A Review on Mycobacteriophages: From Classification to Applications
Abstract
1. Introduction
2. An Explanation of Discovered Mycobacteriophages Infective to M. smegmatis, M. tuberculosis, M. bovis, M. avium Subspecies, and M. abscessus
| Mycobacteriophage/Family | Description | Cluster/Sub Cluster | Origin | CG% Content | Infect | Life Cycle | Completely Sequenced |
|---|---|---|---|---|---|---|---|
| Bxz1/Myoviridae [97] | Generalized transduction, Bxz1-specific tRNA [27] | C [27] | Soil [26] | 64.8 [27] | M. smegmatis mc2155, M. vaccae [26] | Lytic (Clear plaques) [26] | Yes [27] |
| L5/Siphoviridae [97] | Superinfection-stable lysogens, transformation of slow-growing mycobacteria, immobilized tail protein (Gp6) [40,41,42], three tRNA genes [27] | A/A2 [40,41,42] | Isolated from lysogenic strain of M. smegmatis [98] | 63.2 [98] | M. smegmatis mc2155, M. tuberculosis [44] | Temperate [99] | Yes [98] |
| PDRPv/Siphoviridae [100] | Antimicrobial profiles [100], circular permuted dsDNA [100] | B/B1 | Soil | 66 [100] | M. smegmatis mc2155, M. tuberculosis | Lytic [100] | No |
| D29/ Siphoviridae [49] | Lytic activity, inactivation by M. smegmatis extracted mycoside C [34,35,36,37], adsorption [31,32], Lysin B [56] | A/A2 [40] | Soil [101] | 63.6 [102] | M. smegmatis mc2155, M. tuberculosis [83], MAP, M. bovis, M. fortitum [88] | Lytic [83] | Yes [83] |
| BPs/Siphoviridae [57] | Ultra-small genetic elements | G [57,103] | Soil | 66.6 [57] | M. smegmatis mc2155 [57], M. tuberculosis [57] | Temperate [57] | Yes [57] |
| Angel/Siphoviridae [57] | Ultra-small genetic elements | G [57,103] | Soil | 66.6 [57] | M. smegmatis mc2155 [57], M. tuberculosis [57] | Temperate [57] | Yes [57] |
