Improving Drug Sensitivity of HIV-1 Protease Inhibitors by Restriction of Cellular Efflux System in a Fission Yeast Model
Abstract
1. Introduction
2. Results
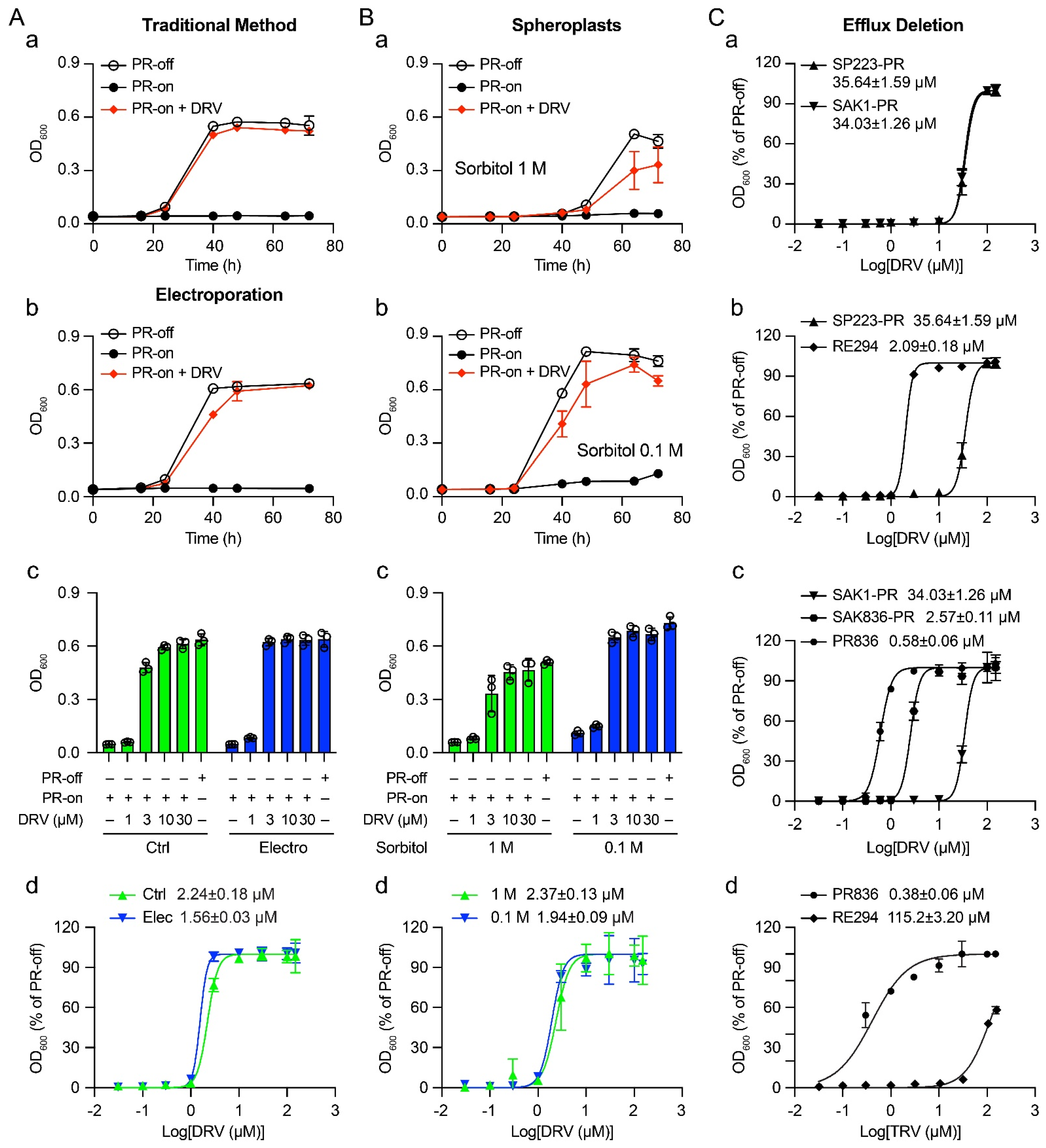
2.1. Determination of EC50 of DRV against HIV-1 PR by Adding Drug Directly to Cell Culture through Passive Diffusion
| Strains or Plasmids | Genotype and Characters | Source or Reference |
|---|---|---|
| Fission yeast strains | ||
| SP223 | Wild-type; h-, ade6-216, leu1-32, ura4-294 | Lab collection |
| RE294 | h-, ade6-216, leu1-32, ura4-294::nmt1-PR(NL4-3)-kanMX; a derivative of SP223 carrying a single integrated copy of HIV-1 PR gene under a nmt1 promoter at the ura4 locus | [9,28] |
| SAK1 | Wild-type, h90, ade6-M216, leu1-32, ura4-D18; a derivative of JY878 | [27] |
| SAK836 | Also known as MDR-supML; h90, ade6-M216, leu1-32, ura4-D18, caf5::bsd, ∆pap1, ∆pmd1, ∆mfs1, ∆bfr1, ∆dnf2, erg5::ura4+ | [27] |
| PR836 | h90, ade6-M216, leu1-32, ura4-D18::nmt1-PR (NL4-3)-ble1, caf5::bsd, ∆pap1, ∆pmd1, ∆mfs1, ∆bfr1, ∆dnf2, erg5::ura4+; a derivative of SAK836 carrying a single integrated copy of HIV-1 PR gene under a nmt1 promoter at the ura4 locus | This study |
| Plasmids | ||
| pYZ1N | A fission yeast expression vector with an inducible nmt1 promoter and a leu1 selectable marker | [32] |
| pYZ1N-PR | Wild-type HIV-1 PR gene cloned in pYZ1N | [28] |
| pJZBle1-PR | A modified pClone1Ble1 vector carrying ura4 flanking sequence and wild-type HIV-1 PR gene driven by a nmt1 promoter. This plasmid was used to create the PR836 strain. | This study |
2.2. Effect of Electroporation on the Drug Sensitivity of DRV
2.3. Determination of the Drug Sensitivity of DRV in Spheroplasts
2.4. Enhanced Drug Sensitivity of FDA-Approved PI Drugs in a Fission Yeast Mutant Strain That Is Defective in Efflux Pumps
3. Discussion
4. Materials and Methods
4.1. Fission Yeast Strains, Plasmids, and Growth Media
4.2. Generation of the Mutant PR836 Strain
4.3. Inducible Gene Expression of HIV-1 PR Gene in Fission Yeast
4.4. Fission Yeast Growth Assay
4.5. Determination of EC50 in Fission Yeast
4.6. Electroporation of SMDs in Fission Yeast
4.7. Transfer of SMDs into Spheroplasts of Fission Yeast
4.8. Small Molecule Drugs
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhao, Y.; Lieberman, H.B. Schizosaccharomyces pombe: A model for molecular studies of eukaryotic genes. DNA Cell Biol. 1995, 14, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Hohmann, S. Nobel Yeast Research. FEMS Yeast Res 2016, 16, fow094. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.Y. Yeast for virus research. Microb. Cell 2017, 4, 311–330. [Google Scholar] [CrossRef] [PubMed]
