Abstract
Foot-and-mouth disease virus (FMDV) can persistently infect pharyngeal epithelia in ruminants but not in pigs. Our previous studies demonstrated that persistent FMDV infection in cattle was associated with under-expression of several chemokines that recruit immune cells. This report focuses on the analysis of differentially expressed genes (DEG) identified during the transitional phase of infection, defined as the period when animals diverge between becoming carriers or terminators. During this phase, Th17-stimulating cytokines (IL6 and IL23A) and Th17-recruiting chemokines (CCL14 and CCL20) were upregulated in animals that were still infected (transitional carriers) compared to those that had recently cleared infection (terminators), whereas chemokines recruiting neutrophils and CD8+ T effector cells (CCL3 and ELR+CXCLs) were downregulated. Upregulated Th17-specific receptor, CCR6, and Th17-associated genes, CD146, MIR155, and ThPOK, suggested increased Th17 cell activity in transitional carriers. However, a complex interplay of the Th17 regulatory axis was indicated by non-significant upregulation of IL17A and downregulation of IL17F, two hallmarks of TH17 activity. Other DEG suggested that transitional carriers had upregulated aryl hydrocarbon receptor (AHR), non-canonical NFκB signaling, and downregulated canonical NFκB signaling. The results described herein provide novel insights into the mechanisms of establishment of FMDV persistence. Additionally, the fact that ruminants, unlike pigs, produce a large amount of AHR ligands suggests a plausible explanation of why FMDV persists in ruminants, but not in pigs.
1. Introduction
Foot-and-mouth disease (FMD) is one of the most contagious and economically devastating viral diseases of livestock; the disease is caused by FMD virus (FMDV), a positive-sense single-stranded RNA virus of the family Picornaviridae (genus Aphthovirus). Susceptible hosts include domestic and wild cloven-hoofed animals such as ruminants and pigs. Infection in cattle begins in the respiratory tract. During this primary infection, the virus replicates locally in the nasopharynx or lungs, depending on the route of exposure [1,2]. The infection subsequently spreads via systemic circulation (viremia) to secondary replication sites causing typical vesicles in the oral cavity, on the feet, and other sites of non-haired skin. Mortality is generally low in adult animals, but persistent infection can occur for long periods (30 days–5 years) in 50–80% of infected ruminants [3,4,5,6,7]. In contrast, persistent infection does not occur in pigs [8].
The site of persistent FMDV infection in cattle has been localized to the epithelial cells of the nasopharyngeal mucosa, including the dorsal soft palate and roof of the nasopharynx [6,9,10,11]. Existing FMD vaccines do not prevent or cure persistent infection of pharyngeal epithelial cells [12]. Interestingly, in one set of experiments, FMDV in the oesophageal–pharyngeal fluid of persistently infected cattle was undetectable after dexamethasone treatment; however, virus levels returned to pretreatment levels after cessation of dexamethasone treatment [13]. Currently, the causal mechanisms of FMDV persistence are unknown.
To understand the mechanisms involved in FMDV persistence, a previous study applied an experimental, hypothesis-free functional genomics and bioinformatics approach to identify candidate mechanisms based on genes differentially expressed in tissues targeted and not targeted for persistent FMDV infection [14] and between the targeted tissues of carriers and non-carriers [15,16,17]. In that previous work, differential gene expression in nasopharyngeal tissues of carriers and non-carriers provided early evidence that type 1 regulatory T cells (Tr1) might play a role in persistent infection [15]. Further transcriptomic investigation using micro-dissected nasopharyngeal epithelia suggested that persistent FMDV infection was associated with compromised apoptosis and a reduced cellular immune response [16]. The continued analysis of the differentially expressed genes (DEG) in micro-dissected epithelia during persistent infection indicated that differential gene expression could affect the recruitment of neutrophils, antigen-experienced T cells and/or dendritic cells (DC), natural killer (NK) cell cytotoxicity, and the Th17 response in persistently FMDV-infected carriers [17]. The lung (a non-targeted tissue) was found to express significantly higher levels of TNF cytokines and their receptors than the pharyngeal tissues [14].
The current study provides further analysis of DEG from previously published data [16] collected during the transitional phase of infection that spans the period from acute to persistent infection. The main objective of this study was to infer potential causative factors and mechanisms of establishing FMDV persistent infection in cattle. Using a systems biology approach, we describe several hypothetical mechanisms for the establishment of persistent FMDV infection based on DEG in nasopharyngeal tissues, including contributory roles for aryl hydrocarbon receptor (AHR) ligands, leukocyte function, signaling pathways, and cytokines, chemokines, and their associated receptors.
2. Results
2.1. Pathway and Gene Ontology Term Analysis
The probes with differential expression at FDR ≤ 0.1 showed that 1274 and 598 known genes were upregulated and downregulated, respectively, in nasopharyngeal epithelia of transitional carriers compared to terminators. The functional analysis of the upregulated DEG using DAVID tools detected significant enrichment in an immune-related gene ontology (GO) term (GO:0006955) and seven KEGG immune processes related to infection in T cells and epithelia, immune cell migration, phagocytosis, and four KEGG signaling pathways involved in immune regulation including (1) PI3K-Akt, (2) NFκB, (3) HIF-1, and (4) Wnt signaling pathways (Table 1). The downregulated genes did not reveal any significant pattern in the same analyses.

Table 1.
Gene ontology (GO) term associated with biological processes (GOTERM_BP) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathways significantly (p value < 0.05 with Benjamini correction for multiple tests) over-represented in transitional carriers compared to terminators.
Analysis of the DEG gene list using the IPA pathways analysis identified the top five inferred upstream regulators for differential expression as (1) estrogen receptor 1 (ESR1), (2) beta-estradiol (an estrogen hormone), (3) KRAS, (4) dexamethasone, and (5) TNF (Figure 1A). Similarly, the top five toxicity-inducing biological processes or signaling pathways were (1) Nrf2-mediated oxidative stress response, (2) hepatic stellate cell activation, (3) PPAR-RXR activation, (4) hypoxia-inducible factor (HIF) signaling, and (5) aryl hydrocarbon receptor (AHR) signaling (Figure 1B). Among the top five upstream regulators, dexamethasone is widely used as an immunosuppressive/anti-inflammatory corticosteroid. Among these top five toxicity-inducing pathways, HIF1α and AHR signaling are mediated by two transcription factors that compete to form heterodimers with ARNT and play a critical role in regulating mucosal immunity [18,19]. NFκB signaling is crucial for the immune response [20]. On this basis, AHR, HIF1A, and NFκB signaling pathways were explored in more detail.
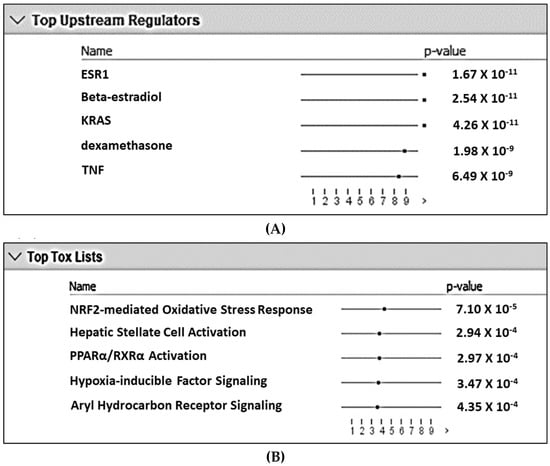
Figure 1.
Top five upstream regulators (A) and top five biological processes/signaling pathways involved in toxicity (B) with the lowest likelihoods (i.e., p-value) of the associations/overlaps between the differentially expressed gene set (both up- and downregulated) and the pathways/biological processes by random chances in the Qiagen Ingenuity Pathway Analysis using the list containing ENTREZ numbers and up- and downregulated DEG. The dots in horizontal lines are the negative log transformation of p-values.
2.2. AHR and HIF1α Signaling
AHR and HIF1α compete to form heterodimers with aryl hydrocarbon receptor nuclear translocators (ARNTs). AHR was expressed at a significantly higher level (8.1-fold), and HIF1α expression was significantly downregulated by 1.8-fold in transitional carriers compared to terminators (Table 2). Among three ARNTs, ARNTL expression was the highest and was at a higher, although not significantly increased level (p = 0.05) in transitional carriers than terminators. The expression level of eight genes known to be induced by AHR signaling including B7H4 [21], CCL20 [22], CD8A [23,24], CD39 [25,26], CYP1B1 [19], IL6 [22,27], IL23A [28], and STAT3 [28] were significantly higher in transitional carriers than in terminators. A transcript variant of CD39 with a longer 3′ non-coding sequence was also expressed at a higher level very close to significance (FDR = 0.11). Three AHR target genes, CYP1A1, CYP1A2, and IL33 [29], were also expressed at higher gene levels (p ≤ 0.05).

Table 2.
Average expression signal intensity (ESI), false discovery rates (FDR), and fold differences in AHR and HIF1A signaling-related genes that were differentially expressed between the nasopharynx epithelia of transitional carriers and terminators.
HIF1 α expression is inducible by STAT3 and NFκB [30]. Three inhibitors of STATs (PIAS2, PIAS3, and PIAS4) were significantly upregulated at or close to significant levels in transitional carriers (DEG in NFκB signaling are listed in Table 3). PDK1 expression level, inducible by HIF1A [31,32], was 37.6 times lower in transitional carriers than in terminators, whereas three key inhibitors of HIF1α, HIF1AN, LIMD1, and VHL [30,33] were expressed at higher levels (p = 0.04, FDR= 0.05 and p = 0.03, respectively) in carriers than in terminators. In contrast to enhancing glucose uptake and glycolysis of HIF1A, an enzyme (ACSS3) that catalyzes the first reaction of fatty acid metabolism was upregulated 9.6-fold. HIF1A is activated by the AKT-mTOR signaling pathway via extracellular ATP and TCR signaling [34]. Although AKT1 and AKT2 were upregulated in transitional carriers, two inhibitory genes (PIK3IP1 and TSC1) of this signaling pathway [35,36] were also significantly upregulated. These results indicate reduced HIF1α signaling but increased AHR signaling in the epithelia of transitional carriers, which could impact mucosal immune response.

Table 3.
Mean expression signal intensities (ESI) and expression differences (fold) in genes of canonical and non-canonical NFκB signaling pathways between of transitional FMDV carriers and terminators.
2.3. NFκB Signaling
Several genes playing a critical role in NFκB signaling were differentially expressed in nasopharyngeal tissues between transitional carriers and terminators (Table 3). IKBKB is an indispensable Iκ kinase of the trimeric IκB kinase (IKK) complex in the canonical NFκB pathway [20]. IKBIP is an IKBKB interacting protein. OTUB1 enhances canonical NFκB signaling [37] but inhibits activation of the non-canonical signaling by de-ubiquitination of TRAF3 [38,39]. TGFB2-OT1 increases the LARP1 level to promote the activation of canonical transcription factors [40]. The expression of IKBKB, IKBIP, NOD2, OTUB1, and TGFB2-OT1 was significantly downregulated in transitional carriers compared to terminators. On the other hand, NFKBIA is an inhibitor of the dimerization of transcription factors p50 and RELA in the canonical pathway [20,41]. Additionally, IFRD2 [42], LCOR [43,44], LRRC33 [45], NLK [46], PGRN [47], SIGLEC11 [48], and TRAF1 [49] have suppressive effects on canonical NFκB signaling. The expression of these genes was significantly upregulated in transitional carriers (Table 3).
