Variability among the Isolates of Broad Bean Mottle Virus and Encapsidation of Host RNAs
Abstract
:1. Introduction
2. Results and Discussion
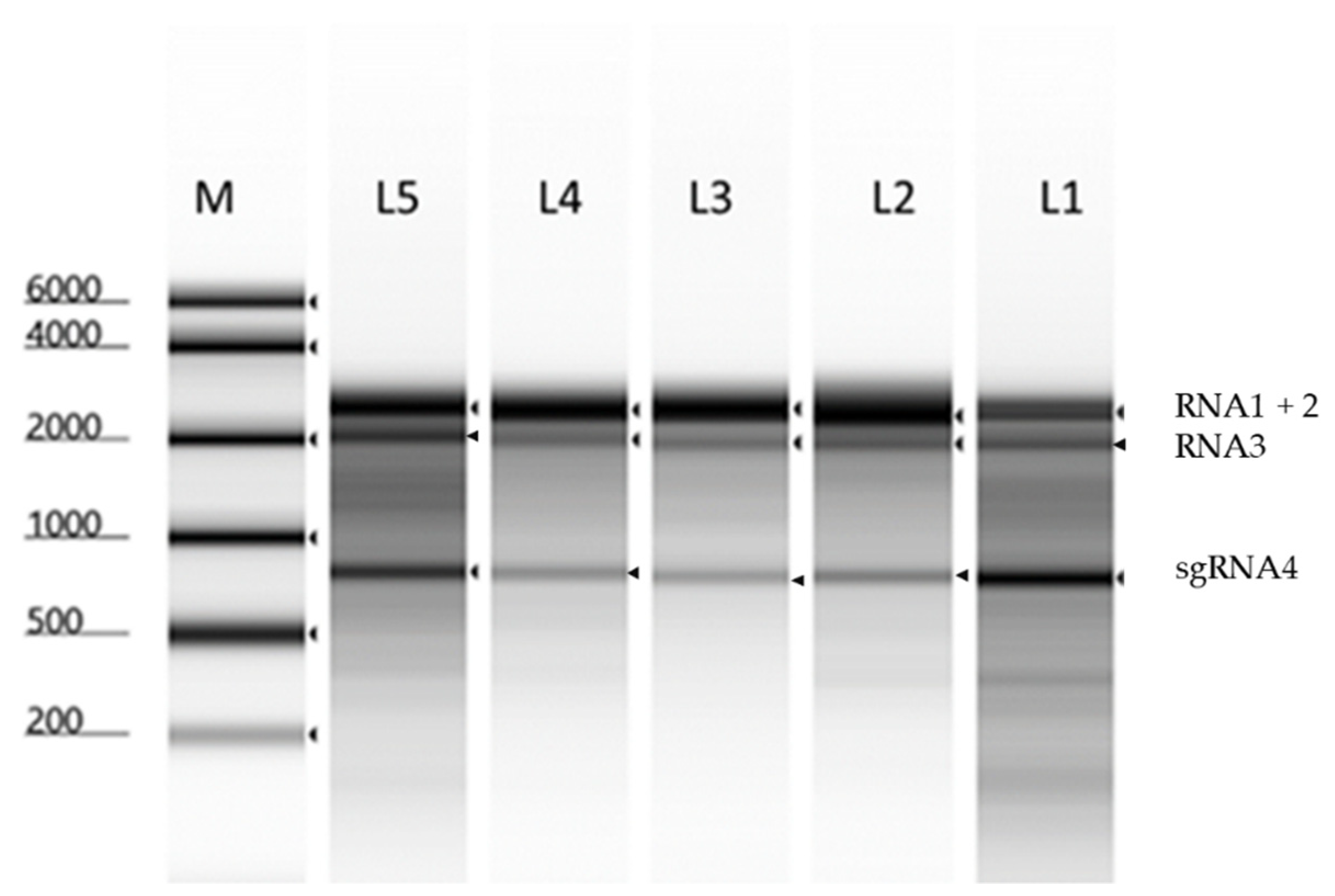
2.1. Propagation and Purification of BBMV RNAs
2.2. Sequencing with the NGS and Sanger Protocols
2.2.1. The Particle Heterogeneity
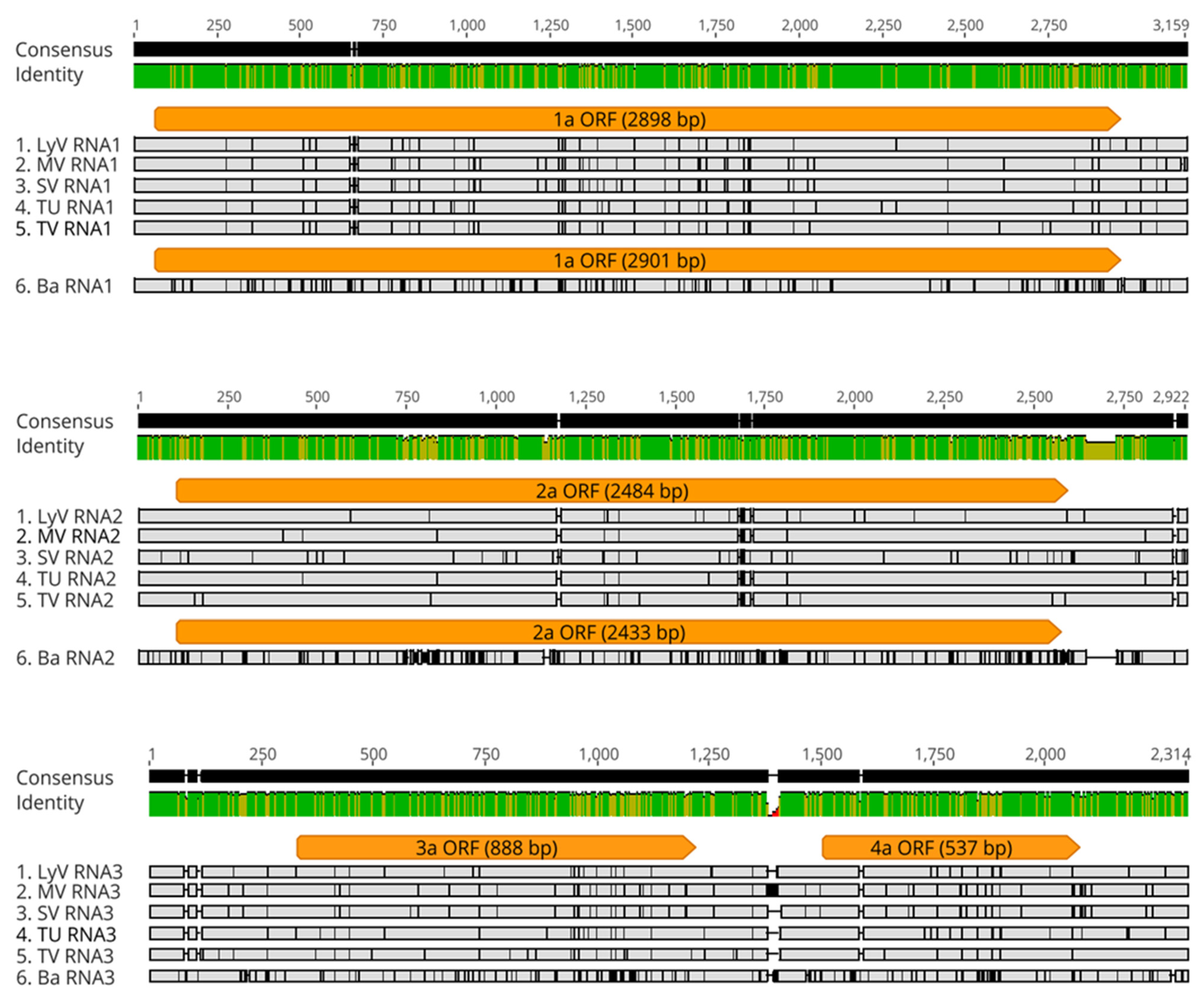
2.2.2. Polymorphism at the Nucleotide Level
2.2.3. The Noncoding Regions
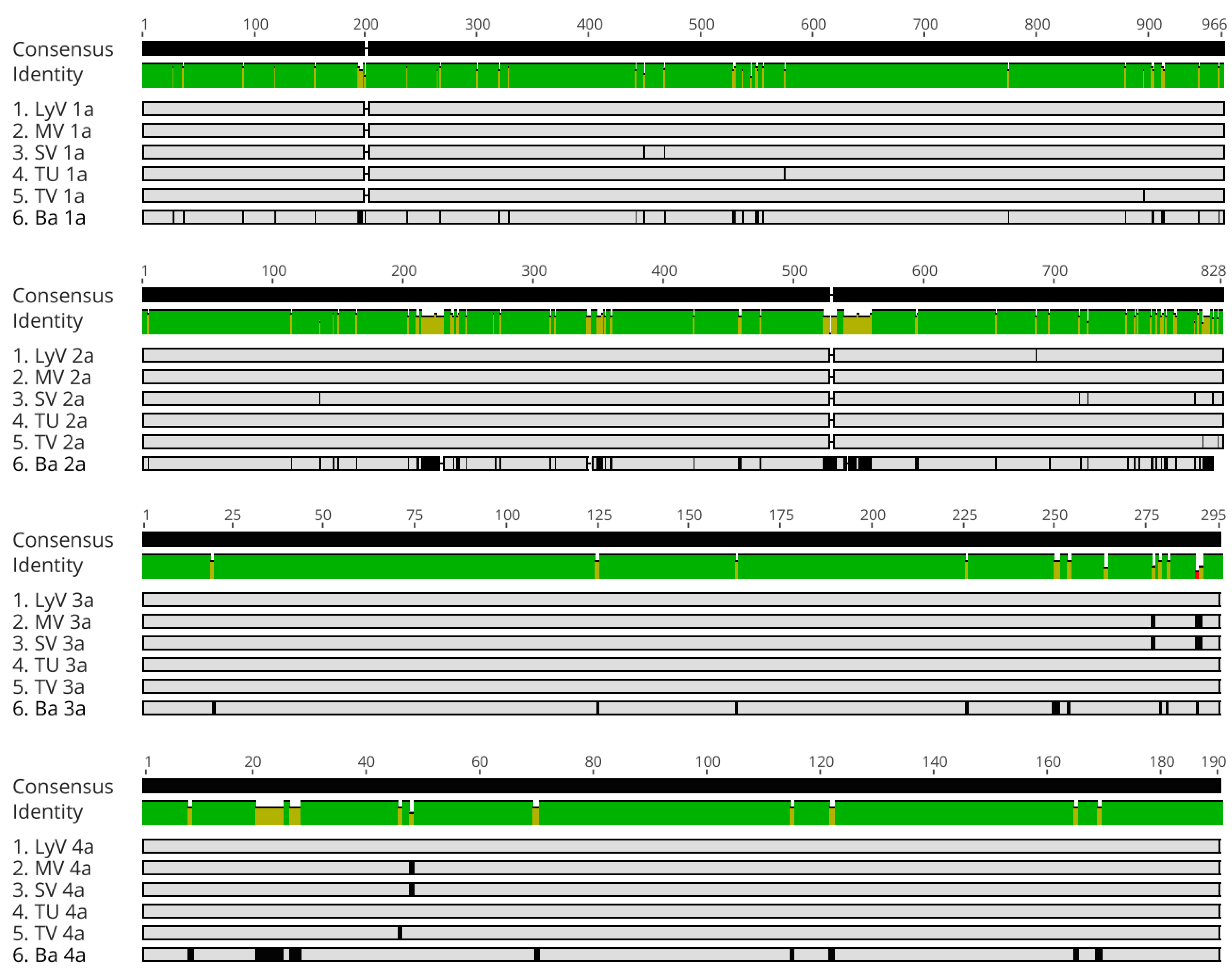
2.2.4. Polymorphism at the Amino Acid Level
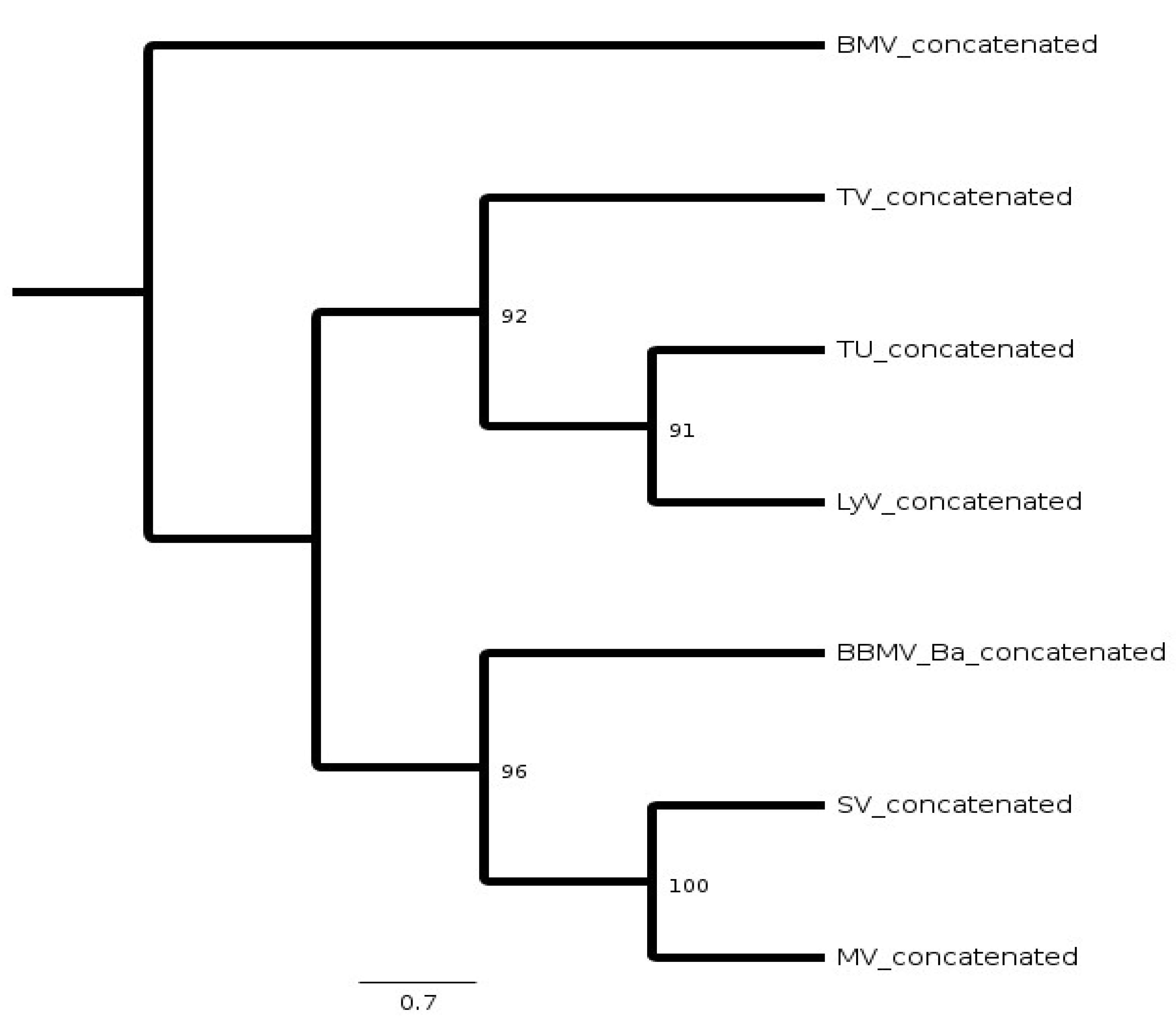
2.3. Phylogeny of BBMV Variants
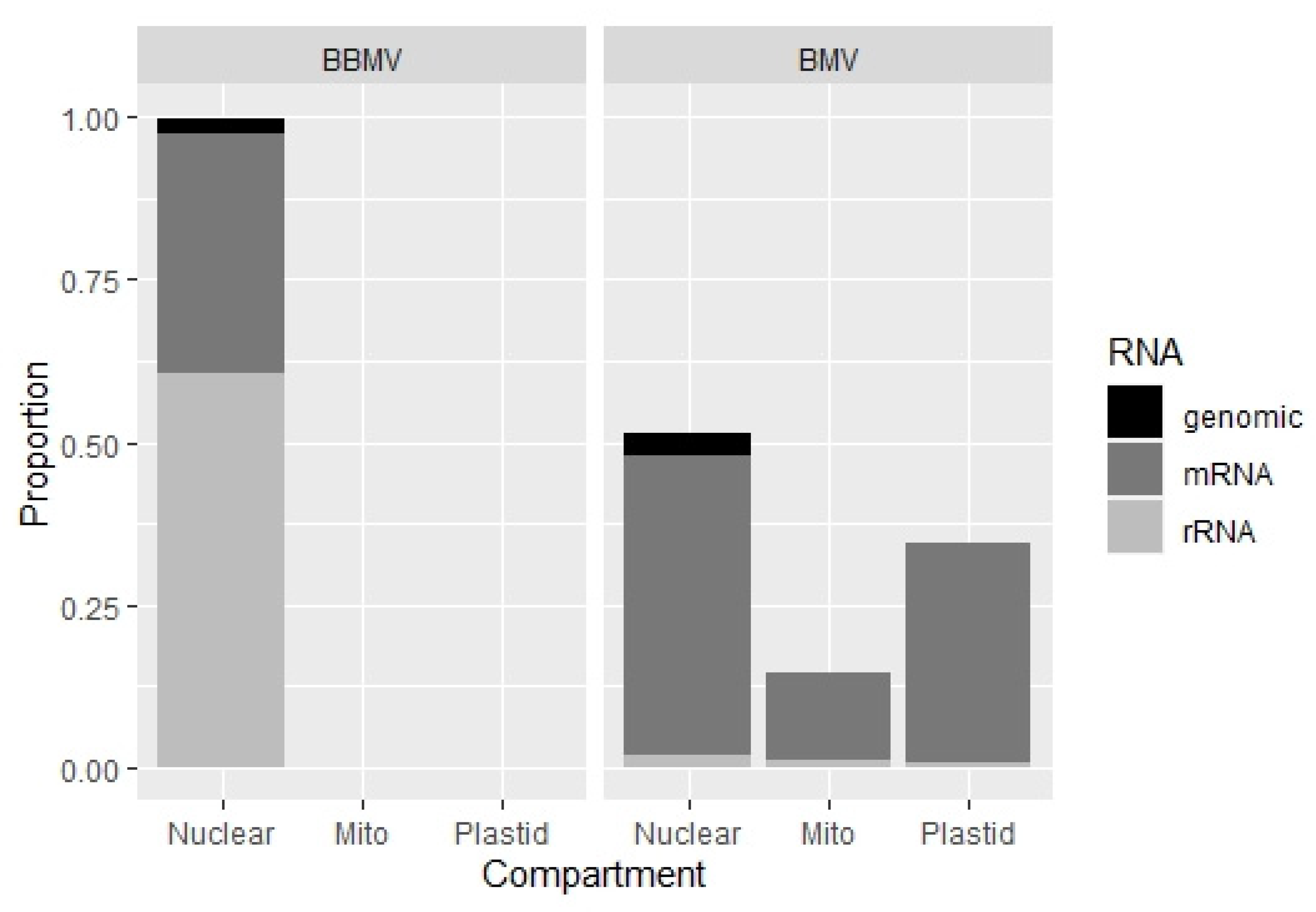
2.4. BBMV Encapsidate Host RNAs
3. Summary and Conclusions
4. Materials and Methods
4.1. Virus Material and Purification
4.2. RNA Extraction and NGS Sequencing
4.3. Rapid Amplification of cDNA Ends (RACE) and Sanger Sequencing
4.4. De Novo Genome Assembly and Sequence Comparisons
4.5. Phylogeny Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fortass, M.; Bos, L. Broad bean mottle virus in Morocco; variability, interaction with food legume species, and seed transmission in faba bean, pea, and chickpea. Eur. J. Plant Pathol. 1992, 98, 329–342. [Google Scholar] [CrossRef]
- Fortass, M.; Diallo, S. Broad bean mottle virus in Morocco; curculionid vectors, and natural occurrence in food legumes other than faba bean (Vicia faba). Eur. J. Plant Pathol. 1993, 99, 219–226. [Google Scholar] [CrossRef]