- Nkeze, J.; Li, L.; Benko, Z.; Li, G.; Zhao, R.Y. Molecular characterization of HIV-1 genome in fission yeast Schizosaccharomyces pombe. Cell Biosci. 2015, 5, 47. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Poulsen, M.; Fenyvuesvolgyi, C.; Yashiroda, Y.; Yoshida, M.; Simard, J.M.; Gallo, R.C.; Zhao, R.Y. Characterization of cytopathic factors through genome-wide analysis of the Zika viral proteins in fission yeast. Proc. Natl. Acad. Sci. USA 2017, 114, E376–E385. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Du, Z.; Zhang, Y.; Antal, J.; Xia, Z.; Wang, Y.; Gao, Y.; Zhao, X.; Han, X.; Cheng, Y.; et al. A distinct class of plant and animal viral proteins that disrupt mitosis by directly interrupting the mitotic entry switch Wee1-Cdc25-Cdk1. Sci. Adv. 2020, 6, eaba3418. [Google Scholar] [CrossRef]
- Zhang, J.; Li, Q.; Cruz Cosme, R.S.; Gerzanich, V.; Tang, Q.; Simard, J.M.; Zhao, R.Y. Genome-Wide Characterization of SARS-CoV-2 Cytopathogenic Proteins in the Search of Antiviral Targets. mBio 2022, 13, e0016922. [Google Scholar] [CrossRef]
- Benko, Z.; Elder, R.T.; Liang, D.; Zhao, R.Y. Fission yeast as a HTS platform for molecular probes of HIV-1 Vpr-induced cell death. Int. J. High Throughput Screen. 2010, 1, 151–162. [Google Scholar]
- Benko, Z.; Zhang, J.; Zhao, R.Y. Development of A Fission Yeast Cell-Based Platform for High Throughput Screening of HIV-1 Protease Inhibitors. Curr. HIV Res. 2019, 17, 429–440. [Google Scholar] [CrossRef]
- Rallis, C.; Bahler, J. Cell-based screens and phenomics with fission yeast. Crit. Rev. Biochem. Mol. Biol. 2016, 51, 86–95. [Google Scholar] [CrossRef]
- Zhang, J.; Vernon, K.; Li, Q.; Benko, Z.; Amoroso, A.; Nasr, M.; Zhao, R.Y. Single-Agent and Fixed-Dose Combination HIV-1 Protease Inhibitor Drugs in Fission Yeast (Schizosaccharomyces pombe). Pathogens 2021, 10, 804. [Google Scholar] [CrossRef] [PubMed]
- Cocucci, E.; Kim, J.Y.; Bai, Y.; Pabla, N. Role of Passive Diffusion, Transporters, and Membrane Trafficking-Mediated Processes in Cellular Drug Transport. Clin. Pharm. 2017, 101, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Perez, P.; Ribas, J.C. Fission Yeast Cell Wall Analysis. Cold Spring Harb. Protoc. 2017, 2017, pdb-top079897. [Google Scholar] [CrossRef] [PubMed]
- Perez, P.; Cortes, J.C.G.; Cansado, J.; Ribas, J.C. Fission yeast cell wall biosynthesis and cell integrity signalling. Cell Surf. 2018, 4, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Hopkins, K.M.; Lieberman, H.B. A method for the preparation and storage of frozen, competent Schizosaccharomyces pombe spheroplasts. Biotechniques 1993, 15, 238. [Google Scholar]
