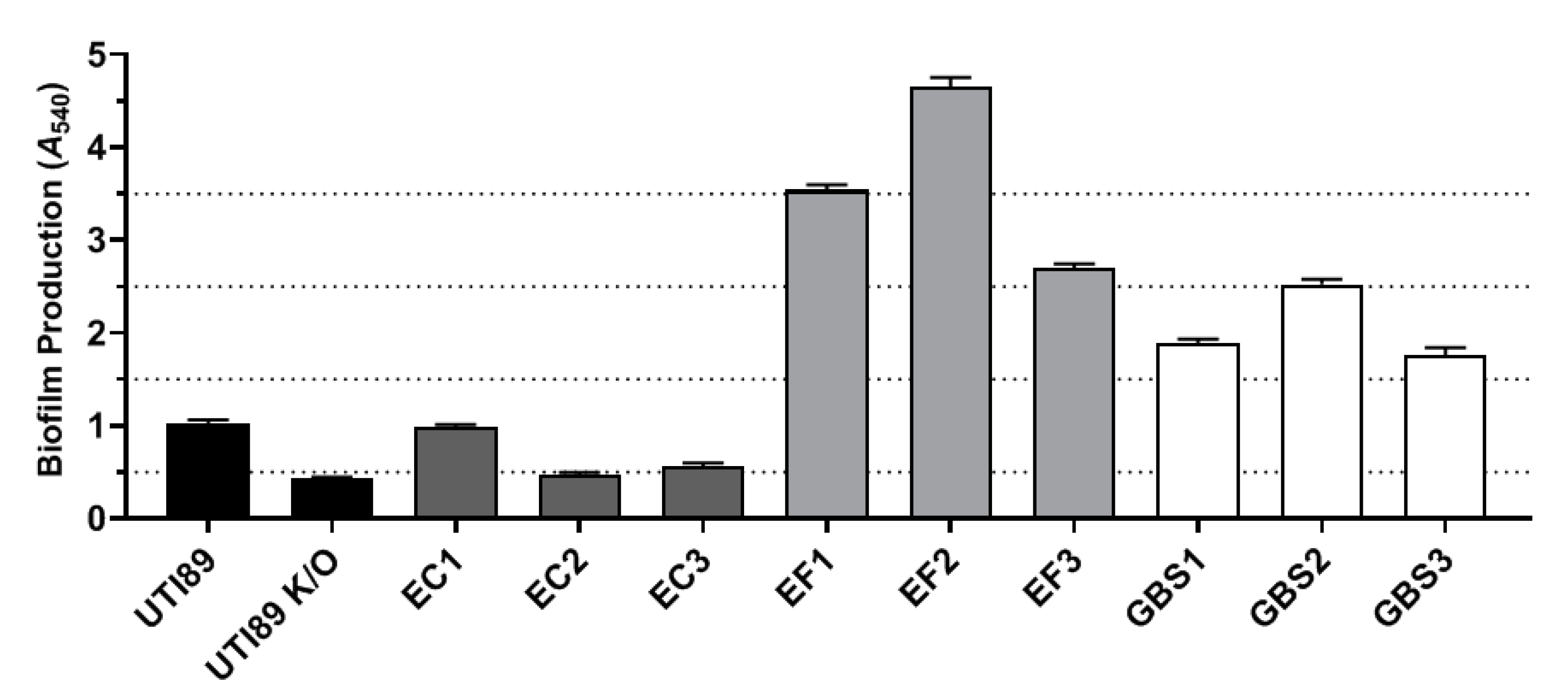
2.2. Part 2: In Vitro Examination of Uropathogen Virulence Mechanisms
3. Discussion
4. Materials and Methods
4.1. Collection and Processing of Urine Samples
4.2. Uropathogen Isolation and Culture
4.3. Cell Culture and Invasion Assay
4.4. Immunofluorescence Staining and Confocal Imaging
4.5. Yeast Cell Agglutination
4.6. Biofilm Assay
4.7. Haemolytic Assay
4.8. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Al-Badr, A.; Al-Shaikh, G. Recurrent urinary tract infections management in women: A review. Sultan Qaboos Univ. Med. J. 2013, 13, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Suskind, A.M.; Saigal, C.S.; Hanley, J.M.; Lai, J.; Setodji, C.M.; Clemens, J.Q. Incidence and management of uncomplicated recurrent urinary tract infections in a national sample of women in the United States. Urology 2016, 90, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Medina, M.; Castillo-Pino, E. An introduction to the epidemiology and burden of urinary tract infections. Ther. Adv. Urol. 2019, 11, 1756287219832172. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.L.; Boyko, E.J.; Scholes, D.; Abraham, L.; Gupta, K.; Fihn, S.D. Predictors of urinary tract infection after menopause: A prospective study. Am. J. Med. 2004, 117, 903–911. [Google Scholar] [CrossRef]
- Wang, C.; Symington, J.W.; Ma, E.; Cao, B.; Mysorekar, I.U. Estrogenic modulation of uropathogenic Escherichia coli infection pathogenesis in a murine menopause model. Infect. Immun. 2013, 81, 733–739. [Google Scholar] [CrossRef] [PubMed]
- Stapleton, A.E. The Vaginal Microbiota and Urinary Tract Infection. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Mulvey, M.A.; Schilling, J.D.; Hultgren, S.J. Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect. Immun. 2001, 69, 4572–4579. [Google Scholar] [CrossRef]
- Justice, S.S.; Lauer, S.R.; Hultgren, S.J.; Hunstad, D.A. Maturation of intracellular Escherichia coli communities requires SurA. Infect. Immun. 2006, 74, 4793–4800. [Google Scholar] [CrossRef]
- Rosen, D.A.; Hooton, T.M.; Stamm, W.E.; Humphrey, P.A.; Hultgren, S.J. Detection of intracellular bacterial communities in human urinary tract infection. PLoS Med. 2007, 4, e329. [Google Scholar] [CrossRef]