| Halo/Siphoviridae [57] | Ultra-small genetic elements | G [57,103] | Soil | 66.7 [61] | M. smegmatis mc2155 [57], M. tuberculosis [57] | Temperate [57] | Yes [57] |
| ZoeJ/Siphoviridae [59] | Superinfection immunity [59] | K/K2 [59] | Soil [104] | Unpublished | M. smegmatis mc2155, M. tuberculosis, M. avium, M. bovis [59] | Temperate [59] | Yes [59] |
| TM4/Siphoviridae [73] | Genetic tools [62,72], diagnostic application, unusual lysogenic pattern, production of proteins similar to transcriptional regulators, generation shuttle plasmid [62] | K/K2 [59] | Unknown | 68.1 [27] | M. smegmatis mc2155 [62], M. tuberculosis H37Rv [59,62], M. avium, MAP | Temperate | Yes [27] |
| FRAT1/Unknown | Integrase gene [105]; carries Kanamycin resistance gene; therapeutic tools [60] | Unknown | Unknown | Unknown | M. smegmatis ATCC607, M. bovis BCG 1173/P2 [60] | Temperate [105] | No |
| D32/Siphoviridae [106] | Lytic activity against M. tuberculosis | Unpublished | Soil | 64 [107] | M. tuberculosis H37Rv [108], M. smegmatis ATCC607, M. smegmatis mc2155 [106] | Lytic [101] | Yes [106] |
| Bo4/Siphoviridae [109] | Lytic activity, active in bloodstream and lysosomal macrophages [74] | G [74] | Unknown | 66.76 [74] | M. smegmatis CMCC93202, M. tuberculosis H37Rv [74] | Lytic [74] | Yes [74] |
| 33D/Siphoviridae [109] | Lytic activity, therapeutic purposes [75] | Unknown | Unknown | M. tuberculosis H37Rv and M. bovis (TMC410) [75] | Lytic [75] | No | |
| SWU1/Siphoviridae [109] | Lytic activity, modification of cell signaling, bull’s eye morphology [42] | A2 [42] | Soil [110] | 62.4 [110] | M. smegmatis mc2155 [42], M. tuberculosis [79]. | Lytic [80] | Yes [80] |
| Che12/Siphoviridae [111] | Diagnosis of tuberculosis [82] | A/A2 [61] | Soil [82] | 62.9 [97] | M. tuberculosis H37Rv [112] and M. smegmatis mc2155 [82] | Temperate [82] | Yes [97] |