- Flor-Parra, I.; Zhurinsky, J.; Bernal, M.; Gallardo, P.; Daga, R.R. A Lallzyme MMX-based rapid method for fission yeast protoplast preparation. Yeast 2014, 31, 61–66. [Google Scholar] [CrossRef]
- Murray, J.M.; Watson, A.T.; Carr, A.M. Transformation of Schizosaccharomyces pombe: Protoplast Procedure. Cold Spring Harb. Protoc. 2016, 2016, pdb-prot090977. [Google Scholar] [CrossRef]
- Sipiczki, M.; Ferenczy, L. Protoplast fusion of Schizosaccharomyces pombe Auxotrophic mutants of identical mating-type. Mol. Gen. Genet. 1977, 151, 77–81. [Google Scholar] [CrossRef]
- Jimenez, J. Cryopreservation of competent Schizosaccharomyces pombe protoplasts. Trends Genet. 1991, 7, 40. [Google Scholar] [CrossRef]
- Gehl, J. Electroporation: Theory and methods, perspectives for drug delivery, gene therapy and research. Acta Physiol. Scand. 2003, 177, 437–447. [Google Scholar] [CrossRef]
- Luft, C.; Ketteler, R. Electroporation Knows No Boundaries: The Use of Electrostimulation for siRNA Delivery in Cells and Tissues. J. Biomol. Screen. 2015, 20, 932–942. [Google Scholar] [CrossRef]
- Suga, M.; Hatakeyama, T. High efficiency transformation of Schizosaccharomyces pombe pretreated with thiol compounds by electroporation. Yeast 2001, 18, 1015–1021. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.M.; Watson, A.T.; Carr, A.M. Transformation of Schizosaccharomyces pombe: Electroporation Procedure. Cold Spring Harb. Protoc. 2016, 2016, pdb-prot090951. [Google Scholar] [CrossRef] [PubMed]
- Iwaki, T.; Giga-Hama, Y.; Takegawa, K. A survey of all 11 ABC transporters in fission yeast: Two novel ABC transporters are required for red pigment accumulation in a Schizosaccharomyces pombe adenine biosynthetic mutant. Microbiology 2006, 152, 2309–2321. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Arita, Y.; Nishimura, S.; Matsuyama, A.; Yashiroda, Y.; Usui, T.; Boone, C.; Yoshida, M. Microarray-based target identification using drug hypersensitive fission yeast expressing ORFeome. Mol. Biosyst. 2011, 7, 1463–1472. [Google Scholar] [CrossRef]
- Kawashima, S.A.; Takemoto, A.; Nurse, P.; Kapoor, T.M. Analyzing fission yeast multidrug resistance mechanisms to develop a genetically tractable model system for chemical biology. Chem. Biol. 2012, 19, 893–901. [Google Scholar] [CrossRef]
- Aoi, Y.; Sato, M.; Sutani, T.; Shirahige, K.; Kapoor, T.M.; Kawashima, S.A. Dissecting the first and the second meiotic divisions using a marker-less drug-hypersensitive fission yeast. Cell Cycle 2014, 13, 1327–1334. [Google Scholar] [CrossRef]