On the other hand, the transcription factors, receptors, or receptor ligands in non-canonical NFκB signaling such as RELB, NFKB2/p100, CD27, LTB, LTBR, and TNFRSF8/CD30 [20] were expressed at significantly higher levels in transitional carriers. Other genes involved in non-canonical NFκB signaling such as MAP3K14/NIK, CD40LG, RANKL, and TNFRSF1B [20] were also upregulated at p ≤ 0.05. Complement membrane attack complexes can activate non-canonical NFκB by forming an Akt+ NIK+ signalosome on Rab5+ endosomes [50]. These results indicate increased non-canonical NFκB signaling and suppressed canonical signaling in the epithelia of transitional carriers. This may promote immune tolerance by inducing tolerogenic DC and Treg cells and suppressing the Th17 response [20,41,51].
2.4. Wnt Signaling
The expression of five Wnts (WNT4, WNT5A, WNT7A, WNT10B, and WNT16) was significantly higher in transitional carriers than in terminators (Table 4), and WNT3 was at a higher level (p = 0.02). These results, together with the Wnt signaling pathway significantly overrepresented by DEG in the KEGG pathway analysis (Table 1), indicated increased Wnt signaling in carriers. This may indicate induction of tolerogenic DC that can inhibit Th17 and CD8+ cytotoxic T cell activity and promote Treg development, as described previously [52,53,54].

Table 4.
Average expression signal intensities (ESI) and expression differences (fold) in WNT genes between transitional carriers and terminators.
2.5. Cytokines and Cytokine Receptors
Expression levels of IL6, IL16, IL23A, IL34, and TNFSF15 were significantly higher (2.5-, 7.2-, 9.1-, 4.4-, and 10.8-fold higher, respectively) in transitional carriers compared to terminators (Table 5). Of these cytokines, IL6 and IL23 promote Th17 differentiation and inhibit the induction of Treg cells in the mucosal immune response [55,56,57]. However, the expression of three cytokines (IL17A, IL17F, and IL22) produced by Th17 cells was not upregulated. IL16 had the highest expression level based on average signal intensities. IL16 recruits CD4-expressing immune cells, preferentially Treg cells [58,59], enhancing the immunosuppressive effect of IL-10 [60], and inducing tolerogenic DC. IL34 is a cytokine of Treg cells [61] and promotes pathogen persistence [62], organ transplant tolerance, immunosuppressive macrophages, and macrophage-M2 polarization [62,63,64,65]. TNFSF15 has diverse functions, including promoting Th2 and Treg response [66,67]. Three Th17 suppressing cytokines (IL21, IL24, and IL33) were also upregulated at the gene levels (p < 0.05). These results suggest that the transitional carriers expressed higher levels of both Th17 stimulatory and Th17 suppressive cytokines, which may explain why three Th17 cytokines were not upregulated in carriers.

Table 5.
Average expression signal intensities (ESI), false discovery rates (FDR), and fold differences in cytokines and receptors that were differentially expressed between transitional carriers and terminators.
There were also eight cytokine receptors (ACVR1B, IL18RAP, IL17RB, IL27RA, sIL10RB, TGFBR3, TNFRSF6B, and TNFRSF8) that had significantly upregulated expression and two (ACVR2B and SIGIRR) that were downregulated in transitional carriers (Table 5). Among these receptors, IL27RA was expressed at a very high level. IL27, the ligand of IL27RA, inhibits the differentiation of Th17 cells and IL-17 production [68] and, together with AHR, promotes Tr1 cell differentiation [69,70,71,72]. ACVR1B, ACVR2B, and TGFBR3 are receptors of the TGFβ superfamily, which promote differentiation of Treg, Th17, and/or follicular helper T cells (Tfh) [73,74]. IL17RB, part of the IL25 receptor complex, promotes differentiation of Th2 cells [75]. IL10RB is a co-receptor for IL-10 and IFNλ signaling, but the function of IL10RB without a transmembrane domain (sIL10RB) is unknown. SIGIRR inhibits IL-33-mediated signaling [76]. TNFRSF6B, a soluble decoy receptor, can skew T cell and macrophage differentiation towards Th2 and M2 phenotypes, respectively, and suppress Th17 immune response [77,78]. TNFRSF8 inhibits the proliferation of CD8+ effector T cells [79]. The results of expression of these receptors suggest that the transitional carriers could have suppressive effects on Th17 and CD8+ effector cells and stimulatory effects on Th2 and Treg cells in the epithelia compared to terminators.
2.6. Chemokines and Chemokine Receptors
There was significant differential expression of chemokines in epithelia of transitional carriers compared to terminators. Six chemokines, CCL11, CCL14, CCL20, CXCL12, CXCL14, and CXCL13, were significantly upregulated in nasopharyngeal epithelia of transitional carriers, and one (CCL3) was downregulated (Table 6). CCL11, CCL20, CXCL12, and CXCL14 were expressed at >13-fold higher level in transitional carriers than terminators, whereas the remainders were < 3-fold higher. CCL11 selectively recruits eosinophils and mast cells [80]. CCL14 recruits a specific subset of CD4+CD146+CCR5+ Th17 cells [81], whereas CCL20 primarily recruits Th17 cells via the CCR6 receptor [82,83]. CXCL12 polarizes Th to Treg cells and macrophages to M2 activation and recruits antiviral CD8+ T cells [84,85,86]. DPP4, which converts CXCL12 to a CXCL12 antagonist [87], was significantly upregulated in transitional carriers. CXCL13 is a chemoattractant for B cells [88]. CXCL14 was the most differentially expressed chemokine with the highest signal intensity among chemokine DEGs (Table 4). CXCL14 primarily chemoattracts monocytes [89,90]. It also recruits immature DC, M2 macrophages, neutrophils, NK cells and B cells [90,91], and Treg cells [92] and has anti-inflammatory and anti-CXCL12 activities [93,94,95]. CCL3 attracts neutrophils, macrophages, naive CD8+ T cells, and NK cells through binding to the receptors CCR1 and CCR5 [96,97,98]. The total expression of neutrophil-recruiting ELR+ CXCLs, including CXCL1, 2, 3, 5, 8, and 15, was 3.2-fold lower in transitional carriers than in terminators (Table 6).

Table 6.
Average expression signal intensities (ESI), false discovery rates (FDR), and fold differences in chemokine and the receptor genes differentially expressed between transitional carriers and terminators.
Three chemokine receptors, CCR1, CCR6, and CCR7, were upregulated in transitional carriers compared to terminators (Table 6). CCR1, CCR3, and CCR5 are expressed on monocyte and macrophages [97]. Upregulated CCR1, together with increased CCR3 and CCR5 (p = 0.01 and 0.06, respectively), supports the increased recruitment of monocytes. Upregulated CCR6, the receptor of CCL20, strongly suggests increased Th17 recruitment in transitional carriers. Similarly, CCR3 (high-affinity receptor of CCL11) [99] and CXCR4 (the receptors of CXCL12 and CXCL14) [94,95,100] also showed upregulated expression (p ≤ 0.05) in transitional carriers. CCR7 is expressed on naive T and B cells, central memory T cells (Tcm), and mature DC [97]. The expression of two receptors, ACKR3 and XCR1, was significantly downregulated in transitional carriers (Table 4). ACKR3 binds and degrades CXCL12 [101], while XCR1 enhances CD8+ DCs in activating CD8+ T cell-mediated defense via antigen cross-presentation [102]. The receptors of ELR+ CXCLs, CXCR1/CXCR2, were also downregulated in transitional carriers (p = 0.03). Therefore, the results of chemokines and their receptors suggest that the epithelia of transitional carriers recruited more monocytes, eosinophils, and Th17 cells, reduced recruitment of neutrophils and CD8+ T cells, decreased antigen-cross presentation to CD8+T cells, and promoted Th17 to Treg transition and macrophage M2 activation compared to terminators.
2.7. T-Cell-Associated Factors
It appears that CD4+ T cells especially CD4+ CD8αα+ T cells, but not CD8αβ+ T cells, were specifically increased in transitional carriers based on higher expression levels of CD4 (p = 0.02), CD8A [103], CD40L (p = 0.01) [104], and ThPOK (CD4 T cell-specific transcription factor) and lower levels of CD8B (p = 0.08) (Table 7). A marker gene of a specific subset of Th17 cells (CD146) [105] and a microRNA gene highly expressed in Th17 (BIC/mir155) [106,107,108,109] were expressed at significantly higher levels in transitional carriers than in terminators, whereas the Th17-specific transcription factor RORC was expressed at a higher gene level (p = 0.03). However, there are three Th17-suppressing DEG (LXRA; STAT5A and TNFRSF6B) [110,111,112] whose expression was significantly upregulated in transitional carriers compared to terminators. Another Th17-suppressing gene (CD69) [111] was also expressed at a higher gene level (p = 0.01) in transitional carriers (Table 7).

Table 7.
Average expression signal intensities (ESI), false discovery rates (FDR), and fold differences in T cell-associated genes differentially expressed between transitional carriers and terminators.
Treg marker genes such as FOXP3 (Table 7) and IL10 (Table 5) were not differentially expressed between transitional carriers and terminators; however, several marker genes of type 1 regulatory (Tr1) T cells (GITR, IRF4, IL21, LAG3, and TNFRSF9) were expressed at higher gene levels (p ≤ 0.05) in transitional carriers (Table 5 and Table 7). Four upregulated DEG mediating Treg immunosuppressive activities (ADCY4, BTLA, CD39, and GIMAP5) were expressed at significantly higher levels in transitional carriers (Table 2 and Table 7). ADCY4 catalyzes the production of cAMP, an immune suppressive mediator of Treg cells [113,114]. BTLA is a marker of exhausted T cells [115] and promotes peripheral Treg cell differentiation and immune tolerance [116]. GIMAP5 plays a central role in maintaining peripheral tolerance and T cell homeostasis in the gut [117,118,119]. These results suggest that Th17 cell activity may be suppressed in transitional carriers despite increased cell recruitment to the epithelial region.
2.8. Myeloid Cell-Associated Factors
The expression of eight genes with immune inhibitory effects on macrophages or antigen-presenting cells (APC) (CD83, CD300D, EMR1, MFSD6, SIGLEC11, SIGLEC15, TIMD4, and TLR2) was significantly upregulated in transitional carriers compared to terminators (Table 8). Signaling through cell-membrane-associated CD83 appears to suppress functions in various immune cell populations [120], and soluble CD83 inhibits human monocyte differentiation into dendritic cells [121]. CD300 proteins are macrophage-specific receptors with regulatory effects [122,123]. CLEC1A dampens dendritic cell activation and downstream Th17 responses [124]. EMR1 mediates the induction of antigen-specific efferent regulatory T cells in peripheral tolerance [125]. MFSD6 is a mediator of MHC haplotype-dependent but not MHC-unrestricted cytotoxicity of macrophages [126]. SIGLEC11 and SIGLEC15 are mainly expressed on macrophages and have an immunosuppressive effect on macrophages [48,127]. TIMD4, expressed only on APC including macrophages, mediates the removal of antigen-specific T cells during the contraction phase of the adaptive immune response [128,129]. TLR2 is a Toll-like receptor that can also induce immune tolerance [130,131,132,133,134]. On the other hand, MFSD6 recognizes certain MHC-I molecules and mediates MHC-I restricted killing by macrophages [126]. MFSD6 expression was 4.1 times lower in transitional carriers compared to terminators (Table 6). These results suggest increased activity of immunosuppressive macrophages and/or dendritic cells in transitional carriers.

Table 8.
Average expression signal intensities (ESI), false discovery rates (FDR), and fold differences in dendritic cells (DC)- and macrophage (Mφ)-expressing genes differentially expressed between transitional carriers and terminators.