- Makkouk, K.M.; Bos, L.; Rizkallah, A.; Azzam, O.I.; Katul, L. Broad bean mottle virus: Identification, host range, serology, and occurrence on faba bean (Vicia faba) in West Asia and North Africa. Eur. J. Plant Pathol. 1989, 94, 195–212. [Google Scholar] [CrossRef] [Green Version]
- Bawden, F.C.; Chaudhuri, R.P.; Kassanis, B. Some properties of broad-bean mottle virus. Ann. Appl. Biol. 1951, 38, 774–784. [Google Scholar] [CrossRef]
- Dzianott, A.M.; Bujarski, J.I. The nucleotide sequence and genome organization of the RNA-1 Segment in two bromoviruses: Broad bean mottle virus and cowpea chlorotic mottle virus. Virology 1991, 185, 553–562. [Google Scholar] [CrossRef]
- Romero, J.; Dzianott, A.M.; Bujarski, J.J. The nucleotide sequence and genome organization of the RNA2 and RNA3 segments in broad bean mottle virus. Virology 1992, 187, 671–681. [Google Scholar] [CrossRef]
- Romero, J.; Huang, Q.; Pogany, J.; Bujarski, J.J. Characterization of Defective Interfering RNA Components That Increase Symptom Severity of Broad Bean Mottle Virus Infections. Virology 1993, 194, 576–584. [Google Scholar] [CrossRef]
- Ahlquist, P.; Dasgupta, R.; Kaesberg, P. Nucleotide sequence of the brome mosaic virus genome and its implications for viral replication. J. Mol. Biol. 1984, 172, 369–383. [Google Scholar] [CrossRef]
- Allison, R.F.; Janda, M.; Ahlquist, P. Sequence of cowpea chlorotic mottle virus RNAs 2 and 3 and evidence of a recombination event during bromovirus evolution. Virology 1989, 172, 321–330. [Google Scholar] [CrossRef]