- Horsley, H.; Malone-Lee, J.; Holland, D.; Tuz, M.; Hibbert, A.; Kelsey, M.; Kupelian, A.; Rohn, J.L. Enterococcus faecalis subverts and invades the host urothelium in patients with chronic urinary tract infection. PLoS ONE 2013, 8, e83637. [Google Scholar] [CrossRef]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef]
- Leclercq, S.Y.; Sullivan, M.J.; Ipe, D.S.; Smith, J.P.; Cripps, A.W.; Ulett, G.C. Pathogenesis of Streptococcus urinary tract infection depends on bacterial strain and β-hemolysin/cytolysin that mediates cytotoxicity, cytokine synthesis, inflammation and virulence. Sci. Rep. 2016, 6, 29000. [Google Scholar] [CrossRef] [PubMed]
- Totsika, M.; Kostakioti, M.; Hannan, T.J.; Upton, M.; Beatson, S.A.; Janetka, J.W.; Hultgren, S.J.; Schembri, M.A. A FimH inhibitor prevents acute bladder infection and treats chronic cystitis caused by multidrug-resistant uropathogenic Escherichia coli ST131. J. Infect. Dis. 2013, 208, 921–928. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Chen, Z.; Gawthorne, J.A.; Mukerjee, C.; Varettas, K.; Mansfield, K.J.; Schembri, M.A.; Moore, K.H. Detection of intracellular bacteria in exfoliated urothelial cells from women with urge incontinence. Pathog. Dis. 2016, 74, ftw067. [Google Scholar] [CrossRef]
- Hannan, T.J.; Totsika, M.; Mansfield, K.J.; Moore, K.H.; Schembri, M.A.; Hultgren, S.J. Host-pathogen checkpoints and population bottlenecks in persistent and intracellular uropathogenic Escherichia coli bladder infection. FEMS Microbiol. Rev. 2012, 36, 616–648. [Google Scholar] [CrossRef] [PubMed]
- Mobley, H.L.T. Measuring Escherichia coli gene expression during human urinary tract infections. Pathogens 2016, 5, 7. [Google Scholar] [CrossRef]
- Rosen, D.A.; Pinkner, J.S.; Walker, J.N.; Elam, J.S.; Jones, J.M.; Hultgren, S.J. Molecular variations in Klebsiella pneumoniae and Escherichia coli FimH affect function and pathogenesis in the urinary tract. Infect. Immun. 2008, 76, 3346–3356. [Google Scholar] [CrossRef]
- Thumbikat, P.; Berry, R.E.; Zhou, G.; Billips, B.K.; Yaggie, R.E.; Zaichuk, T.; Sun, T.T.; Schaeffer, A.J.; Klumpp, D.J. Bacteria-induced uroplakin signaling mediates bladder response to infection. PLoS Pathog. 2009, 5, e1000415. [Google Scholar] [CrossRef]
- Terlizzi, M.E.; Gribaudo, G.; Maffei, M.E. UroPathogenic Escherichia coli (UPEC) Infections: Virulence factors, bladder responses, antibiotic, and non-antibiotic antimicrobial strategies. Front. Microbiol. 2017, 8, 1566. [Google Scholar] [CrossRef]