2.9. Innate Immunity
Transitional carriers had a generally downregulated expression of defensin genes with two genes (DEFB1 and DEFB103A) significantly downregulated by 3.4- and 10.7-fold and one defensin gene (DEFB4B) significantly upregulated by 10.8-fold in transitional carriers (Table 8). DEFB103A is a broad-spectrum antimicrobial and has anti-picornavirus activity [135,136], which played a role in FMDV persistence. NID1 (a soluble NCR2 ligand with NK cell suppressing activity) [137] and MADD (an apoptosis-inhibiting gene) [138] were expressed at significantly higher levels (12.1-and 9.7-fold higher, respectively) in transitional carriers than those in terminators (Table 9).

Table 9.
Average expression signal intensities (ESI), false discovery rates (FDR), and fold differences of innate and humoral immunity-related genes differentially expressed between the nasopharynx epithelia of transitional carriers and terminators.
3. Discussion
Historically, it has been reported that approximately 50% of FMDV-infected ruminants remain persistently infected 28 days after infection [3,4,5]; however, experimental studies have shown that the proportion of carriers is often substantially higher [6,11].
Persistent infection does not occur in pigs [8], indicating the involvement of host-specific factors in determining the divergence between FMDV carriers and terminators. The immune mechanisms inferred in this study are consistent with several hypothesized mechanisms identified in nasopharyngeal tissues during persistent infection [17], including (1) reduced recruitment of neutrophils and CD8+ T effector cells, (2) suppressed NK and macrophage cytotoxicity via downregulated MFSD6 and NID1, and (3) suppression of the Th17 response and canonical NFκB signaling pathway. Additionally, previous work demonstrated that expression of chemokines that recruit neutrophils and CD8+ T effector cells was reported to be significantly lower in pharyngeal tissue than in the lung, where primary, but not persistent, infection occurs [14,139], indirectly supporting the involvement of these chemokines in preventing FMDV persistent infection.
The differential expression of ELR+ CXCLs and CCL3 is consistent with the microscopic analyses of the nasopharynx of the animals included in this study, wherein there were reduced quantities of CD8+ T cells in the epithelia of transitional carriers compared to terminators [16]. These results suggested the importance of neutrophil and CD8+ T effector cell recruiting chemokines in FMDV clearance, given that CD8+ cytotoxic T cells kill infected cells and neutrophils can clear virus infection via phagocytosis and extracellular traps [140]. This also agrees in part with the finding from one study that dexamethasone injection inhibited FMDV production in the oesophageal–pharyngeal fluid of persistently infected cattle but did not cure the infection [13]. This is based on dexamethasone treatment causing neutrophilia, lymphopenia, and eosinopenia in ruminants [141,142,143].
IL-17RA signaling in the epithelium, activated by two Th17-specific cytokines, IL17A and IL17F, is required for neutrophil recruitment. During the transitional phase, the involvement of Th17 cells in the FMDV infection of pharyngeal epithelia was strongly supported by upregulated expression of Th17-promoting cytokines and chemokines (IL6, IL23A, CCL14, CCL20, and CCR6 listed in Table 5 and Table 6) and Th17-associated genes (CD4, CD146, MIR155, TGFBR3, and ThPOK shown in Table 7) in transitional carriers, suggesting that the Th17 response was needed to clear FMDV. However, the expression of Th17 cytokines (IL17A, IL17F, and IL22) was not upregulated in transitional carriers, indicating that the activity of Th17 cells was suppressed, potentially as a result of upregulated Th17 suppressing genes CD39, CD69, IL16; LXRA, STAT5A, TIMD4, and TNFRSF6B (Table 7). The suppression could also be mediated by several upregulated immune suppressive genes including ADCY4, BTLA, GIMAP5, and IL34 based on publications cited herein.
AHR plays a key role in regulating Th17 differentiation and activity [19]. Among CD4+ T cells, AHR expression is restricted to the Th17 cell subset, including Treg cells [144]. Natural AHR agonists enhance Th17 differentiation [145]. Different AHR ligands, such as TCDD and FICZ, induce different effects on Th17 and Treg [146]. Dietary AHR ligands (indole-3-carbinol and 3,3′-diindolylmethane) can cause trans-differentiation of Th17 cells into T cells with regulatory phenotypes during the resolution of inflammation, reduce IL17 expression [147] and induce immune tolerance [148]. Tryptophan derivatives such as indole-3-lactic acid produced by Lactobacillus inducted CD4+CD8αα double positive intraepithelial lymphocytes (DP IELs), which display regulatory functions associated with immune tolerance [149]. Our results suggest increased activity of regulatory T cells, which was supported by upregulated cytokine (IL34) and effector genes (ADCY4, B7H4, CD8A, and CD39) of regulatory T cells. B7H4, CD8A, and CD39 are known to be AHR target genes. Several type 1 regulatory (Tr1) T cell genes (GITR, IL21, IRF4, LAG3, and TNFRSF9) [112], but not Treg marker genes (IL10 and FOXP3), were upregulated (p ≤ 0.05) in transitional carriers (Table 5 and Table 7), supporting AHR-induced Th17 trans-differentiation into regulatory T cells. AHR also induces suppressive macrophages and tolerogenic DC to promote the differentiation of Treg cells [21,150,151,152,153,154,155,156]. Our results (Table 8) indicated the induction of suppressive macrophages and tolerogenic DC in carriers.
Effects of AHR signaling can also be mediated through dimerization with other transcription factors such as estrogen receptors [157,158,159], HIF1A [18,160,161,162,163]; NFκB [164], PPAR [165,166]. Interestingly, estrogen receptor, ESR1, was detected as a top up-stream regulator, and DEG were over-represented in the PPAR signaling pathway in this study. HIF1A and AHR compete to form heterodimers with AHR nuclear translocators (ARNTs) and mutually inhibit each other. Reduced HIF1A signaling inhibits IL17 production in CD4+ T cells and cytotoxicity of CD8+ T cells contributing to T cell exhaustion in chronic infections [167]. AHR:RelA dimerization antagonizes the classical NFκB pathway, whereas AHR:RelB enhances non-canonical pathway signaling [164,168]. The DEG listed in Table 3 indicate higher non-canonical and lower canonical NFκB signaling in transitional carriers compared to terminators, according to the review article by Sun (2017), which could suppress the Th17 response according to IL17 signaling in the canonical NFκB pathway [169]. The non-canonical NFκB pathway plays an important role in promoting immune tolerance by inducing tolerogenic DC and Treg cells [20,41], suppressing the Th17 response [51]. Additionally, AHR signaling enhances Wnt signaling [103], and both Wnt and AHR signaling can induce tolerogenic DC [52,54].
Our results demonstrated upregulated AHR and its several target genes and downregulated HIF1A and its target genes in transitional carriers (listed in Table 2). AHR is a promiscuous xenobiotic receptor and ligand-dependent transcription factor that binds to various chemicals such as plant flavonoids, polyphenolics, and indoles as well as to pollutants such as synthetic polycyclic aromatic hydrocarbons and dioxin-like compounds [170]. Interestingly, some short chain fatty acids (SCFA), e.g., propionate and butyrate, are also AHR ligands [171] and can induce AHR expression [172,173] and increase cell response to AHR ligand stimulation [174]. Some of the AHR ligands are produced in the rumen as part of normal ruminant physiology, suggesting an interesting hypothetical mechanism to explain why ruminants, but not pigs are prone to persistent FMDV infections.
It is well-established that B-cell function is altered during the FMDV carrier state. Specifically, anti-FMDV IgA detection in secretions has been reported to be significantly higher in carriers than in non-carriers [175,176]. This indicates chronic stimulation of B cells and suggests that antibodies alone cannot clear the FMDV carrier state, but rather cell-mediated immunity is required. In the current study, B-cell induction was indicated by upregulation of CD21, CD19, and CD81 (the B cell co-receptor complex) and 16 immunoglobulin probes in transitional carriers (data not shown).
In summary, this work supports previous studies that indicated that the establishment and maintenance of the FMDV carrier state are associated with differential gene expression in the nasopharyngeal tissues, known to be the site of persistent infection. Specifically, pathway analysis of DEG suggested that several immune regulatory mechanisms are associated with FMDV persistence. DEG of cytokines, chemokines, and the receptors suggest an increased differentiation and migration of Th17 cells but reduced recruitment of neutrophils and CD8+ T effector cells to the infected tissues of transitional carriers compared to terminators. However, IL17A and IL17F expression were not increased in carriers, indicating complex regulation of the Th17 response. The Th17 response is known to play a key role in inducing the expression of neutrophil recruiting chemokines, which is regulated, in part, by AHR signaling. Upregulated AHR signaling in carriers was also supported by DEG in NFκB and Wnt signaling pathways. This leads us to speculate that AHR ligands produced in the rumen and their effect on various physiological functions might play a role in establishing FMDV persistent infection, which could also explain why FMDV persists in some ruminants but not in pigs.
4. Materials and Methods
4.1. Study Design and Gene Expression Data
The microarray data used in this study and the design of the animal experiments have been reported previously [16]. All data utilized in this study were derived from microarray-based gene expression profiles of micro-dissected nasopharyngeal epithelia from FMDV-infected cattle during the transitional phase of infection spanned from 12 to 21 dpi. The previous works defined the transitional phase as the period after acute infection but before the defined carrier phase; animals that remained infected during the transitional phase (transitional carriers) consistently progressed to becoming carriers [6].
The data were produced using a custom bovine gene expression 60-mer oligonucleotide microarray as described by Zhu et al. [14]. Microarrays and reagents were manufactured by Agilent Technologies (San Jose, CA, USA), and the lab procedures were conducted based on the protocols and equipment recommended by the manufacturer. For comparison of the gene expression levels between transitional carriers (animals that were still infected) and terminators (animals that had recently cleared infection) during the transitional phase of infection, microarray expression data from the micro-dissected nasopharyngeal epithelia of three animals from each cohort were compared, as previously reported by Stenfeldt et al. [16].
4.2. Statistical Analysis
R scripts implemented with the LIMMA package [177] were used to normalize and analyze the microarray data as previously described [15]. All signal intensities (averaged photons per pixel in the microarray images) used in the statistical analysis were Log2 transformed. Genes differentially expressed between transitional carriers and terminators with a false discovery rate (FDR) of 0.10 or smaller and an expression difference of at least 50% were considered statistically significant genes in this study. This FDR threshold increases the detection power (fewer false negatives/type II errors) with a false positive (type I error) rate of 0.10 in declared DEG, or one expected false positive in ten DEG, compared to FDR at 0.05, which balances type I and type II errors.
4.3. Pathway Analysis
The methods of pathway analysis of DEG have been described [17]. All bovine genes included in the microarray design were mapped to human reference genes using computer analysis via NCBI BLAST and/or manual annotation by aligning the microarray probe sequences on bovine genome sequences on the UCSC Genome Browser using BLAT (https://genome.ucsc.edu/index.html (accessed on 1 June 2022)). The list of upregulated and downregulated genes associated with each human Entrez Gene ID was analyzed with Ingenuity Pathway Analysis (IPA) (Qiagen, Germantown, MD, USA) and the NCBI Functional Annotation Bioinformatics Microarray Analysis program (DAVID Bioinformatics Resources version 6.8) to identify the biological pathways significantly over-represented among DEG. The biological functions of DEG were determined based on scientific publications (included as cited references) or on the NCBI Gene database (https://www.ncbi.nlm.nih.gov/gene/ (accessed on 1 June 2022)).