| DS6A/Siphoviridae [109] | Formation of plaque only on MTBC, loss of acid fastness, generation of shuttle plasmid [62] | Singleton [3] | Unknown | 68.4 [63] | M. tuberculosis H37Rv [113], M. tuberculosis complex [63] | Temperate | Yes [114] |
| CRB2/Siphoviridae [76] | Non-transducing profile; ORFs in its genome have a probable function [76] | B/B9 [76] | Unknown | 69.78 [76] | M. smegmatis mc2155 and M. tuberculosis | Lytic [76] | Yes [76] |
| vB_MapS_FF47/Siphoviridae [5] | Lytic activity, no virulent or temperate genes [5] | Unpublished | Bovine feces [5] | 58.6 [5] | MAP ATCC19698 [5] and M. smegmatis mc2155 [5] | Lytic [5] | Yes [5] |
| AN3/Siphoviridae [115] | Used for typing of M. avium intracellular scrofulaceum [116] | Unpublished | Unknown | Unpublished | M. smegmatis mc2155 [115] and M. avium intracellular scrofulaceum [116] | Unpublished | Yes [115] |
| AN9/Siphoviridae [117] | Used for typing of M. avium intracellular scrofulaceum [116] | Unpublished | Unknown | Unpublished | M. smegmatis mc2155 [117] and M. avium-intracellulare-scrofulaceum complex [116,118] | Unpublished | Yes [117] |
| ANI8/Siphoviridae [119] | Used for phage typing of M. avium intracellular scrofulaceum (MAIS) [118] | Unpublished | Unknown | Unpublished | M. smegmatis mc2155 [119] and M. avium intracellular scrofulaceum (MAIS) [118] | Unpublished | Yes [119] |
| VA6/Siphoviridae [120] | Used for typing of M. avium intracellular scrofulaceum [116] | Unpublished | Unknown | Unpublished | M. smegmatis mc2155 [120] and M. avium intracellular scrofulaceum [116] | Unpublished | Yes [120] |
| VC3/Siphoviridae [121] | Used for typing of M. avium intracellular scrofulaceum [116] | Unpublished | Unknown | Unpublished | M. smegmatis mc2155 [121] and M. avium intracellular scrofulaceum [116] | Unpublished | Yes [121] |