- Baldassarri, L.; Bertuccini, L.; Creti, R.; Filippini, P.; Ammendolia, M.G.; Koch, S.; Huebner, J.; Orefici, G. Glycosaminoglycans mediate invasion and survival of Enterococcus faecalis into macrophages. J. Infect. Dis. 2005, 191, 1253–1262. [Google Scholar] [CrossRef]
- Daw, K.; Baghdayan, A.S.; Awasthi, S.; Shankar, N. Biofilm and planktonic Enterococcus faecalis elicit different responses from host phagocytes in vitro. FEMS Immunol. Med. Mic. 2012, 65, 270–282. [Google Scholar] [CrossRef] [PubMed]
- Elhadidy, M.; Zahran, E. Biofilm mediates Enterococcus faecalis adhesion, invasion and survival into bovine mammary epithelial cells. Lett. Appl. Microbiol. 2014, 58, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Doran, K.S.; Chang, J.C.; Benoit, V.M.; Eckmann, L.; Nizet, V. Group B streptococcal beta-hemolysin/cytolysin promotes invasion of human lung epithelial cells and the release of interleukin-8. J. Infect. Dis. 2002, 185, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Rajagopal, L. Understanding the regulation of Group B Streptococcal virulence factors. Future Microbiol. 2009, 4, 201–221. [Google Scholar] [CrossRef] [PubMed]
- Kodner, C.M.; Thomas Gupton, E.K. Recurrent urinary tract infections in women: Diagnosis and management. Am. Fam. Physician 2010, 82, 638–643. [Google Scholar]
- Wallace, K.M.; Drake, M.J. Overactive bladder. F1000Research 2015, 4, F1000. [Google Scholar] [CrossRef]
- Abrams, P. Describing bladder storage function: Overactive bladder syndrome & detrusor overactivity. Urology 2003, 62, 28–37. [Google Scholar]
- Morris, A.R.; Westbrook, J.I.; Moore, K.H. A longitudinal study over 5 to 10 years of clinical outcomes in women with idiopathic detrusor overactivity. BJOG-Int. J. Obstet. Gynaecol. 2008, 115, 239–246. [Google Scholar] [CrossRef]
- Walsh, C.A.; Moore, K.H. Overactive bladder in women: Does low-count bacteriuria matter? A review. Neurourol. Urodyn. 2011, 30, 32–37. [Google Scholar] [CrossRef]
- Walsh, C.A.; Siddins, A.; Parkin, K.; Mukerjee, C.; Moore, K.H. Prevalence of “low-count” bacteriuria in female urinary incontinence versus continent female controls: A cross-sectional study. Int. Urogynecol. J. 2011, 22, 1267–1272. [Google Scholar] [CrossRef]