4.4. Biological Inferences
The biological inferences have been described [17], which were based on (i) reported biological functions of DEG, (ii) differential gene expression including averaged signal intensity and magnitudes (fold difference) of upregulated or downregulated expression, assuming that (1) genes with a higher signal intensity and larger differential expression have a more substantial biological role in their gene group and (2) upregulated expression enhances gene activities and vice versa. Differential expression of genes with cell-specific expression was also used to infer the differences in the number of the cells. Genes with no significant differential expression (FDR > 0.10) but known to play important roles in the relevant biological pathways/processes associated with other DEG were also used as references or supporting results for DEG-related mechanisms. Probabilities of differential expression at gene levels are listed as p-values along with the FDR. Expression levels of genes downregulated or upregulated in transitional carriers compared to terminators are shown as negative and positive values (fold changes), respectively. Immune regulatory mechanisms especially involved in mucosal immunity and its association with ruminant physiology were also taken into consideration in the formulation of the hypothesis.
Author Contributions
Data curation, J.J.Z., J.A.C. and M.E.; Formal analysis, J.J.Z., E.A.B. and J.A.C.; Funding acquisition, L.L.R. and J.A.; Investigation, C.S., E.A.B., J.A.C., M.E. and J.A.; Methodology, J.J.Z., C.S. and M.E.; Project administration, L.L.R. and J.A.; Resources, L.L.R. and J.A.; Supervision, L.L.R. and J.A.; Visualization, J.J.Z., E.A.B. and J.A.C.; Writing—original draft, J.J.Z., J.A.C. and J.A.; Writing—review and editing, C.S., J.A.C., M.E., L.L.R. and J.A. All authors have read and agreed to the published version of the manuscript.
Funding
The data used in this study are derived from research funded by the U.S. Department of Agriculture, Agricultural Research Service-CRIS project 1940-32000-061-00D, and an interagency agreement with the Science and Technology Directorate of the U.S. Department of Homeland Security under award No. HSHQDC-11-X-00131.
Institutional Review Board Statement
All experimental procedures were approved by the Plum Island Animal Disease Center institutional animal care and usage committee (protocol 209-17-R), which ensures humane and ethical treatment of animals.
Informed Consent Statement
Not applicable.
Data Availability Statement
The animal experiments of the microarray studies have been published, and the original raw data are available in the NCBI databases (accession number: GSE104058) (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE104058 (accessed on 1 June 2022)). The datasets generated for this study are located at: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE104058 (accessed on 1 June 2022).
Acknowledgments
The authors thank Theresa Aponte for her checking and formatting the references and George Smoliga for sample processing during the animal experiment.
Conflicts of Interest
The authors declare no competing interest.
References
- Arzt, J.; Pacheco, J.M.; Rodriguez, L.L. The early pathogenesis of foot-and-mouth disease in cattle after aerosol inoculation. Vet. Pathol. 2010, 47, 1048–1063. [Google Scholar] [CrossRef]
- Arzt, J.; Juleff, N.; Zhang, Z.; Rodriguez, L.L. The pathogenesis of foot-and-mouth disease I: Viral pathways in cattle. Transbound. Emerg. Dis. 2011, 58, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Burrows, R. Studies on the carrier state of cattle exposed to foot-and-mouth disease virus. Epidemiol. Infect. 1966, 64, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Burrows, R. The persistence of foot-and-mouth disease virus in sheep. Epidemiol. Infect. 1968, 66, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Moonen, P.; Schrijver, R. Carriers of foot-and-mouth disease virus: A review. Vet. Q. 2000, 22, 193–197. [Google Scholar] [CrossRef] [PubMed]
- Stenfeldt, C.; Eschbaumer, M.; Rekant, S.I.; Pacheco, J.M.; Smoliga, G.R.; Hartwig, E.J.; Rodriguez, L.L.; Arzt, J. The foot-and-mouth disease carrier state divergence in cattle. J. Virol. 2016, 90, 6344–6364. [Google Scholar] [CrossRef] [PubMed]
- Bertram, M.R.; Vu, L.T.; Pauszek, S.J.; Brito, B.P.; Hartwig, E.J.; Smoliga, G.R.; Hoang, B.H.; Phuong, N.T.; Stenfeldt, C.; Fish, I.H.; et al. Lack of transmission of foot-and-mouth disease virus from persistently infected cattle to naïve cattle under field conditions in Vietnam. Front. Vet. Sci. 2018, 5, 174. [Google Scholar] [CrossRef]
- Stenfeldt, C.; Pacheco, J.M.; Smoliga, G.R.; Bishop, E.; Pauszek, S.J.; Hartwig, E.J.; Rodriguez, L.L.; Arzt, J. Detection of foot-and-mouth disease virus RNA and capsid protein in lymphoid tissues of convalescent pigs does not indicate existence of a carrier state. Transbound. Emerg. Dis. 2016, 63, 152–164. [Google Scholar] [CrossRef]
- Zhang, Z.D.; Kitching, R.P. The localization of persistent foot and mouth disease virus in the epithelial cells of the soft palate and pharynx. J. Comp. Pathol. 2001, 124, 89–94. [Google Scholar] [CrossRef]
- Pacheco, J.M.; Smoliga, G.R.; O’Donnell, V.; Brito, B.P.; Stenfeldt, C.; Rodriguez, L.L.; Arzt, J. Persistent foot-and-mouth disease virus infection in the nasopharynx of cattle; tissue-specific distribution and local cytokine expression. PLoS ONE 2015, 10, e0125698. [Google Scholar] [CrossRef]
- Stenfeldt, C.; Hartwig, E.J.; Smoliga, G.R.; Palinski, R.; Silva, E.B.; Bertram, M.R.; Fish, I.H.; Pauszek, S.J.; Arzt, J. Contact challenge of cattle with foot-and-mouth disease virus validates the role of the nasopharyngeal epithelium as the site of primary and persistent infection. Msphere 2018, 3, e00493-18. [Google Scholar] [CrossRef] [PubMed]
- Stenfeldt, C.; Arzt, J. The carrier conundrum; a review of recent advances and persistent gaps regarding the carrier state of Foot-and-mouth disease virus. Pathogens 2020, 9, 167. [Google Scholar] [CrossRef] [PubMed]
- Ilott, M.C.; Salt, J.S.; Gaskell, R.M.; Kitching, R.P. Dexamethasone inhibits virus production and the secretory IgA response in oesophageal-pharyngeal fluid in cattle persistently infected with foot-and-mouth disease virus. Epidemiol. Infect. 1997, 118, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.J.; Arzt, J.; Puckette, M.C.; Smoliga, G.R.; Pacheco, J.M.; Rodriguez, L.L. Mechanisms of foot-and-mouth disease virus tropism inferred from differential tissue gene expression. PLoS ONE 2013, 8, e64119. [Google Scholar] [CrossRef] [PubMed]
- Eschbaumer, M.; Stenfeldt, C.; Smoliga, G.R.; Pacheco, J.M.; Rodriguez, L.L.; Li, R.W.; Zhu, J.; Arzt, J. Transcriptomic analysis of persistent infection with foot-and-mouth disease virus in cattle suggests impairment of apoptosis and cell-mediated immunity in the nasopharynx. PLoS ONE 2016, 11, e0162750. [Google Scholar] [CrossRef]
- Stenfeldt, C.; Eschbaumer, M.; Smoliga, G.R.; Rodriguez, L.L.; Zhu, J.J.; Arzt, J. Clearance of a persistent picornavirus infection is associated with enhanced pro-apoptotic and cellular immune responses. Sci. Rep. 2017, 7, 17800–17814. [Google Scholar] [CrossRef]
- Zhu, J.J.; Stenfeldt, C.; Bishop, E.A.; Canter, J.A.; Eschbaumer, M.; Rodriguez, L.L.; Arzt, J. Mechanisms of maintenance of foot-and-mouth disease virus persistence inferred from genes differentially expressed in nasopharyngeal epithelia of virus carriers and non-carriers. Front. Vet. Sci. 2020, 7, 340. [Google Scholar] [CrossRef]
- Wagage, S.; Hunter, C.A. Interrelated roles for the aryl hydrocarbon receptor and hypoxia inducible factor-1α in the immune response to infection. Curr. Immunol. Rev. 2015, 11, 43–54. [Google Scholar] [CrossRef][Green Version]
- Gutiérrez-Vázquez, C.; Quintana, F.J. Regulation of the immune response by the aryl hydrocarbon receptor. Immunity 2018, 48, 19–33. [Google Scholar] [CrossRef]
- Sun, S.C. The non-canonical NF-κB pathway in immunity and inflammation. Nat. Rev. Immunol. 2017, 17, 545–558. [Google Scholar] [CrossRef]
- Bruhs, A.; Haarmann-Stemmann, T.; Frauenstein, K.; Krutmann, J.; Schwarz, T.; Schwarz, A. Activation of the arylhydrocarbon receptor causes immunosuppression primarily by modulating dendritic cells. J. Invest. Dermatol. 2015, 135, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Lahoti, T.S.; Boyer, J.A.; Kusnadi, A.; Gulsum, M.E.; Murray, I.A.; Perdew, G.H. Aryl hydrocarbon receptor activation synergistically induces lipopolysaccharide-mediated expression of proinflammatory chemokine (c-c motif) ligand 20. Toxicol. Sci. 2013, 1, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Innocentin, S.; Withers, D.R.; Roberts, N.A.; Gallagher, A.R.; Grigorieva, E.F.; Willhelm, C.; Veldhoen, M. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell 2011, 147, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, K.; Maekawa, Y.; Kataoka, K.; Ishifune, C.; Nishida, J.; Arimochi, H.; Kitamura, A.; Yoshimoto, T.; Tomita, S.; Nagahiro, S.; et al. The ARNT–STAT3 axis regulates the differentiation of intestinal intraepithelial TCRαβ+ CD8αα+ cells. Nat. Commun. 2013, 4, 2112. [Google Scholar] [CrossRef] [PubMed]
- Apetoh, L.; Quintana, F.J.; Pot, C.; Joller, N.; Xiao, S.; Kumar, D.; Burns, E.J.; Sherr, D.H.; Weiner, H.L.; Kuchroo, V.K. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat. Immunol. 2010, 11, 854–861. [Google Scholar] [CrossRef]