- Benko, Z.; Elder, R.T.; Li, G.; Liang, D.; Zhao, R.Y. HIV-1 Protease in the Fission Yeast Schizosaccharomyces pombe. PLoS ONE 2016, 11, e0151286. [Google Scholar] [CrossRef]
- Benko, Z.; Liang, D.; Li, G.; Elder, R.T.; Sarkar, A.; Takayama, J.; Ghosh, A.K.; Zhao, R.Y. A fission yeast cell-based system for multidrug resistant HIV-1 proteases. Cell Biosci. 2017, 7, 5. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Anderson, D.D.; Weber, I.T.; Mitsuya, H. Enhancing protein backbone binding--a fruitful concept for combating drug-resistant HIV. Angew. Chem. Int. Ed. Engl. 2012, 51, 1778–1802. [Google Scholar] [CrossRef]
- Maundrell, K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 1993, 123, 127–130. [Google Scholar] [CrossRef]
- Zhao, Y.; Elder, R.T.; Chen, M.; Cao, J. Fission yeast expression vectors adapted for positive identification of gene insertion and green fluorescent protein fusion. Biotechniques 1998, 25, 438–444. [Google Scholar] [CrossRef] [PubMed]
- Neumann, E.; Kakorin, S.; Toensing, K. Fundamentals of electroporative delivery of drugs and genes. Bioelectrochem. Bioenerg. 1999, 48, 3–16. [Google Scholar] [CrossRef]
- Diamond, R.J.; Rose, A.H. Osmotic properties of spheroplasts from Saccharomyces cerevisiae grown at different temperatures. J. Bacteriol. 1970, 102, 311–319. [Google Scholar] [CrossRef]
- Brunton, L.L.; Lazo, J.S.; Parker, K.L. Goodman and Gilman’s The Pharmacological Basis of Therapeutics, 11th ed.; McGraw-Hill: New York, NY, USA, 2006. [Google Scholar]
- Ghosh, A.K.; Osswald, H.L.; Prato, G. Recent Progress in the Development of HIV-1 Protease Inhibitors for the Treatment of HIV/AIDS. J. Med. Chem. 2016, 16, 5172–5208. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Reddy, G.S.; Nalam, M.N.; Anjum, S.G.; Cao, H.; Schiffer, C.A.; Rana, T.M. Structure-based design, synthesis, and structure-activity relationship studies of HIV-1 protease inhibitors incorporating phenyloxazolidinones. J. Med. Chem. 2010, 53, 7699–7708. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, L.; Jiang, W.; Lu, Y. A Modified Gibson Assembly Method for Cloning Large DNA Fragments with High GC Contents. Methods Mol. Biol. 2018, 1671, 203–209. [Google Scholar] [CrossRef]
- Grimm, C.; Kohli, J.; Murray, J.; Maundrell, K. Genetic engineering of Schizosaccharomyces pombe: A system for gene disruption and replacement using the ura4 gene as a selectable marker. Mol. Gen. Genet. 1988, 215, 81–86. [Google Scholar] [CrossRef]
- Basi, G.; Schmid, E.; Maundrell, K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 1993, 123, 131–136. [Google Scholar] [CrossRef]