| Muddy/Siphoviridae [122] | Lytic activity [123] | AB [123] | Soil [122] | Unpublished | M. smegmatis mc2155 [16], M. abscessus (GD01) [16] | Lytic [123] | Yes [122] |
| Araucaria/Siphoviridae [90] | Infection via adhesion to cell wall saccharide and protein [90] | Dori-like [90] | Respiratory tract sample [90] | 64.41 [90] | M. abscessus subsp. bolletii CIP108541 [90] | Temperate [90] | Yes [90] |
| Prophage phiT46-1/Siphoviridae [95] | Polymorphic toxin-immunity cassette [95] | Unpublished | It was isolated by spontaneous release from M. abscessus strain Taiwan-46 [95] | 64 [95] | M. abscessus strain BWH-C [95] | Temperate [95] | Yes [95] |
| Prophage phT45/Siphoviridae [124] | Polymorphic toxin-immunity cassette associated with type VII secretion systems [124] | Unpublished | It was isolated by spontaneous release from M. abscessus strain Taiwan-45 [124] | 65 [124] | M. abscessus strain BWH-C [124] | Lytic [124] | Yes [124] |
| Adler [125] | Genes encoding cytochrome P450 (heme protein) catalyze monooxygenase activity [125] | Unpublished | Unknown | Unknown | M. abscessus subspecies bolletii F1660 [125] | Unknown | No |
| Chancellor/Siphoviridae [126] | Virion structure and assembly genes, lytic activity, Lysin A, Lysin B, holin genes, ability to infect M. tuberculosis [126] | K/K4 [126] | Soil [126] | 68 [126] | M. smegmatis mc2155 [126], predicted to infect M. tuberculosis [126] | Temperate [126] | Yes [126] |
| Mitti/Siphoviridae [126] | Virion structure and assembly genes, lytic activity, Lysin A, Lysin B, holin genes, ability to infect M. tuberculosis [126] | K/K4 [126] | Soil [126] | 68 [126] | M. smegmatis mc2155 [126], predicted to infect M. tuberculosis [126] | Temperate [126] | Yes [126] |