- Longhi, M.S.; Vuerich, M.; Kalbasi, A.; Kenison, J.E.; Yeste, A.; Csizmadia, E.; Vaughn, B.; Feldbrugge, L.; Mitsuhashi, S.; Wegiel, B.; et al. Bilirubin suppresses Th17 immunity in colitis by upregulating CD39. JCI Insight 2017, 2, e92791. [Google Scholar] [CrossRef]
- DiNatale, B.C.; Schroeder, J.C.; Francey, L.J.; Kusnadi, A.; Perdew, G.H. Mechanistic insights into the events that lead to synergistic induction of interleukin 6 transcription upon activation of the aryl hydrocarbon receptor and inflammatory signaling. J. Biol. Chem. 2010, 285, 24388–24397. [Google Scholar] [CrossRef]
- Cibrian, D.; Saiz, M.L.; de la Fuente, H.; Sánchez-Díaz, R.; Moreno-Gonzalo, O.; Jorge, I.; Ferrarini, A.; Vázquez, J.; Punzón, C.; Fresno, M.; et al. CD69 controls the uptake of L-tryptophan through LAT1-CD98 and AhR-dependent secretion of IL-22 in psoriasis. Nat. Immunol. 2016, 17, 985–996. [Google Scholar] [CrossRef]
- Tajima, H.; Tajiki-Nishino, R.; Watanabe, Y.; Kurata, K.; Fukuyama, T. Activation of aryl hydrocarbon receptor by benzo [a] pyrene increases interleukin 33 expression and eosinophil infiltration in a mouse model of allergic airway inflammation. J. Appl. Toxicol. 2020, 40, 1545–1553. [Google Scholar] [CrossRef]
- D’Ignazio, L.; Bandarra, D.; Rocha, S. NF-κB and HIF crosstalk in immune responses. FEBS J. 2016, 283, 413–424. [Google Scholar] [CrossRef]
- Papandreou, I.; Cairns, R.A.; Fontana, L.; Lim, A.L.; Denko, N.C. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006, 3, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Tchernyshyov, I.; Semenza, G.L.; Dang, C.V. HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006, 3, 177–185. [Google Scholar] [CrossRef]
- Foxler, D.E.; Bridge, K.S.; James, V.; Webb, T.M.; Mee, M.; Wong, S.C.K.; Feng, Y.; Constantin-Teodosiu, D.; Petursdottir, T.E.; Bjornsson, J.; et al. The LIMD1 protein bridges an association between the prolyl hydroxylases and VHL to repress HIF-1 activity. Nat. Cell Biol. 2012, 14, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.C.; Quintin, J.; Cramer, R.A.; Shepardson, K.M.; Saeed, S.; Kumar, V.; Giamarellos-Bourboulis, E.J.; Matens, J.H.A.; Rao, N.A.; Aghajanirefah, A.; et al. mTOR-and HIF-1α–mediated aerobic glycolysis as metabolic basis for trained immunity. Science 2014, 345, 1250684. [Google Scholar] [CrossRef] [PubMed]
- Uche, U.U.; Piccirillo, A.R.; Kataoka, S.; Grebinoski, S.J.; D’Cruz, L.M.; Kane, L.P. PIK3IP1/TrIP restricts activation of T cells through inhibition of PI3K/Akt. J. Exp. Med. 2018, 215, 3165–3179. [Google Scholar] [CrossRef]
- Howie, D.; Waldmann, H.; Cobbold, S. Nutrient sensing via mTOR in T Cells maintains a tolerogenic microenvironment. Front. Immunol. 2014, 5, 409. [Google Scholar] [CrossRef]
- Goncharov, T.; Niessen, K.; de Almagro, M.C.; Izrael-Tomasevic, A.; Fedorova, A.V.; Varfolomeev, E.; Arnott, D.; Deshayes, K.; Kirkpatrivk, D.S.; Vucic, D. OTUB1 modulates c-IAP1 stability to regulate signaling pathways. EMBO J. 2013, 32, 1103–1114. [Google Scholar] [CrossRef]
- Peng, Y.; Xu, R.; Zheng, X. HSCARG negatively regulates the cellular antiviral RIG-I like receptor signaling pathway by inhibiting TRAF3 ubiquitination via recruiting OTUB1. PLoS Pathog. 2014, 10, e1004041. [Google Scholar] [CrossRef]
- Häcker, H.; Tseng, P.H.; Karin, M. Expanding TRAF function: TRAF3 as a tri-faced immune regulator. Nat. Rev. Immunol. 2011, 11, 457–468. [Google Scholar] [CrossRef]
- Huang, S.; Lu, W.; Ge, D.; Meng, N.; Li, Y.; Su, L.; Zhang, S.; Zhang, Y.; Zhao, B.; Miao, J. A new microRNA signal pathway regulated by long noncoding RNA TGFB2-OT1 in autophagy and inflammation of vascular endothelial cells. Autophagy 2015, 11, 2172–2183. [Google Scholar] [CrossRef]
- van Delft, M.A.; Huitema, L.F.; Tas, S.W. The contribution of NF-κB signalling to immune regulation and tolerance. Eur. J. Clin. Investig. 2015, 45, 529–539. [Google Scholar] [CrossRef]
- Micheli, L.; Leonardi, L.; Conti, F.; Maresca, G.; Colazingari, S.; Mattei, E.; Lira, S.A.; Farioli-Vecchioli, S.; Caruso, M.; Tirone, F. PC4/Tis7/IFRD1 stimulates skeletal muscle regeneration and is involved in myoblast differentiation as a regulator of MyoD and NF-kappaB. J. Biol. Chem. 2011, 286, 5691–5707. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.R.; Gonzales, N.; Aronowski, J. Pleiotropic role of PPARγ in intracerebral hemorrhage: An intricate system involving Nrf2, RXR, and NF-κB. CNS Neurosci. Ther. 2015, 21, 357–366. [Google Scholar] [CrossRef]
- Shalom-Barak, T.; Liersemann, J.; Memari, B.; Flechner, L.; Devor, C.E.; Bernardo, T.M.; Kim, S.; Matsumoto, N.; Friedman, S.L.; Evans, R.M.; et al. Ligand-dependent corepressor (LCoR) is a rexinoid-inhibited peroxisome proliferator-activated receptor γ-retinoid X receptor α coactivator. Mol. Cell Biol. 2018, 38, e00107–e00117. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, Z.; Chai, L.; Che, Y.; Min, S.; Yang, R. Identification and characterization of a unique leucine-rich repeat protein (LRRC33) that inhibits Toll-like receptor-mediated NF-κB activation. Biochem. Biophys. Res. Commun. 2013, 434, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Li, S.Z.; Zhang, H.H.; Liang, J.B.; Song, Y.; Jin, B.X.; Xing, N.N.; Fan, G.C.; Du, R.L.; Zhang, X.D. Nemo-like kinase (NLK) negatively regulates NF-kappa B activity through disrupting the interaction of TAK1 with IKKβ. Biochim. Biophys. Acta. 2014, 1843, 1365–1372. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jian, J.; Li, G.; Hettinghouse, A.; Liu, C. Progranulin: A key player in autoimmune diseases. Cytokine 2018, 101, 48–55. [Google Scholar] [CrossRef]
- Shahraz, A.; Kopatz, J.; Mathy, R.; Kappler, J.; Winter, D.; Kapoor, S.; Schütza, V.; Scheper, T.; Gieselmann, V.; Neumann, H. Anti-inflammatory activity of low molecular weight polysialic acid on human macrophages. Sci. Rep. 2015, 5, 16800. [Google Scholar] [CrossRef]
- Abdul-Sater, A.A.; Edilova, M.I.; Clouthier, D.L.; Mbanwi, A.; Kremmer, E.; Watts, T.H. The signaling adaptor TRAF1 negatively regulates Toll-like receptor signaling and this underlies its role in rheumatic disease. Nat. Immunol. 2017, 18, 26–35. [Google Scholar] [CrossRef]
- Jane-wit, D.; Surovtseva, Y.V.; Qin, L.; Li, G.; Liu, R.; Clark, P.; Manes, T.D.; Wang, C.; Kashgarian, M.; Kirkiles-Smith, N.C.; et al. Complement membrane attack complexes activate noncanonical NF-κB by forming an Akt+ NIK+ signalosome on Rab5+ endosomes. Proc. Natl. Acad. Sci. USA 2015, 112, 9686–9691. [Google Scholar] [CrossRef]
- Koliesnik, I.O.; Andreas, N.; Romanov, V.S.; Sreekantapuram, S.; Krljanac, B.; Haenold, R.; Weih, F. RelB regulates Th17 differentiation in a cell-intrinsic manner. Immunobiology 2018, 223, 191–199. [Google Scholar] [CrossRef]
- Suryawanshi, A.; Tadagavadi, R.K.; Swafford, D.; Manicassamy, S. Modulation of inflammatory responses by Wnt/β-Catenin signaling in dendritic cells: A novel immunotherapy target for autoimmunity and cancer. Front. Immunol. 2016, 7, 460. [Google Scholar] [CrossRef] [PubMed]
- Manicassamy, S.; Reizis, B.; Ravindran, R.; Nakaya, H.; Salazar-Gonzalez, R.M.; Wang, Y.C.; Pulendran, B. Activation of b-catenine in dendritic cells regulates immunity versus tolerance in the intestine. Science 2010, 329, 849–853. [Google Scholar] [CrossRef] [PubMed]
- Martin-Orozco, E.; Sanchez-Fernandez, A.; Ortiz-Parra, I.; Ayala-San Nicolas, M. WNT signaling in tumors: The way to evade drugs and immunity. Front. Immunol. 2019, 10, 2854. [Google Scholar] [CrossRef] [PubMed]
- Hunter, C.A.; Jones, S.A. IL-6 as a keystone cytokine in health and disease. Nat. Immunol. 2015, 16, 448–457. [Google Scholar] [CrossRef]
- Burkholder, B.; Huang, R.Y.; Burgess, R.; Luo, S.; Jones, V.S.; Zang, W.; Lv, Z.; Gao, C.; Wang, B.; Zhang, Y.; et al. Tumor-induced perturbations of cytokines and immune cell networks. Biochim. Biophys. Acta. 2014, 1845, 182–201. [Google Scholar] [CrossRef]
- McGeachy, M.J.; Cua, D.J. The link between IL-23 and Th17 cell-mediated immune pathologies. Semin. Immunol. 2007, 19, 372–376. [Google Scholar] [CrossRef]
- Cruikshank, W.W.; Kornfeld, H.; Center, D.M. Interleukin-16. J. Leukoc. Biol. 2000, 67, 757–766. [Google Scholar] [CrossRef]
- Skundric, D.S.; Cruikshank, W.W.; Montgomery, P.C.; Lisak, R.P.; Tse, H.Y. Emerging role of IL-16 in cytokine-mediated regulation of multiple sclerosis. Cytokine 2015, 75, 234–248. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Fujita, T.; Hirai, I.; Sahara, H.; Torigoe, T.; Ezoe, K.; Saito, T.; Cruikshank, W.W.; Yotsuyanagi, T.; Sato, N. Immunosuppressive effect on T cell activation by interleukin-16-and interleukin-10-cDNA-double-transfected human squamous cell line. Burns 2009, 35, 383–389. [Google Scholar] [CrossRef]
- Bézie, S.; Picarda, E.; Ossart, J.; Tesson, L.; Usual, C.; Renaudin, K.; Anegon, I.; Guillonneau, C. IL-34 is a Treg-specific cytokine and mediates transplant tolerance. J. Clin. Investig. 2015, 125, 3952–3964. [Google Scholar] [CrossRef] [PubMed]
- Guillonneau, C.; Bézie, S.; Anegon, I. Immunoregulatory properties of the cytokine IL-34. Cell Mol. Life Sci. 2017, 74, 2569–2586. [Google Scholar] [CrossRef] [PubMed]
- Foucher, E.D.; Blanchard, S.; Preisser, L.; Garo, E.; Ifrah, N.; Guardiola, P.; Delneste, Y.; Jeannin, P. IL-34 induces the differentiation of human monocytes into immunosuppressive macrophages. antagonistic effects of GM-CSF and IFNγ. PLoS ONE 2013, 8, e56045. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Pan, G.; Tang, C.; Li, Z.; Zheng, D.; Wei, X.; Wu, Z. IL-34 inhibits acute rejection of rat liver transplantation by inducing Kupffer cell M2 polarization. Transplantation 2018, 102, e265–e274. [Google Scholar] [CrossRef] [PubMed]