- Ni, P.; Vaughan, R.C.; Tragesser, B.; Hoover, H.; Kao, C.C. The Plant Host Can Affect the Encapsidation of Brome Mosaic Virus (BMV) RNA: BMV Virions Are Surprisingly Heterogeneous. J. Mol. Biol. 2013, 426, 1061–1076. [Google Scholar] [CrossRef] [Green Version]
- Hoover, H.S.; Wang, J.C.-Y.; Middleton, S.; Ni, P.; Zlotnick, A.; Vaughan, R.C.; Kao, C.C. Phosphorylation of the Brome Mosaic Virus Capsid Regulates the Timing of Viral Infection. J. Virol. 2016, 90, 7748–7760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sacher, R.; Ahlquist, P. Effects of deletions in the N-terminal basic arm of brome mosaic virus coat protein on RNA packaging and systemic infection. J. Virol. 1989, 63, 4545–4552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marsh, L.; Hall, T. Evidence Implicating a tRNA Heritage for the Promoters of Positive-strand RNA Synthesis in Brome Mosaic and Related Viruses. Cold Spring Harb. Symp. Quant. Biol. 1987, 52, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Marsh, L.E.; Huntley, C.C.; Pogue, G.P.; Connell, J.P.; Hall, T.C. Regulation of (+):(-)-strand asymmetry in replication of brome mosaic virus RNA. Virology 1991, 182, 76–83. [Google Scholar] [CrossRef]
- Dinant, S.; Janda, M.; Kroner, P.A.; Ahlquist, P. Bromovirus RNA replication and transcription require compatibility between the polymerase- and helicase-like viral RNA synthesis proteins. J. Virol. 1993, 67, 7181–7189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacobs, A.; Hoover, H.; Smith, E.; Clemmer, D.E.; Kim, C.