- Brubaker, L.; Nager, C.W.; Richter, H.E.; Visco, A.; Nygaard, I.; Barber, M.D.; Schaffer, J.; Meikle, S.; Wallace, D.; Shibata, N.; et al. Urinary bacteria in adult women with urgency urinary incontinence. Int. Urogynecol. J. 2014, 25, 1179–1184. [Google Scholar] [CrossRef] [PubMed]
- Balachandran, A.A.; Wildman, S.S.; Strutt, M.; Duckett, J. Is chronic urinary infection a cause of overactive bladder? Eur. J. Obstet. Gynecol. Reprod. Biol. 2016, 201, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, P.; Barclay, D.; Zamora, R.; Yoshimura, N.; Peters, K.; Vodovotz, Y.; Chancellor, M. Urine cytokines suggest an inflammatory response in the overactive bladder: A pilot study. Int. Urol. Nephrol. 2010, 42, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Ghoniem, G.; Faruqui, N.; Elmissiry, M.; Mahdy, A.; Abdelwahab, H.; Oommen, M.; Abdel-Mageed, A.B. Differential profile analysis of urinary cytokines in patients with overactive bladder. Int. Urogynecol. J. 2011, 22, 953–961. [Google Scholar] [CrossRef]
- Ma, E.; Vetter, J.; Bliss, L.; Lai, H.H.; Mysorekar, I.U.; Jain, S. A multiplexed analysis approach identifies new association of inflammatory proteins in patients with overactive bladder. Am. J. Physiol. Renal Physiol. 2016, 311, F28–F34. [Google Scholar] [CrossRef]
- Tyagi, P.; Tyagi, V.; Qu, X.; Chuang, Y.C.; Kuo, H.C.; Chancellor, M. Elevated CXC chemokines in urine noninvasively discriminate OAB from UTI. Am. J. Physiol. Renal Physiol. 2016, 311, F548–F554. [Google Scholar] [CrossRef]
- Thomas-White, K.J.; Hilt, E.E.; Fok, C.; Pearce, M.M.; Mueller, E.R.; Kliethermes, S.; Jacobs, K.; Zilliox, M.J.; Brincat, C.; Price, T.K.; et al. Incontinence medication response relates to the female urinary microbiota. Int. Urogynecol. J. 2016, 27, 723–733. [Google Scholar] [CrossRef]
- Chen, Z.; Phan, M.D.; Bates, L.J.; Peters, K.M.; Mukerjee, C.; Moore, K.H.; Schembri, M.A. The urinary microbiome in patients with refractory urge incontinence and recurrent urinary tract infection. Int. Urogynecol. J. 2018, 29, 1775–1782. [Google Scholar] [CrossRef]
- Brierley, S.M.; Goh, K.G.K.; Sullivan, M.J.; Moore, K.H.; Ulett, G.C.; Grundy, L. Innate immune response to bacterial urinary tract infection sensitises high-threshold bladder afferents and recruits silent nociceptors. Pain 2020, 161, 202–210. [Google Scholar] [CrossRef]