- Boulakirba, S.; Pfeifer, A.; Mhaidly, R.; Obba, S.; Goulard, M.; Schmitt, T.; Chaintreuil, P.; Calleja, A.; Furstoss, N.; Orange, F.; et al. IL-34 and CSF-1 display an equivalent macrophage differentiation ability but a different polarization potential. Sci. Rep. 2018, 8, 256. [Google Scholar] [CrossRef] [PubMed]
- Sidhu-Varma, M.; Shih, D.Q.; Targan, S.R. Differential levels of Tl1a affect the expansion and function of regulatory T cells in modulating murine colitis. Inflamm. Bowel Dis. 2016, 22, 548–559. [Google Scholar] [CrossRef]
- Valatas, V.; Kolios, G.; Bamias, G. TL1A (TNFSF15) and DR3 (TNFRSF25): A co-stimulatory system of cytokines with diverse functions in gut mucosal immunity. Front. Immunol. 2019, 10, 583. [Google Scholar] [CrossRef]
- Liu, H.; Rohowsky-Kochan, C. Interleukin-27-mediated suppression of human Th17 cells is associated with activation of STAT1 and suppressor of cytokine signaling protein 1. J. Interferon Cytokine Res. 2011, 31, 459–469. [Google Scholar] [CrossRef]
- Aparicio-Siegmund, S.; Garbers, C. The biology of interleukin-27 reveals unique pro- and anti-inflammatory functions in immunity. Cytokine Growth Factor Rev. 2015, 26, 579–586. [Google Scholar] [CrossRef]
- Yoshida, H.; Hunter, C.A. The immunobiology of interleukin-27. Annu. Rev. Immunol. 2015, 33, 417–443. [Google Scholar] [CrossRef]
- Mascanfroni, I.D.; Yeste, A.; Vieira, S.M.; Burns, E.J.; Patel, B.; Sloma, I.; Wu, Y.; Mayo, L.; Ben-Hamo, R.; Efroni, S.; et al. IL-27 acts on DCs to suppress the T cell response and autoimmunity by inducing expression of the immunoregulatory molecule CD39. Nat. Immunol. 2013, 14, 1054–1063. [Google Scholar] [CrossRef] [PubMed]
- Pot, C.; Jin, H.; Awasthi, A.; Lie, S.M.; Lai, C.Y.; Madan, R.; Sharpe, A.H.; Karp, C.L.; Miaw, S.C.; Ho, I.C.; et al. Cutting edge: IL-27 induces the transcription factor c-Maf, cytokine IL-21, and the costimulatory receptor ICOS that coordinately act together to promote differentiation of IL-10-producing Tr1 cells. J. Immunol. 2009, 183, 797–801. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S. The role of transforming growth factor β in T helper 17 differentiation. Immunology. 2018, 155, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Morianos, I.; Papadopoulou, G.; Semitekolou, M.; Xanthou, G. Activin-A in the regulation of immunity in health and disease. J. Autoimmun. 2019, 104, 102314. [Google Scholar] [CrossRef]
- Angkasekwinai, P.; Park, H.; Wang, Y.H.; Wang, Y.H.; Chang, S.H.; Corry, D.B.; Liu, Y.J.; Zhu, Z.; Dong, C. Interleukin 25 promotes the initiation of proallergic type 2 responses. J. Exp. Med. 2007, 204, 1509–1517. [Google Scholar] [CrossRef]
- Bulek, K.; Swaidani, S.; Qin, J.; Lu, Y.; Gulen, M.F.; Herjan, T.; Min, B.; Kastelein, R.A.; Aronica, M.; Kosz-Vnenchak, M.; et al. The essential role of single Ig IL-1 receptor-related molecule/toll IL-1R8 in regulation of Th2 immune response. J. Immunol. 2009, 182, 2601–2609. [Google Scholar] [CrossRef]
- Hsieh, S.L.; Lin, W.W. Decoy receptor 3: An endogenous immunomodulator in cancer growth and inflammatory reactions. J. Biomed. Sci. 2017, 24, 39. [Google Scholar] [CrossRef]
- Mueller, A.M.; Pedré, X.; Killian, S.; David, M.; Steinbrecher, A. The decoy receptor 3 (DcR3, TNFRSF6B) suppresses Th17 immune responses and is abundant in human cerebrospinal fluid. J. Neuroimmunol. 2009, 209, 57–64. [Google Scholar] [CrossRef]
- Kurts, C.; Carbone, F.R.; Krummel, M.F.; Koch, K.M.; Miller, J.F.; Heath, W.R. Signaling through CD30 protects against autoimmune diabetes mediated by CD8 T cells. Nature 1999, 398, 341–344. [Google Scholar] [CrossRef]
- Menzies-Gow, A.; Ying, S.; Sabroe, I.; Stubbs, V.L.; Soler, D.; Williams, T.J.; Kay, A.B. Eotaxin (CCL11) and eotaxin-2 (CCL24) induce recruitment of eosinophils, basophils, neutrophils, and macrophages as well as features of early- and late-phase allergic reactions following cutaneous injection in human atopic and nonatopic volunteers. J. Immunol. 2002, 169, 2712–2718. [Google Scholar] [CrossRef]
- Li, W.; Liu, L.; Gomez, A.; Zhang, J.; Ramadan, A.; Zhang, Q.; Choi, S.W.; Zhang, P.; Greenson, J.K.; Liu, C.; et al. Proteomics analysis reveals a Th17-prone cell population in presymptomatic graft-versus-host disease. JCI Insight 2016, 1, e86660. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.Y.; Eri, R.; Lyons, A.B.; Grimm, M.C.; Korner, H. CC Chemokine ligand 20 and its cognate receptor CCR6 in mucosal T cell immunology and inflammatory bowel disease: Odd couple or axis of evil? Front. Immunol. 2013, 4, 194. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.Y.S.; Körner, H. The CCR6-CCL20 axis in humoral immunity and T-B cell immunobiology. Immunobiology 2019, 224, 449–454. [Google Scholar] [CrossRef] [PubMed]
- Meiron, M.; Zohar, Y.; Anunu, R.; Wildbaum, G.; Karin, N. CXCL12 (SDF-1alpha) suppresses ongoing experimental autoimmune encephalomyelitis by selecting antigen-specific regulatory T cells. J. Exp. Med. 2008, 205, 2643–2655. [Google Scholar] [CrossRef] [PubMed]
- Comito, G.; Giannoni, E.; Segura, C.P.; Barcellos-de-Souza, P.; Raspollini, M.R.; Baroni, G.; Lanciotti, M.; Serni, S.; Chiarugi, P. Cancer-associated fibroblasts and M2-polarized macrophages synergize during prostate carcinoma progression. Oncogene 2014, 33, 2423–2431. [Google Scholar] [CrossRef]
- Lim, K.; Hyun, Y.M.; Lambert-Emo, K.; Capece, T.; Bae, S.; Miller, R.; Topham, D.J.; Kim, M. Neutrophil trails guide influenza-specific CD8+ T cells in the airways. Science 2015, 349, aaa4352. [Google Scholar] [CrossRef]
- Zhong, J.; Rajagopalan, S. Dipeptidyl peptidase-4 regulation of SDF-1/CXCR4 axis: Implications for cardiovascular disease. Front. Immunol. 2015, 6, 477. [Google Scholar] [CrossRef]
- Ansel, K.M.; Ngo, V.N.; Hyman, P.L.; Luther, S.A.; Förster, R.; Sedgwick, J.D.; Browning, J.L.; Lipp, M.; Cyster, J.G. A chemokine-driven positive feedback loop organizes lymphoid follicles. Nature 2000, 406, 309–314. [Google Scholar] [CrossRef]
- Kurth, I.; Willimann, K.; Schaerli, P.; Hunziker, T.; Clark-Lewis, I.; Moser, B. Monocyte selectivity and tissue localization suggests a role for breast and kidney-expressed chemokine (BRAK) in macrophage development. J. Exp. Med. 2001, 194, 855–861. [Google Scholar] [CrossRef]
- Lu, J.; Chatterjee, M.; Schmid, H.; Beck, S.; Gawaz, M. CXCL14 as an emerging immune and inflammatory modulator. J. Inflamm. 2016, 13, 1–8. [Google Scholar] [CrossRef]
- Cereijo, R.; Gavaldà-Navarro, A.; Cairó, M.; Quesada-López, T.; Villarroya, J.; Morón-Ros, S.; Sánchez-Infantes, D.; Peyrou, M.; Iglesias, R.; Mampel, T.; et al. CXCL14, a brown adipokine that mediates brown-fat-to-macrophage communication in thermogenic adaptation. Cell Metab. 2018, 28, 750–763. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.T.; Liu, S.P.; Lin, C.H.; Lee, S.W.; Hsu, C.Y.; Sytwu, H.K.; Hsieh, C.H.; Shyu, W.C. A crucial role of CXCL14 for promoting regulatory T cells activation in stroke. Theranostics 2017, 7, 855–875. [Google Scholar] [CrossRef] [PubMed]
- Rajasekaran, G.; Dinesh Kumar, S.; Nam, J.; Jeon, D.; Kim, Y.; Lee, C.W.; Park, I.S.; Shin, S.Y. Antimicrobial and anti-inflammatory activities of chemokine CXCL14-derived antimicrobial peptide and its analogs. Biochim. Biophys. Acta. Biomembr. 2019, 1861, 256–267. [Google Scholar] [CrossRef] [PubMed]
- Tanegashima, K.; Suzuki, K.; Nakayama, Y.; Tsuji, K.; Shigenaga, A.; Otaka, A.; Hara, T. CXCL14 is a natural inhibitor of the CXCL12-CXCR4 signaling axis. FEBS Lett. 2013, 587, 1731–1735. [Google Scholar] [CrossRef] [PubMed]
- Tanegashima, K.; Tsuji, K.; Suzuki, K.; Shigenaga, A.; Otaka, A.; Hara, T. Dimeric peptides of the C-terminal region of CXCL14 function as CXCL12 inhibitors. FEBS Lett. 2013, 587, 3770–3775. [Google Scholar] [CrossRef]
- Ramos, C.D.; Canetti, C.; Souto, J.T.; Silva, J.S.; Hogaboam, C.M.; Ferreira, S.H.; Cunha, F.Q. MIP-1alpha[CCL3] acting on the CCR1 receptor mediates neutrophil migration in immune inflammation via sequential release of TNF-alpha and LTB4. J. Leukoc Biol. 2005, 78, 167–177. [Google Scholar] [CrossRef]
- Griffith, J.W.; Sokol, C.L.; Luster, A.D. Chemokines and chemokine receptors: Positioning cells for host defense and immunity. Annu. Rev. Immunol. 2014, 32, 659–702. [Google Scholar] [CrossRef]
- Castellino, F.; Huang, A.Y.; Altan-Bonnet, G.; Stoll, S.; Scheinecker, C.; Germain, R.N. Chemokines enhance immunity by guiding naive CD8+ T cells to sites of CD4+ T cell-dendritic cell interaction. Nature 2006, 440, 890–895. [Google Scholar] [CrossRef]
- Ye, J.; Kohli, L.L.; Stone, M.J. Characterization of binding between the chemokine eotaxin and peptides derived from the chemokine receptor CCR3. J. Biol. Chem. 2000, 275, 27250–27257. [Google Scholar] [CrossRef]
- Collins, P.J.; McCully, M.L.; Martínez-Muñoz, L.; Santiago, C.; Wheeldon, J.; Caucheteux, S.; Thelen, S.; Cecchinato, V.; Laufer, J.M.; Purvanov, V.; et al. Epithelial chemokine CXCL14 synergizes with CXCL12 via allosteric modulation of CXCR4. FASEB J. 2017, 31, 3084–3097. [Google Scholar] [CrossRef]