-H.; Kao, C.C. The intrinsically disordered N-terminal arm of the brome mosaic virus coat protein specifically recognizes the RNA motif that directs the initiation of viral RNA replication. Nucleic Acids Res. 2017, 46, 324–335. [Google Scholar] [CrossRef] [Green Version]
- Rao, A.; Grantham, G.L. Molecular studies on bromovirus capsid protein: II. Functional analysis of the amino-terminal arginine-rich motif and its role in encapsidation, movement, and pathology. Virology 1996, 226, 294–305. [Google Scholar] [CrossRef] [Green Version]
- Lucas, R.W.; Larson, S.B.; McPherson, A. The crystallographic structure of brome mosaic virus. J. Mol. Biol. 2002, 317, 95–108. [Google Scholar] [CrossRef] [Green Version]
- Speir, J.A.; Munshi, S.; Wang, G.; Baker, T.S.; Johnson, J.E. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy. Structure 1995, 3, 63–78. [Google Scholar] [CrossRef] [Green Version]
- Fujita, Y.; Mise, K.; Okuno, T.; Ahlquist, P.; Furusawa, I. A Single Codon Change in a Conserved Motif of a Bromovirus Movement Protein Gene Confers Compatibility with a New Host. Virology 1996, 223, 283–291. [Google Scholar] [CrossRef] [Green Version]
- Mise, K.; Allison, R.F.; Janda, M.; Ahlquist, P. Bromovirus movement protein genes play a crucial role in host specificity. J. Virol. 1993, 67, 2815–2823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeda, A.; Kaido, M.; Okuno, T.; Mise, K. The C terminus of the movement protein of Brome mosaic virus controls the requirement for coat protein in cell-to-cell movement and plays a role in long-distance movement. J. Gen. Virol. 2004, 85, 1751–1761. [Google Scholar] [CrossRef] [PubMed]