| Wintermute/Siphoviridae [126] | Virion structure and assembly genes, lytic activity, Lysin A, Lysin B, holin genes, ability to infect M. tuberculosis [126] | K/K4 [126] | Soil [126] | 68 [126] | M. smegmatis mc2155 [126], predicted to infect M. tuberculosis [126] | Temperate [126] | Yes [126] |
| ShedlockHolmes /Siphoviridae [127] | Ability to infect M. tuberculosis, having tRNA [127] | K/K3 [127] | Soil [127] | 67.3 [127] | M. smegmatis MC2155 [127], predicted to infect M. tuberculosis [127] | Temperate [127] | Yes [127] |
| Deby/Siphoviridae [128] | Ability to infect M. tuberculosis, structural and assembly genes, having tRNA [128] | K/K1 [128] | Soil [128] | 66.5 [128] | M. smegmatis mc2155 [128], predicted to infect M. tuberculosis [128] | Temperate [128] | Yes [128] |
| LaterM/Siphoviridae [128] | Ability to infect M. tuberculosis, structural and assembly genes, having tRNA [128] | K/K1 [128] | Soil [128] | 66.5 [128] | M. smegmatis mc2155 [128], predicted to infect M. tuberculosis [128] | Temperate [128] | Yes [128] |
| LilPharaoh/Siphoviridae [128] | Ability to infect M. tuberculosis, structural and assembly genes, having tRNA [128] | K/K1 [128] | Soil [128] | 67.1 [128] | M. smegmatis mc2155 [128], predicted to infect M. tuberculosis [128] | Temperate [128] | Yes [128] |
| SgBeansprout/Siphoviridae [128] | Ability to infect M. tuberculosis, structural and assembly genes, having tRNA [128] | K/K1 [128] | Soil [128] | 67.1 [128] | M. smegmatis mc2155 [128], predicted to infect M. tuberculosis [128] | Temperate [128] | Yes [128] |
| Sulley/Siphoviridae [128] | Ability to infect M. tuberculosis, structural and assembly genes, having tRNA [128] | K/K1 [128] | Soil [128] | 66.4 [128] | M. smegmatis mc2155 [128], predicted to infect M. tuberculosis [128] | Temperate [128] | Yes [128] |