- Janssens, R.; Struyf, S.; Proost, P. Pathological roles of the homeostatic chemokine CXCL12. Cytokine Growth Factor Rev. 2018, 44, 51–68. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Takahama, Y. XCL1 and XCR1 in the immune system. Microbes Infect. 2012, 14, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Moon, S.J.; Lee, S.W. Lineage re-commitment of CD4CD8αα intraepithelial lymphocytes in the gut. BMB Rep. 2016, 49, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Lederman, S.; Yellin, M.J.; Krichevsky, A.; Belko, J.; Lee, J.J.; Chess, L. Identification of a novel surface protein on activated CD4+ T cells that induces contact-dependent B cell differentiation (help). J. Exp. Med. 1992, 175, 1091–1101. [Google Scholar] [CrossRef]
- Flanagan, K.; Fitzgerald, K.; Baker, J.; Regnstrom, K.; Gardai, S.; Bard, F.; Mocci, S.; Seto, P.; You, M.; Larochelle, C.; et al. Laminin-411 is a vascular ligand for MCAM and facilitates TH17 cell entry into the CNS. PLoS ONE 2012, 7, e40443. [Google Scholar] [CrossRef]
- Kohlhaas, S.; Garden, O.A.; Scudamore, C.; Turner, M.; Okkenhaug, K.; Vigorito, E. Cutting edge: The Foxp3 target miR-155 contributes to the development of regulatory T cells. J. Immunol. 2009, 182, 2578–2582. [Google Scholar] [CrossRef]
- Lu, L.F.; Thai, T.H.; Calado, D.P.; Chaudhry, A.; Kubo, M.; Tanaka, K.; Loeb, G.B.; Lee, H.; Yoshimura, A.; Rajewski, K.; et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity 2009, 30, 80–91. [Google Scholar] [CrossRef]
- Escobar, T.M.; Kanellopoulou, C.; Kugler, D.G.; Kilaru, G.; Nguyen, C.K.; Nagarajan, V.; Bhairavabhotla, R.K.; Northrup, D.; Zahr, R.; Burr, P.; et al. miR-155 activates cytokine gene expression in Th17 cells by regulating the DNA-binding protein Jarid2 to relieve polycomb-mediated repression. Immunity 2014, 40, 865–879. [Google Scholar] [CrossRef]
- Yao, R.; Ma, Y.L.; Liang, W.; Li, H.; Ma, Z.; Yu, X.; Liao, Y. MicroRNA-155 modulates Treg and Th17 cells differentiation and Th17 cell function by targeting SOCS1. PLoS ONE 2012, 7, e46082. [Google Scholar] [CrossRef]
- Cui, G.; Qin, X.; Wu, L.; Zhang, Y.; Sheng, X.; Yu, Q.; Sheng, H.; Xi, B.; Zhang, J.Z.; Zang, Y.Q. Liver X receptor (LXR) mediates negative regulation of mouse and human Th17 differentiation. J. Clin. Investig. 2011, 121, 658–670. [Google Scholar] [CrossRef]
- Martin, P.; Gomez, M.; Lamana, A.; Cruz-Adalia, A.; Ramirez-Huesca, M.; Ursa, M.A.; Yanez-Mo, M.; Sanchez-Madrid, F. CD69 association with Jak3/Stat5 proteins regulates Th17 cell differentiation. Mol. Cell Biol. 2010, 30, 4877–4889. [Google Scholar] [CrossRef]
- Zeng, H.; Zhang, R.; Jin, B.; Chen, L. Type 1 regulatory T cells: A new mechanism of peripheral immune tolerance. Cell Mol Immunol. 2015, 12, 566–571. [Google Scholar] [CrossRef]
- Arce-Sillas, A.; Álvarez-Luquín, D.D.; Tamaya-Domínguez, B.; Gomez-Funtes, S.; Trejo-Garcia, A.; Melo-Salas, M.; Cárdenas, G.; Rodriguez-Ramirez, J.; Adalid-Peralta, L. Regulatory T Cells: Molecular actions on effector cells in immune regulation. J. Immunol. Res. 2016, 2016, 1720827. [Google Scholar] [CrossRef]
- Rueda, C.M.; Jackson, C.M.; Chougnet, C.A. Regulatory T-cell-mediated suppression of conventional T-cells and dendritic cells by different cAMP intracellular pathways. Front. Immunol. 2016, 7, 216. [Google Scholar] [CrossRef]
- Wherry, E.J.; Kurachi, M. Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 2015, 15, 486–499. [Google Scholar] [CrossRef]
- Jones, A.; Bourque, J.; Kuehm, L.; Opejin, A.; Teague, R.M.; Gross, C.; Hawiger, D. Immunomodulatory functions of BTLA and HVEM govern induction of extrathymic regulatory T cells and tolerance by dendritic cells. Immunity 2016, 45, 1066–1077. [Google Scholar] [CrossRef]
- Aksoylar, H.I.; Lampe, K.; Barnes, M.J.; Plas, D.R.; Hoebe, K. Loss of immunological tolerance in Gimap5-deficient mice is associated with loss of Foxo in CD4+ T cells. J. Immunol. 2012, 188, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Endale, M.; Aksoylar, H.I.; Hoebe, K. Central role of gimap5 in maintaining peripheral tolerance and T cell homeostasis in the gut. Mediat. Inflamm. 2015, 2015, 436017. [Google Scholar] [CrossRef] [PubMed]
- Patterson, A.R.; Bolcas, P.; Lampe, K.; Cantrell, R.; Ruff, B.; Lewkowich, I.; Hogan, S.P.; Janssen, E.M.; Bleesing, J.; Hershey, G.K.K.; et al. Loss of GTPase of immunity-associated protein 5 (Gimap5) promotes pathogenic CD4+ T-cell development and allergic airway disease. J. Allergy Clin. Immunol. 2019, 143, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Mao, L.; Xiao, F.; Liao, Z.; Yin, J.; Li, W.; Sun, M.; Liu, M.; Ji, X.; Liu, C.; et al. Interferon-stimulated genes inhibit caprine parainfluenza virus type 3 replication in Madin-Darby bovine kidney cells. Vet. Microbiol. 2020, 241, 108573. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Liang, S.; Zhong, Z.; Wen, J.; Li, W.; Wang, L.; Xu, J.; Zhong, F.; Li, X. Soluble CD83 inhibits human monocyte differentiation into dendritic cells in vitro. Cell Immunol. 2014, 292, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Nakano-Yokomizo, T.; Tahara-Hanaoka, S.; Nakahashi-Oda, C.; Nabekura, T.; Tchao, N.K.; Kadosaki, M.; Totsuka, N.; Kurita, N.; Nakamagoe, K.; Tamaoka, A.; et al. The immunoreceptor adapter protein DAP12 suppresses B lymphocyte-driven adaptive immune responses. J. Exp. Med. 2011, 208, 1661–1671. [Google Scholar] [CrossRef] [PubMed]
- Gasiorowski, R.E.; Ju, X.; Hart, D.N.; Clark, G.J. CD300 molecule regulation of human dendritic cell functions. Immunol. Lett. 2013, 149, 93–100. [Google Scholar] [CrossRef]
- Lopez Robles, M.D.; Pallier, A.; Huchet, V.; Le Texier, L.; Remy, S.; Braudeau, V.; Delbos, L.; Moreau, A.; Louvet, C.; Brosseau, C.; et al. Cell-surface C-type lectin-like receptor CLEC-1 dampens dendritic cell activation and downstream Th17 responses. Blood Adv. 2017, 1, 557–568. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.H.; Faunce, D.E.; Stacey, M.; Terajewicz, A.; Nakamura, T.; Zhang-Hoover, J.; Kerley, M.; Mucenski, M.L.; Gordon, S.; Stein-Streilein, J. The macrophage F4/80 receptor is required for the induction of antigen-specific efferent regulatory T cells in peripheral tolerance. J. Exp. Med. 2005, 201, 1615–1625. [Google Scholar] [CrossRef]
- Yoshida, R. MHC class I recognition by monocyte-/macrophage-specific receptors. Adv. Immunol. 2014, 124, 207–247. [Google Scholar] [CrossRef]
- Wang, J.; Sun, J.; Liu, L.N.; Flies, D.B.; Nie, X.; Toki, M.; Zhang, J.; Song, C.; Zarr, M.; Zhou, X.; et al. Siglec-15 as an immune suppressor and potential target for normalization cancer immunotherapy. Nat. Med. 2019, 25, 656–666. [Google Scholar] [CrossRef]
- Freeman, G.J.; Casasnovas, J.M.; Umetsu, D.T.; DeKruyff, R.H. TIM genes: A family of cell surface phosphatidylserine receptors that regulate innate and adaptive immunity. Immunol. Rev. 2010, 235, 172–189. [Google Scholar] [CrossRef]
- Albackerk, L.A.; Karisola, P.; Chang, Y.J.; Umetsu, S.E.; Zhou, M.; Akbari, O.; Kobayashi, N.; Baumgarth, N.; Freeman, G.J.; Umetsu, D.T.; et al. TIM-4, a receptor for phosphatidylserine, controls adaptive immunity by regulating the removal of antigen-specific T cells. J. Immunol. 2010, 185, 6839–6849. [Google Scholar] [CrossRef]
- Round, J.L.; Lee, S.M.; Li, J.; Tran, G.; Jabri, B.; Chatila, T.A.; Mazmanian, S.K. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 2011, 332, 974–977. [Google Scholar] [CrossRef]
- Xiong, Y.; Pennini, M.; Vogel, S.N.; Medvedev, A.E. IRAK4 kinase activity is not required for induction of endotoxin tolerance but contributes to TLR2-mediated tolerance. J. Leukoc. Biol. 2013, 94, 291–300. [Google Scholar] [CrossRef]
- Manoharan, I.; Hong, Y.; Suryawanshi, A.; Angus-Hill, M.L.; Sun, Z.; Mellor, A.L.; Munn, D.H.; Manicassamy, S. TLR2-dependent activation of beta-catenin pathway in dendritic cells induces regulatory responses and attenuates autoimmune inflammation. J. Immunol. 2014, 193, 4203–4213. [Google Scholar] [CrossRef] [PubMed]
- Bhaskaran, N.; Cohen, S.; Zhang, Y.; Weinberg, A.; Pandiyan, P. TLR-2 signaling promotes IL-17A production in CD4+ CD25+ Foxp3+ regulatory cells during oropharyngeal candidiasis. Pathogens 2015, 4, 90–110. [Google Scholar] [CrossRef] [PubMed]
- Choteau, L.; Vancraeyneste, H.; Le Roy, D.; Dubuquoy, L.; Romani, L.; Jouault, T.; Poulain, D.; Sendid, B.; Calandra, T.; Roger, T.; et al. Role of TLR1, TLR2 and TLR6 in the modulation of intestinal inflammation and Candida albicans elimination. Gut Pathog. 2017, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Schröder, J.M.; Harder, J. Human beta-defensin-2. Int. J. Biochem. Cell Biol. 1999, 31, 645–651. [Google Scholar] [CrossRef]
- Chen, W.; Liu, Z.; Zhang, Q.; Yan, Q.; Jing, S. Induction and antiviral activity of human β-defensin 3 in intestinal cells with picornavirus infection. Acta Virol. 2018, 62, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Gaggero, S.; Bruschi, M.; Petretto, A.; Parodi, M.; Del Zotto, G.; Lavarello, C.; Prato, C.; Santucci, L.; Barbuto, A.; Bottino, C.; et al. Nidogen-1 is a novel extracellular ligand for the NKp44 activating receptor. Oncoimmunology 2018, 7, e1470730. [Google Scholar] [CrossRef] [PubMed]
- Turner, A.; Li, L.C.; Pilli, T.; Qian, L.; Wiley, E.L.; Setty, S.; Christov, K.; Ganesh, L.; Maker, A.V.; Li, P.; et al. MADD knock-down enhances doxorubicin and TRAIL induced apoptosis in breast cancer cells. PLoS ONE 2013, 8, e56817. [Google Scholar] [CrossRef]
- Zhu, J.J.; Canter, J.A.; Rodriguez, L.L.; Arzt, J. A novel bovine CXCL15 gene in the GRO chemokine gene cluster. Vet. Immunol. Immunopathol. 2020, 220, 109990. [Google Scholar] [CrossRef]