- Wu, X.R.; Sun, T.T.; Medina, J.J. In vitro binding of type 1-fimbriated Escherichia coli to uroplakins Ia and Ib: Relation to urinary tract infections. Proc. Natl. Acad. Sci. USA 1996, 93, 9630–9635. [Google Scholar] [CrossRef]
- Guiton, P.S.; Hung, C.S.; Hancock, L.E.; Caparon, M.G.; Hultgren, S.J. Enterococcal biofilm formation and virulence in an optimized murine model of foreign body-associated urinary tract infections. Infect. Immun. 2010, 78, 4166–4175. [Google Scholar] [CrossRef] [PubMed]
- Kline, K.A.; Lewis, A.L. Gram-positive uropathogens, polymicrobial urinary tract infection, and the emerging microbiota of the urinary tract. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Garg, S.; Mohan, B.; Taneja, N. Biofilm formation capability of enterococcal strains causing urinary tract infection vis-a-vis colonisation and correlation with enterococcal surface protein gene. Indian J. Med. Microbiol. 2017, 35, 48–52. [Google Scholar] [CrossRef]
- Lalioui, L.; Pellegrini, E.; Dramsi, S.; Baptista, M.; Bourgeois, N.; Doucet-Populaire, F.; Rusniok, C.; Zouine, M.; Glaser, P.; Kunst, F.; et al. The SrtA Sortase of Streptococcus agalactiae is required for cell wall anchoring of proteins containing the LPXTG motif, for adhesion to epithelial cells, and for colonization of the mouse intestine. Infect. Immun. 2005, 73, 3342–3350. [Google Scholar] [CrossRef] [PubMed]
- Nobbs, A.H.; Rosini, R.; Rinaudo, C.D.; Maione, D.; Grandi, G.; Telford, J.L. Sortase A utilizes an ancillary protein anchor for efficient cell wall anchoring of pili in Streptococcus agalactiae. Infect. Immun. 2008, 76, 3550–3560. [Google Scholar] [CrossRef]
- Rosini, R.; Margarit, I. Biofilm formation by Streptococcus agalactiae: Influence of environmental conditions and implicated virulence factors. Front. Cell. Infect. Microbiol. 2015, 5, 6. [Google Scholar] [CrossRef]
- Yadav, P.; Verma, S.; Bauer, R.; Kumari, M.; Dua, M.; Johri, A.K.; Yadav, V.; Spellerberg, B. Deciphering streptococcal biofilms. Microorganisms 2020, 8, 1835. [Google Scholar] [CrossRef] [PubMed]
- Parker, R.E.; Laut, C.; Gaddy, J.A.; Zadoks, R.N.; Davies, H.D.; Manning, S.D. Association between genotypic diversity and biofilm production in group B Streptococcus. BMC Microbiol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.Y.; Nizet, V. The group B streptococcal β-hemolysin/cytolysin. In The Comprehensive Sourcebook of Bacterial Protein Toxins, 3rd ed.; Alouf, J.E., Popoff, M.R., Eds.; Institut Pasteur: Paris, France, 2006; pp. 737–747. [Google Scholar]