- Citovsky, V.; Knorr, D.; Schuster, G.; Zambryski, P. The P30 movement protein of tobacco mosaic virus is a single-strand nucleic acid binding protein. Cell 1990, 60, 637–647. [Google Scholar] [CrossRef]
- Traynor, P.; Ahlquist, P. Use of bromovirus RNA2 hybrids to map cis- and trans-acting functions in a conserved RNA replication gene. J. Virol. 1990, 64, 69–77. [Google Scholar] [CrossRef] [Green Version]
- Flasinski, S.; Dzianott, A.; Pratt, S.; Bujarski, J.J. Mutational analysis of the coat protein gene of brome mosaic virus: Effects on replication and movement in barley and in Chenopodium hybridum. Molecular plant-microbe interactions. Mol. Plant-Microbe Interact. 1995, 8, 23–31. [Google Scholar] [CrossRef]
- Ni, P.; Wang, Z.; Ma, X.; Das, N.C.; Sokol, P.; Chiu, W.; Dragnea, B.; Hagan, M.; Kao, C.C. An Examination of the Electrostatic Interactions between the N-Terminal Tail of the Brome Mosaic Virus Coat Protein and Encapsidated RNAs. J. Mol. Biol. 2012, 419, 284–300. [Google Scholar] [CrossRef] [Green Version]
- Shrestha, N.; Weber, P.H.; Burke, S.V.; Wysocki, W.P.; Duvall, M.R.; Bujarski, J.J. Next generation sequencing reveals packaging of host RNAs by brome mosaic virus. Virus Res. 2018, 252, 82–90. [Google Scholar] [CrossRef]
- Sims, D.; Sudbery, I.; Ilott, N.E.; Heger, A.; Ponting, C.P. Sequencing depth and coverage: Key considerations in genomic analyses. Nat. Rev. Genet. 2014, 15, 121–132. [Google Scholar] [CrossRef]