| Paola/Siphoviridae [128] | Ability to infect M. tuberculosis, structural and assembly genes, having tRNA [128] | K/K5 [128] | Soil [128] | 65 [128] | M. smegmatis mc2155, predicted to infect M. tuberculosis [128] | Temperate [128] | Yes [128] |
| Joy99/Siphoviridae [129] | Ability to infect M. tuberculosis; its genome contains genes that contribute in virion assembly/structure/lysis proteins/host integration and excision proteins/DNA primase/RusA-like resolvase/RtcB-like integrase genes; having tRNA [129] | K/K1 [129] | Soil [129] | 66.6 [129] | M. smegmatis mc2155 [129], predicted to infect M. tuberculosis [129] | Unpublished, three-ring morphology with clear center spot, thin middle ring, and turbid outer ring [129] | Yes [129] |
| 20ES/Siphoviridae [130] | Capability to infect M. tuberculosis, presence of ParA and ParB genes in its genome [77] | A [77] | Soil [130] | 63.43 [77] | M. smegmatis mc2155 [130], able to infect M. tuberculosis H37Rv and M. bovis var BCG [77] | Temperate [77] | Yes [130] |
| Kerberos/Siphoviridae [131] | Capability to infect M. tuberculosis; presence of virion structure/assembly/nonstructural genes in its genome; having tRNA [131] | A/A2 [131] | Soil [131] | 63.5 [131] | M. smegmatis mc2155 [131] | Temperate [131] | Yes [131] |
| Pomar16/Siphoviridae [131] | Capability to infect M. tuberculosis; virion structure/assembly/nonstructural genes in its genome; having tRNA [131] | A/A2 [131] | Soil [131] | 63.5 [131] | M. smegmatis mc2155 [131] | Temperate [131] | Yes [131] |
| StarStuff/Siphoviridae [131] | Capability to infect M. tuberculosis; virion structure/assembly/nonstructural genes in its genome; having tRNA [131] | A/A2 [131] | Soil [131] | 63.5 [131] | M. smegmatis mc2155 [131] | Temperate [131] | Yes [131] |