- Delcaru, C.; Alexandru, I.; Podgoreanu, P.; Grosu, M.; Stavropoulos, E.; Chifiriuc, M.C.; Lazar, V. Microbial biofilms in urinary tract infections and prostatitis: Etiology, pathogenicity, and combating strategies. Pathogens 2016, 5, 65. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Bjarnsholt, T. The role of bacterial biofilms in chronic infections. APMIS 2013, 121, 1–58. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.H.; Tian, H.L. Microbial interactions in biofilms: Impacts on homeostasis and pathogenesis. In Microbial Biofilms-Importance and Applications; Dhanasekaran, D., Thajuddin, N., Eds.; InTech: Rijeka, Croatia, 2016; pp. 43–62. [Google Scholar]
- Oliveira, N.M.; Martinez-Garcia, E.; Xavier, J.; Durham, W.M.; Kolter, R.; Kim, W.; Foster, K.R. Biofilm formation as a response to ecological competition. PLoS Biol. 2015, 13, e1002191. [Google Scholar] [CrossRef] [PubMed]
- Brubaker, L.; Wolfe, A.J. The female urinary microbiota/microbiome: Clinical and research implications. Rambam Maimonides Med. J. 2017, 8, e0015. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.L.; Hung, C.-S.; Xu, J.; Reigstad, C.S.; Magrini, V.; Sabo, A.; Blasiar, D.; Bieri, T.; Meyer, R.R.; Ozersky, P.; et al. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: A comparative genomics approach. Proc. Natl. Acad. Sci. USA 2006, 103, 5977–5982. [Google Scholar] [CrossRef]
- Wright, K.J.; Seed, P.C.; Hultgren, S.J. Development of intracellular bacterial communities of uropathogenic Escherichia coli depends on type 1 pili. Cell. Microbiol. 2007, 9, 2230–2241. [Google Scholar] [CrossRef]
- Mohamed, J.A.; Huang, W.; Nallapareddy, S.R.; Teng, F.; Murray, B.E. Influence of origin of isolates, especially endocarditis isolates, and various genes on biofilm formation by Enterococcus faecalis. Infect. Immun. 2004, 72, 3658–3663. [Google Scholar] [CrossRef]
- Toledo-Arana, A.; Valle, J.; Solano, C.; Arrizubieta, M.J.; Cucarella, C.; Lamata, M.; Amorena, B.; Leiva, J.; Penadés, J.R.; Lasa, I. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl. Environ. Microbiol. 2001, 67, 4538–4545. [Google Scholar] [CrossRef]