- Kuhn, J.H.; Bao, Y.; Bavari, S.; Becker, S.; Bradfute, S.; Brister, J.R.; Bukreyev, A.A.; Chandran, K.; Davey, R.A.; Dolnik, O.; et al. Virus nomenclature below the species level: A standardized nomenclature for natural variants of viruses assigned to the family Filoviridae. Arch. Virol. 2013, 158, 301–311. [Google Scholar] [CrossRef]
- Takeda, A.; Nakamura, W.; Sasaki, N.; Goto, K.; Kaido, M.; Okuno, T.; Mise, K. Natural isolates of Brome mosaic virus with the ability to move from cell to cell independently of coat protein. J. Gen. Virol. 2005, 86, 1201–1211. [Google Scholar] [CrossRef]
- Bujarski, J.J.; Kaesberg, P. Genetic recombination between RNA components of a multipartite plant virus. Nature 1986, 321, 528–531. [Google Scholar] [CrossRef] [PubMed]
- Dzianott, A.; Rauffer-Bruyere, N.; Bujarski, J.J. Studies on Functional Interaction between Brome Mosaic Virus Replicase Proteins during RNA Recombination, Using Combined Mutants in Vivo and in Vitro. Virology 2001, 289, 137–149. [Google Scholar] [CrossRef] [Green Version]
- Kolondam, B.; Rao, P.; Sztuba-Solinska, J.; Weber, P.H.; Dzianott, A.; Johns, M.A.; Bujarski, J.J. Co-infection with two strains of Brome mosaic bromovirus reveals common RNA recombination sites in different hosts. Virus Evol. 2015, 1, vev021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shahgolzari, M.; Pazhouhandeh, M.; Milani, M.; Khosroushahi, A.Y.; Fiering, S. Plant viral nanoparticles for packaging and in vivo delivery of bioactive cargos. WIREs Nanomed. Nanobiotechnol. 