- Zhao, Y.; Cao, J.; O’Gorman, M.R.; Yu, M.; Yogev, R. Effect of human immunodeficiency virus type 1 protein R (vpr) gene expression on basic cellular function of fission yeast Schizosaccharomyces pombe. J. Virol. 1996, 70, 5821–5826. [Google Scholar] [CrossRef]
- Aoki, M.; Das, D.; Hayashi, H.; Aoki-Ogata, H.; Takamatsu, Y.; Ghosh, A.K.; Mitsuya, H. Mechanism of Darunavir (DRV)’s High Genetic Barrier to HIV-1 Resistance: A Key V32I Substitution in Protease Rarely Occurs, but Once It Occurs, It Predisposes HIV-1 To Develop DRV Resistance. mBio 2018, 9, e02425-17. [Google Scholar] [CrossRef] [PubMed]
| Drug Name | Brand Name | Molecular Weight (g/mol) * | EC50 in RE294 (μM) | EC50 in PR836 (μM) | Difference in EC50 (Fold) | Difference in EC50 (log10) |
|---|---|---|---|---|---|---|
| Ritonavir (RTV) | Norvir | 720.94 | 115.2 ± 3.20 | 0.38 ± 0.06 | 303.2 | 2.48 |
| Amprenavir (APV) | Agenerase | 505.63 | 55.41 ± 3.18 | 0.35 ± 0.06 | 163.0 | 2.21 |
| Lopinavir (LPV) | Kaletra | 628.81 | 4.59 ± 0.06 | 0.10 ± 0.01 | 45.9 | 1.66 |
| Indinavir (IDV) | Crixivan | 613.79 | 48.64 ± 2.51 | 1.52 ± 0.16 | 32.2 | 1.51 |
| Atazanavir (ATV) | Reyataz | 704.86 | 21.08 ± 0.07 | 0.75 ± 0.02 | 28.1 | 1.45 |
| Saquinavir (SQV) | Invirase | 670.84 | 9.88 ± 0.25 | 0.74 ± 0.07 | 13.4 | 1.13 |
| Nelfinavir (NFV) | Viracept | 567.78 | 9.7 ± 0.34 | 0.75 ± 0.03 | 12.9 | 1.11 |
| Tipranavir (TPV) | Aptivus | 602.66 | 7.86 ± 0.32 | 0.74 ± 0.07 | 11.2 | 1.05 |
| Darunavir (DRV) | Prezista | 547.66 | 2.09 ± 0.18 | 0.58 ± 0.06 | 3.6 | 0.56 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Li, Q.; Kawashima, S.A.; Nasr, M.; Xue, F.; Zhao, R.Y. Improving Drug Sensitivity of HIV-1 Protease Inhibitors by Restriction of Cellular Efflux System in a Fission Yeast Model. Pathogens 2022, 11, 804. https://doi.org/10.3390/pathogens11070804
Zhang J, Li Q, Kawashima SA, Nasr M, Xue F, Zhao RY. Improving Drug Sensitivity of HIV-1 Protease Inhibitors by Restriction of Cellular Efflux System in a Fission Yeast Model. Pathogens. 2022; 11(7):804. https://doi.org/10.3390/pathogens11070804
Chicago/Turabian StyleZhang, Jiantao, Qi Li, Shigehiro A. Kawashima, Mohamed Nasr, Fengtian Xue, and Richard Y. Zhao. 2022. "Improving Drug Sensitivity of HIV-1 Protease Inhibitors by Restriction of Cellular Efflux System in a Fission Yeast Model" Pathogens 11, no. 7: 804. https://doi.org/10.3390/pathogens11070804
APA StyleZhang, J., Li, Q., Kawashima, S. A., Nasr, M., Xue, F., & Zhao, R. Y. (2022). Improving Drug Sensitivity of HIV-1 Protease Inhibitors by Restriction of Cellular Efflux System in a Fission Yeast Model. Pathogens, 11(7), 804. https://doi.org/10.3390/pathogens11070804