- Schönrich, G.; Raftery, M.J. Neutrophil extracellular traps go viral. Front. Immunol. 2016, 7, 366. [Google Scholar] [CrossRef]
- Pruett, J.H.; Fisher, W.F.; DeLoach, J.R. Effects of dexamethasone on selected parameters of the bovine immune system. Vet. Res. Commun. 1987, 11, 305–323. [Google Scholar] [PubMed]
- Jain, N.C.; Vegad, J.L.; Shrivastava, A.B.; Jain, N.K.; Garg, U.K.; Kolte, G.N. Haematological changes in buffalo calves inoculated with Escherichia coli endotoxin and corticosteroids. Res. Vet. Sci. 1989, 47, 305–308. [Google Scholar] [CrossRef]
- Thanasak, J.; Jorritsma, R.; Hoek, A.; Noordhuizen, J.P.; Rutten, V.P.; Müller, K.E. The effects of a single injection of dexamethasone-21-isonicotinate on the lymphocyte functions of dairy cows at two weeks post-partum. Vet. Res. 2004, 35, 103–112. [Google Scholar] [CrossRef][Green Version]
- Veldhoen, M.; Hirota, K.; Westendorf, A.M.; Buer, J.; Dumoutier, L.; Renauld, J.C.; Stockinger, B. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature 2008, 453, 106–109. [Google Scholar] [CrossRef]
- Veldhoen, M.; Hirota, K.; Christensen, J.; O’Garra, A.; Stockinger, B. Natural agonists for aryl hydrocarbon receptor in culture medium are essential for optimal differentiation of Th17 T cells. J. Exp. Med. 2009, 206, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Quintana, F.J.; Basso, A.S.; Iglesias, A.H.; Korn, T.; Farez, M.F.; Bettelli, E.; Caccamo, M.; Oukka, M.; Weiner, H.L. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008, 453, 65–71. [Google Scholar] [CrossRef]
- Singh, N.P.; Singh, U.P.; Rouse, M.; Zhang, J.; Chatterje, S.; Nagarkatti, P.S.; Nagarkatti, M. Dietary indoles suppress delayed-type hypersensitivity by inducing a switch from proinflammatory Th17 cells to anti-inflammatory regulatory T cells through regulation of microRNA. J. Immunol. 2016, 196, 1108–1122. [Google Scholar] [CrossRef] [PubMed]
- Hammerschmidt-Kamper, C.; Biljes, D.; Merches, K.; Steiner, I.; Daldrup, T.; Bol-Schoenmakers, M.; Pieters, R.H.H.; Esser, C. Indole-3-carbinol, a plant nutrient and AhR-Ligand precursor, supports oral tolerance against OVA and improves peanut allergy symptoms in mice. PLoS ONE 2017, 12, e0180321. [Google Scholar] [CrossRef]
- Cervantes-Barragan, L.; Chai, J.N.; Tianero, M.D.; Di Luccia, B.; Ahern, P.P.; Merriman, J.; Cortez, V.S.; Caparon, M.G.; Donia, M.S.; Gilfillan, S.; et al. Lactobacillus reuteri induces gut intraepithelial CD4+ CD8αα+ T cells. Science 2017, 357, 806–810. [Google Scholar] [CrossRef]
- Schiering, C.; Wincent, E.; Metidji, A.; Iseppon, A.; Li, Y.; Potocnik, A.J.; Omenetti, S.; Henderson, C.J.; Wolf, C.R.; Nebert, D.W.; et al. Feedback control of AHR signalling regulates intestinal immunity. Nature 2017, 542, 242–245. [Google Scholar] [CrossRef]
- Mezrich, J.D.; Fechner, J.H.; Zhang, X.; Johnson, B.P.; Burlingham, W.J.; Bradfield, C.A. An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J. Immunol. 2010, 185, 3190–3198. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.T.; Kimura, A.; Nakahama, T.; Chinen, I.; Masuda, K.; Nohara, K.; Fujii-Kuriyama, Y.; Kishimoto, T. Aryl hydrocarbon receptor negatively regulates dendritic cell immunogenicity via a kynurenine-dependent mechanism. Proc. Natl. Acad. Sci. USA 2010, 107, 19961–19966. [Google Scholar] [CrossRef] [PubMed]
- Quintana, F.J.; Murugaiyan, G.; Farez, M.F.; Mitsdoerffer, M.; Tukpah, A.M.; Burns, E.J.; Weiner, H.L. An endogenous aryl hydrocarbon receptor ligand acts on dendritic cells and T cells to suppress experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. USA 2010, 107, 20768–20773. [Google Scholar] [CrossRef]
- Platzer, B.; Richter, S.; Kneidinger, D.; Waltenberger, D.; Woisetschläger, M.; Strobl, H. Aryl hydrocarbon receptor activation inhibits in vitro differentiation of human monocytes and Langerhans dendritic cells. J. Immunol. 2009, 183, 66–74. [Google Scholar] [CrossRef]
- Jurado-Manzano, B.B.; Zavala-Reyes, D.; Turrubiartes-Martínez, E.A.; Portales-Pérez, D.P.; González-Amaro, R.; Layseca-Espinosa, E. FICZ generates human tDCs that induce CD4+ CD25high Foxp3+ Treg-like cell differentiation. Immunol. Lett. 2017, 190, 84–92. [Google Scholar] [CrossRef]
- Shinde, R.; McGaha, T.L. The aryl hydrocarbon receptor: Connecting immunity to the microenvironment. Trends Immunol. 2018, 39, 1005–1020. [Google Scholar] [CrossRef] [PubMed]
- Swedenborg, E.; Pongratz, I. AhR and ARNT modulate ER signaling. Toxicology 2010, 268, 132–138. [Google Scholar] [CrossRef]
- Göttel, M.; Le Corre, L.; Dumont, C.; Schrenk, D.; Chagnon, M.C. Estrogen receptor α and aryl hydrocarbon receptor cross-talk in a transfected hepatoma cell line (HepG2) exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol. Rep. 2014, 1, 1029–1036. [Google Scholar] [CrossRef]
- Helle, J.; Bader, M.I.; Keiler, A.M.; Zierau, O.; Vollmer, G.; Chittur, S.V.; Tenniswood, M.; Kretzschmar, G. Cross-talk in the female rat mammary gland: Influence of aryl hydrocarbon receptor on estrogen receptor signaling. Environ. Health Perspect. 2016, 124, 601–610. [Google Scholar] [CrossRef]
- Vorrink, S.U.; Domann, F.E. Regulatory crosstalk and interference between the xenobiotic and hypoxia sensing pathways at the AhR-ARNT-HIF1α signaling node. Chem. Biol. Interact. 2014, 218, 82–88. [Google Scholar] [CrossRef]
- Shi, L.Z.; Wang, R.; Huang, G.; Vogel, P.; Neale, G.; Green, D.R.; Chi, H. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J. Exp. Med. 2011, 208, 1367–1376. [Google Scholar] [CrossRef] [PubMed]
- Dang, E.V.; Barbi, J.; Yang, H.Y.; Jinasena, D.; Yu, H.; Zheng, Y.; Bordman, Z.; Fu, J.; Kim, Y.; Yen, H.R.; et al. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell 2011, 146, 772–784. [Google Scholar] [CrossRef] [PubMed]
- Gagliani, N.; Amezcua Vesely, M.C.; Iseppon, A.; Brockmann, L.; Xu, H.; Palm, N.W.; de Zoete, M.R.; Licona-Limón, P.; Paiva, R.S.; Ching, T.; et al. Th17 cells transdifferentiate into regulatory T cells during resolution of inflammation. Nature 2015, 523, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Vogel, C.F.; Khan, E.M.; Leung, P.S.; Gershwin, M.E.; Chang, W.L.; Wu, D.; Haarmann-Stemmann, T.; Hoffmann, A.; Denison, M.S. Cross-talk between aryl hydrocarbon receptor and the inflammatory response: A role for nuclear factor-κB. J. Biol. Chem. 2014, 289, 1866–1875. [Google Scholar] [CrossRef]
- Liu, P.C.; Phillips, M.A.; Matsumura, F. Alteration by 2,3,7,8-Tetrachlorodibenzo-p-dioxin of CCAAT/enhancer binding protein correlates with suppression of adipocyte differentiation in 3T3-L1 cells. Mol. Pharmacol. 1996, 49, 989–997. [Google Scholar]
- Borland, M.G.; Krishnan, P.; Lee, C.; Albrecht, P.P.; Shan, W.; Bility, M.T.; Marcus, C.B.; Lin, J.M.; Amin, S.; Gonzalez, F.J.; et al. Modulation of aryl hydrocarbon receptor (AHR)-dependent signaling by peroxisome proliferator-activated receptor β/δ (PPARβ/δ) in keratinocytes. Carcinogenesis 2014, 35, 1602–1612. [Google Scholar] [CrossRef]
- Phan, A.T.; Goldrath, A.W. Hypoxia-inducible factors regulate T cell metabolism and function. Mol. Immunol. 2015, 68, 527–535. [Google Scholar] [CrossRef]
- Tian, Y.; Ke, S.; Denison, M.S.; Rabson, A.B.; Gallo, M.A. Ah receptor and NF-kappaB interactions, a potential mechanism for dioxin toxicity. J. Biol. Chem. 1999, 274, 510–515. [Google Scholar] [CrossRef]
- Amatya, N.; Garg, A.V.; Gaffen, S.L. IL-17 Signaling: The Yin and the Yang. Trends Immunol. 2017, 38, 310–322. [Google Scholar] [CrossRef]
- Cella, M.; Colonna, M. Aryl hydrocarbon receptor: Linking environment to immunity. Semin. Immunol. 2015, 27, 310–314. [Google Scholar] [CrossRef]
- Marinelli, L.; Martin-Gallausiaux, C.; Bourhis, J.M.; Béguet-Crespel, F.; Blottière, H.M.; Lapaque, N. Identification of the novel role of butyrate as AhR ligand in human intestinal epithelial cells. Sci. Rep. 2019, 9, 643. [Google Scholar] [CrossRef] [PubMed]
- Garrison, P.M.; Rogers, J.M.; Brackney, W.R.; Denison, M.S. Effects of histone deacetylase inhibitors on the Ah receptor gene promoter. Arch. Biochem. Biophys. 2000, 374, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.H. Immune regulation by microbiome metabolites. Immunology 2018, 154, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Jin, U.H.; Cheng, Y.; Park, H.; Davidson, L.A.; Callaway, E.S.; Chapkin, R.S.; Jayaraman, A.; Asante, A.; Allred, C.; Weaver, E.A.; et al. Short chain fatty acids enhance aryl hydrocarbon (Ah) responsiveness in mouse colonocytes and Caco-2 human colon cancer cells. Sci. Rep. 2017, 7, 10163. [Google Scholar] [CrossRef] [PubMed]
- Salt, J.S.; Mulcahy, G.; Kitching, R.P. Isotype-specific antibody responses to foot-and-mouth disease virus in sera and secretions of ‘carrier’and ‘non-carrier’cattle. Epidemiol. Infect. 1996, 117, 349–360. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Parida, S.; Anderson, J.; Cox, S.J.; Barnett, P.V.; Paton, D.J. Secretory IgA as an indicator of oro-pharyngeal foot-and-mouth disease virus replication and as a tool for post vaccination surveillance. Vaccine 2006, 24, 1107–1116. [Google Scholar] [CrossRef]
- Smyth, G.K. Limma: Linear models for microarray data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Gentleman, R., Carey, V.J., Huber, W., Irizarry, R.A., Dudoit, S., Eds.; Statistics for Biology and Health; Springer: New York, NY, USA, 2015. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).