2020, 12, e1629. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; de Ruiter, M.V.; Maassen, S.J.; Cornelissen, J.J.L.M. Nanoreactors via Encapsulation of Catalytic Gold Nanoparticles within Cowpea Chlorotic Mottle Virus Protein Cages. In Protein Scaffolds 2018; Humana Press: New York, NY, USA, 2018; Volume 1798, pp. 1–9. [Google Scholar] [CrossRef]
- Vaughan, R.; Tragesser, B.; Ni, P.; Ma, X.; Dragnea, B.; Kao, C.C. The Tripartite Virions of the Brome Mosaic Virus Have Distinct Physical Properties That Affect the Timing of the Infection Process. J. Virol. 2014, 88, 6483–6491. [Google Scholar] [CrossRef] [Green Version]
- Routh, A.; Domitrovic, T.; Johnson, J.E. Host RNAs, including transposons, are encapsidated by a eukaryotic single-stranded RNA virus. Proc. Natl. Acad. Sci. USA 2012, 109, 1907–1912. [Google Scholar] [CrossRef] [Green Version]
- Gochhait, S.; Malhotra, D.; Rai, E.; Bamezai, R.N.K. Automated Fluoroscence Sequencing and Troubleshooting. In Advanced Techniques in Soil Microbiology 2007; Springer: Berlin/Heidelberg, Germany, 2007; pp. 35–51. [Google Scholar] [CrossRef]
- Zhu, B. Bacteriophage T7 DNA polymerase–sequenase. Front. Microbiol. 2014, 5, 181. [Google Scholar] [CrossRef] [Green Version]
- Cox, M.P.; Peterson, D.A.; Biggs, P.J. SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinform. 2010, 11, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Jaganathan, D.; Bohra, A.; Thudi, M.; Varshney, R.K. Fine mapping and gene cloning in the post-NGS era: Advances and prospects. Theor. Appl. Genet. 