- Zheng, J.X.; Bai, B.; Lin, Z.W.; Pu, Z.Y.; Yao, W.M.; Chen, Z.; Li, D.Y.; Deng, X.B.; Deng, Q.W.; Yu, Z.J. Characterization of biofilm formation by Enterococcus faecalis isolates derived from urinary tract infections in China. J. Med. Microbiol. 2018, 67, 60–67. [Google Scholar] [CrossRef]
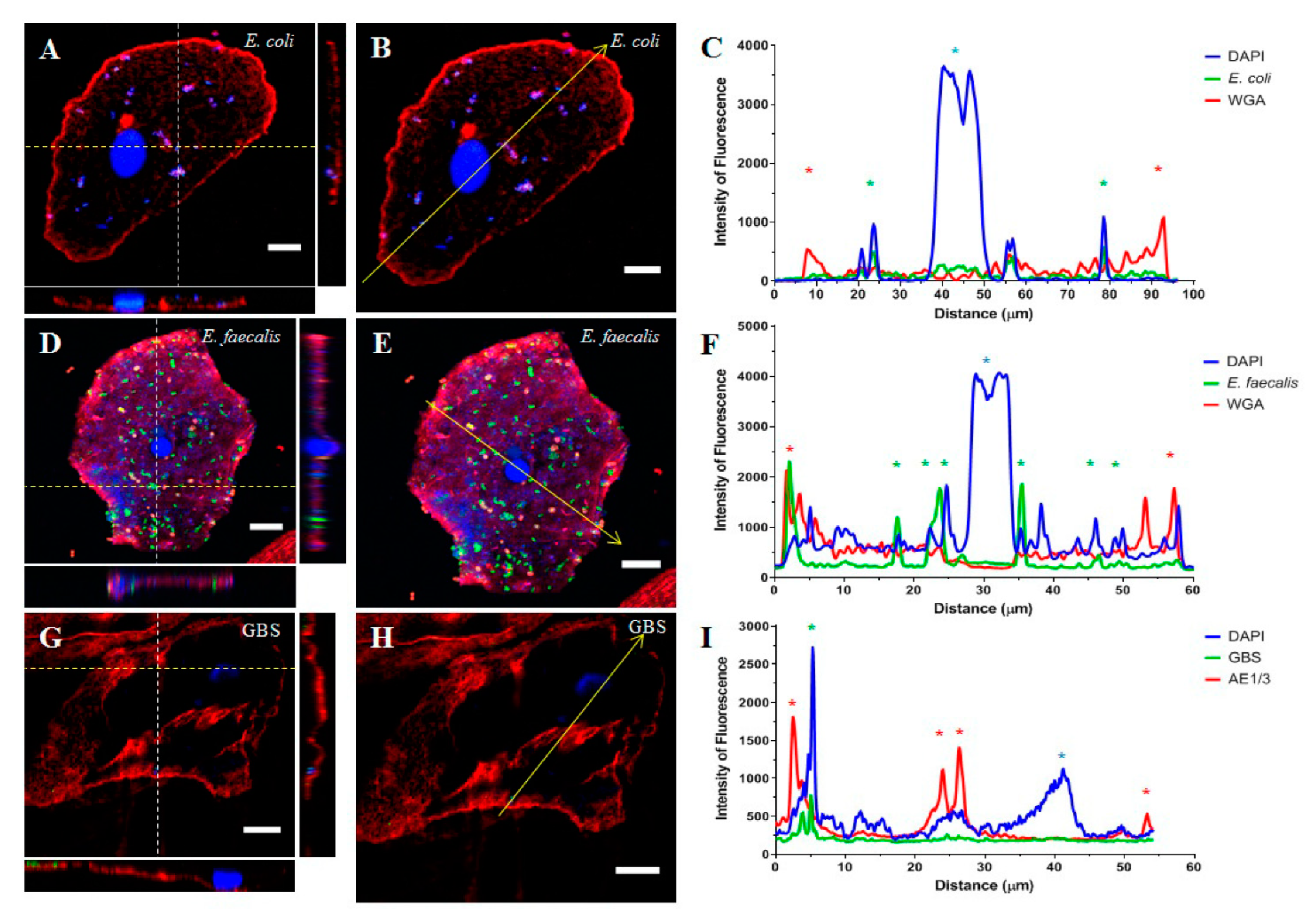
| MSU Result | Samples (n) | Confirmed Intracellular Localisation (%) * |
|---|---|---|
| Escherichia coli | 5 | 76.9% (10/13) |
| Enterococcus faecalis | 4 | 76.9% (10/13) |
| Group B Streptococcus | 6 | 71.4% (5/7) |
| Polymicrobial | 22 | 82.9% (34/41) E. coli (92.3%; 23/25) E. faecalis (68.8%; 11/16) |
| Bacterial Isolate | Localisation within RT4 Cells | Yeast Cell Agglutination | Alpha-Haemolysis | Beta-Haemolysis | Biofilm Formation |
|---|---|---|---|---|---|
| UTI89 | ✓ | ✓ | ✓ | – | Weak |
| UTI89ΔfimH | ✗ | ✗ | ✓ | – | Non-forming |
| E. coli 1 | ✓ | ✓ | ✓ | – | Weak |
| E. coli 2 | ✓ | ✓ | ✓ | – | Non-forming |
| E. coli 3 | ✗ | ✗ | ✓ | – | Weak |
| E. faecalis 1 | ✓ | ✗ | – | ✓ | Very strong |
| E. faecalis 2 | ✓ | ✗ | – | ✓ | Very strong |
| E. faecalis 3 | ✓ | ✗ | ✓ | – | Strong |
| GBS1 | * | ✗ | – | ✓ | Intermediate |
| GBS2 | ✓ | ✗ | – | ✓ | Strong |
| GBS3 | ✓ | ✗ | – | ✓ | Intermediate |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ognenovska, S.; Mukerjee, C.; Sanderson-Smith, M.; Moore, K.H.; Mansfield, K.J. Virulence Mechanisms of Common Uropathogens and Their Intracellular Localisation within Urothelial Cells. Pathogens 2022, 11, 926. https://doi.org/10.3390/pathogens11080926
Ognenovska S, Mukerjee C, Sanderson-Smith M, Moore KH, Mansfield KJ. Virulence Mechanisms of Common Uropathogens and Their Intracellular Localisation within Urothelial Cells. Pathogens. 2022; 11(8):926. https://doi.org/10.3390/pathogens11080926
Chicago/Turabian StyleOgnenovska, Samantha, Chinmoy Mukerjee, Martina Sanderson-Smith, Kate H. Moore, and Kylie J. Mansfield. 2022. "Virulence Mechanisms of Common Uropathogens and Their Intracellular Localisation within Urothelial Cells" Pathogens 11, no. 8: 926. https://doi.org/10.3390/pathogens11080926
APA StyleOgnenovska, S., Mukerjee, C., Sanderson-Smith, M., Moore, K. H., & Mansfield, K. J. (2022). Virulence Mechanisms of Common Uropathogens and Their Intracellular Localisation within Urothelial Cells. Pathogens, 11(8), 926. https://doi.org/10.3390/pathogens11080926