2020, 133, 1791–1810. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Yoon, A.; Lee, S.; Kim, S.; Han, J.; Chung, J. Next-generation sequencing enables the discovery of more diverse positive clones from a phage-displayed antibody library. Exp. Mol. Med. 2017, 49, e308. [Google Scholar] [CrossRef] [Green Version]
- Hunt, M.; Gall, A.; Ong, S.H.; Brener, J.; Ferns, B.; Goulder, P.; Nastouli, E.; Keane, J.A.; Kellam, P.; Otto, T.D. IVA: Accurate de novo assembly of RNA virus genomes. Bioinformatics 2015, 31, 2374–2376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shrestha, N.; Duvall, M.R.; Bujarski, J.J. Variability among the Isolates of Broad Bean Mottle Virus and Encapsidation of Host RNAs. Pathogens 2022, 11, 817. https://doi.org/10.3390/pathogens11070817
Shrestha N, Duvall MR, Bujarski JJ. Variability among the Isolates of Broad Bean Mottle Virus and Encapsidation of Host RNAs. Pathogens. 2022; 11(7):817. https://doi.org/10.3390/pathogens11070817
Chicago/Turabian StyleShrestha, Nipin, Melvin R. Duvall, and Jozef J. Bujarski. 2022. "Variability among the Isolates of Broad Bean Mottle Virus and Encapsidation of Host RNAs" Pathogens 11, no. 7: 817. https://doi.org/10.3390/pathogens11070817
APA StyleShrestha, N., Duvall, M. R., & Bujarski, J. J. (2022). Variability among the Isolates of Broad Bean Mottle Virus and Encapsidation of Host RNAs. Pathogens, 11(7), 817. https://doi.org/10.3390/pathogens11070817