| Omega/Siphoviridae [132] | Lack of DNA ligase gene [133]; possible role in mycobacterial virulence as the phage encodes gene 61 that is a close homolog of tuberculosis Lsr2; may play role in humoral and cellular immune responses [134]. | J [61] | Unknown | 61.4 [61] | Mycobacterium sp. [132] | It is possibly temperate because it forms slightly turbid plaques, and stable lysogens could be recovered [135] | Yes [132] |
| Cjw1/Siphoviridae [136] | A possible role in mycobacterial virulence as the phage encodes gene 39, which is a close homolog of leprosy Lsr2; may play a role in humoral and cellular immune responses [137]. | E [138] | Unknown | 63.7 [61] | Mycobacterium sp. [136] | It is possibly temperate because it produces hazy to turbid plaques at 37 and 42 °C, respectively [139] | Yes [136] |
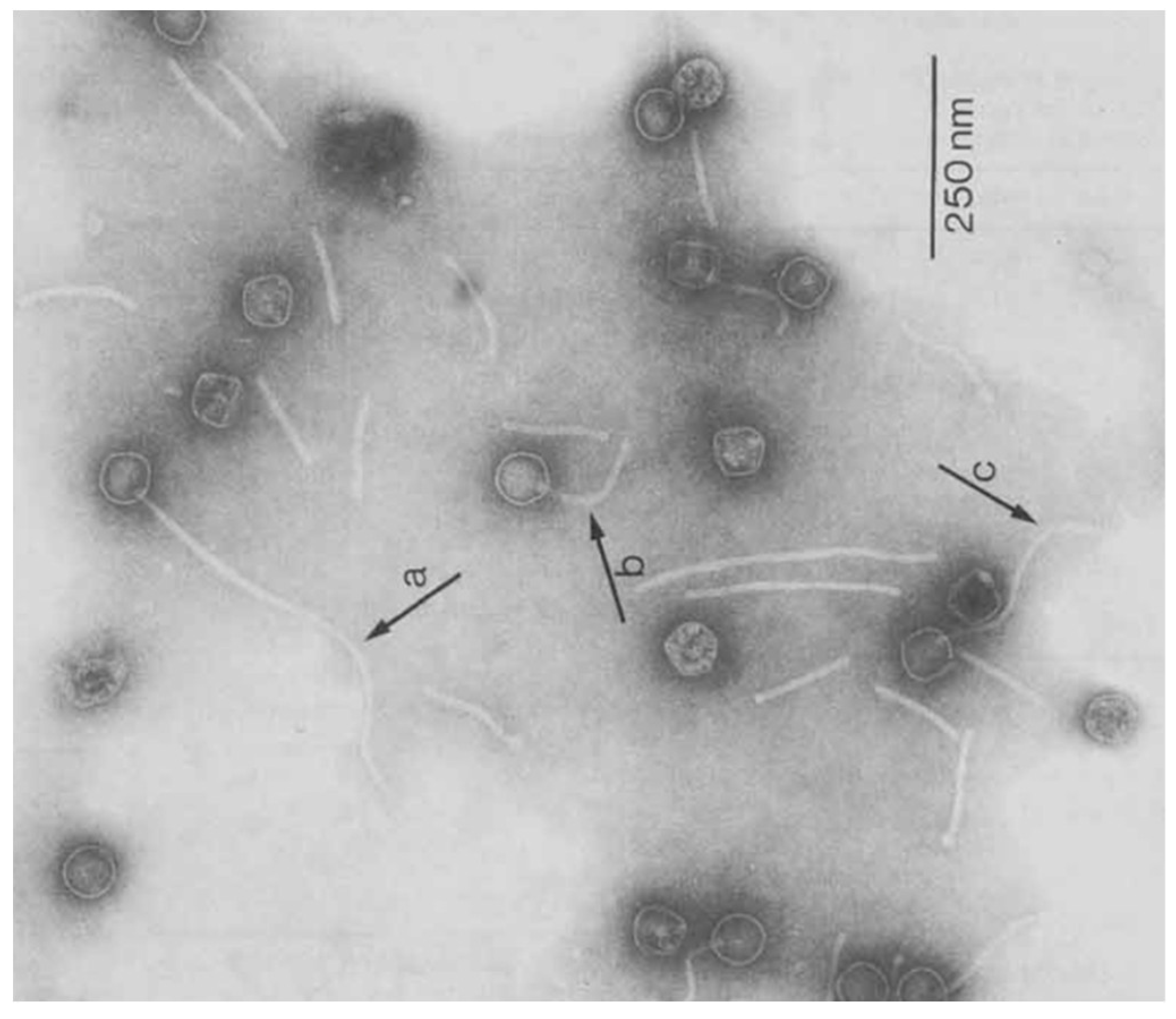
3. Morphology
4. Classification
5. Life Cycle of Mycobacteriophages
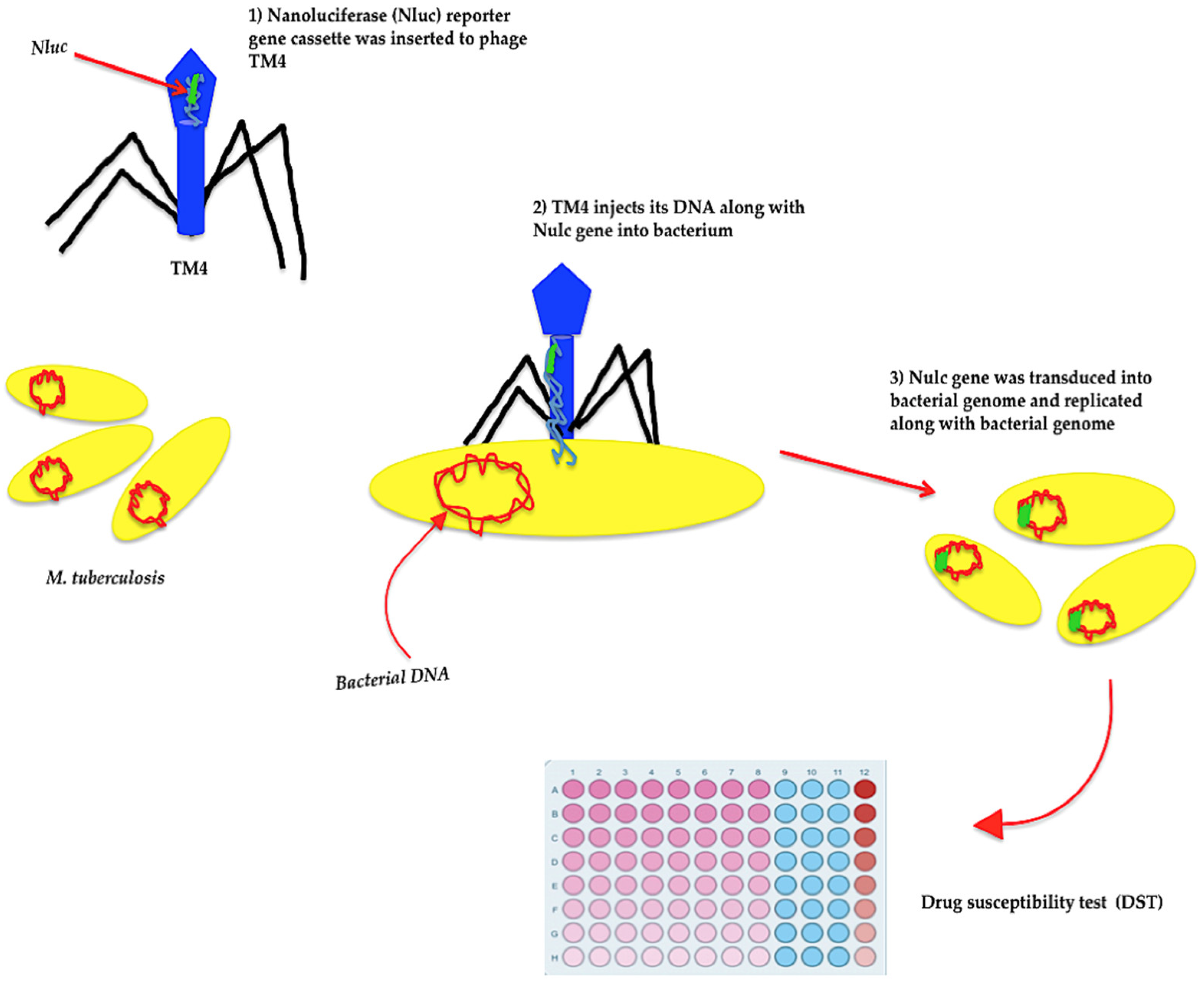
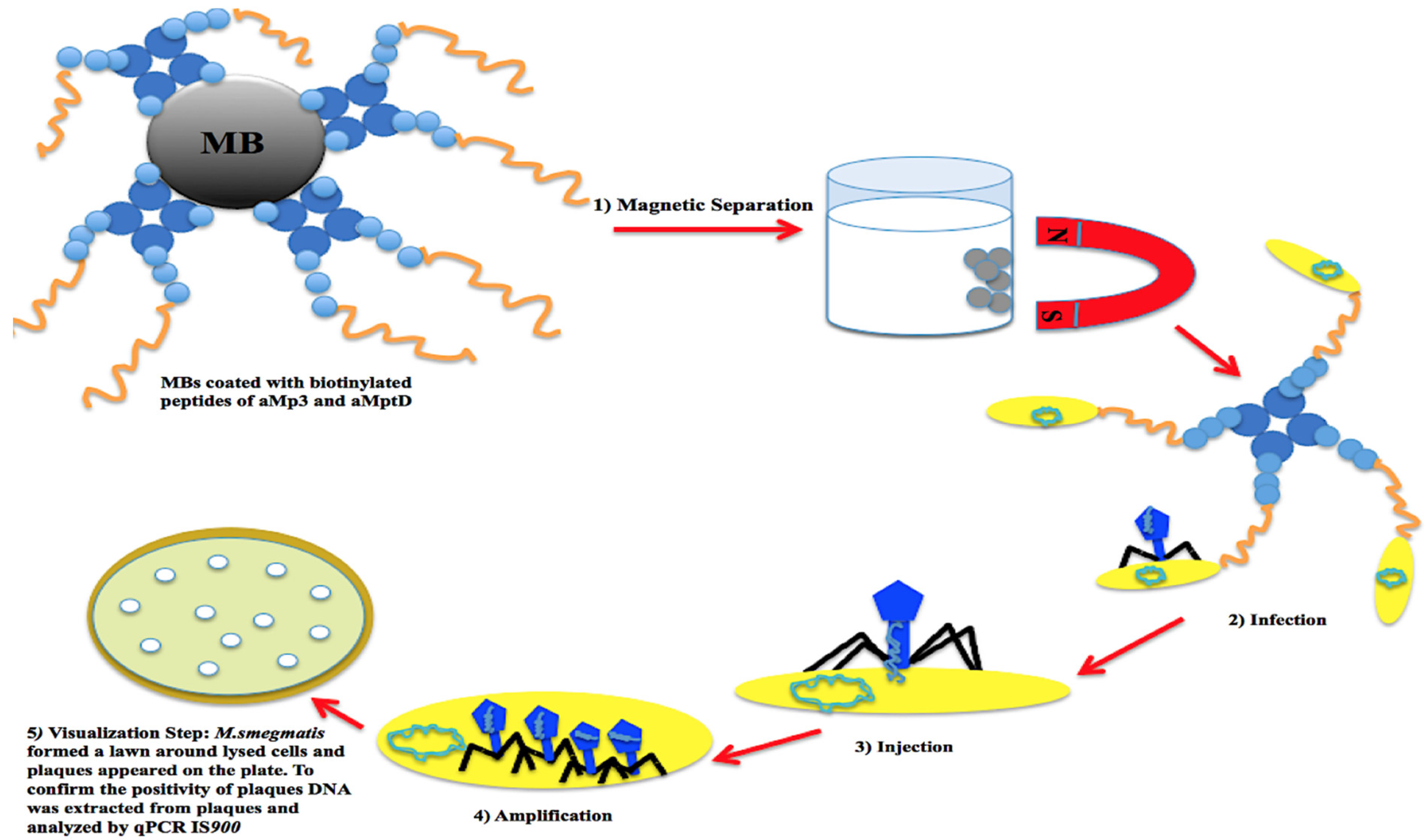
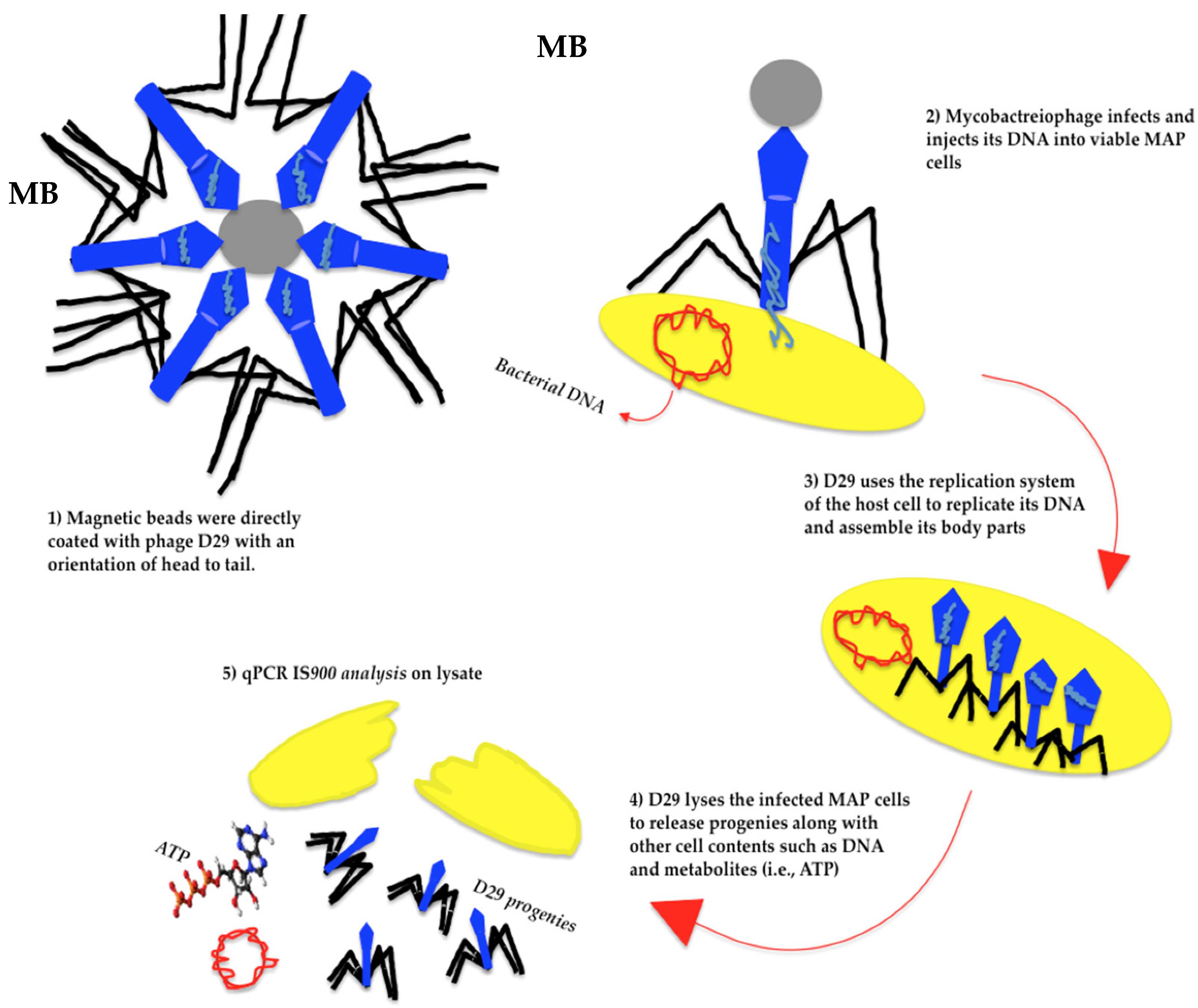
6. Mycobacteriophages and Detection of Pathogenic Mycobacteria
7. Mycobacteriophages and Treatment of Mycobacterial Infections
8. Phage Resistance
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Crane, A.; Versoza, C.J.; Hua, T.; Kapoor, R.; Lloyd, L.; Mehta, R.; Menolascino, J.; Morais, A.; Munig, S.; Patel, Z.; et al. Phylogenetic relationships and codon usage bias amongst cluster K mycobacteriophages. G3 Genes|Genomes|Genet. 2021, 11, jkab291. [Google Scholar] [CrossRef] [PubMed]
- Hatfull, G.F. Actinobacteriophages: Genomics, dynamics, and applications. Annu. Rev. Virol. 2020, 7, 37–61. [Google Scholar] [CrossRef] [PubMed]
- Hatfull, G.F. Mycobacteriophages: Windows into tuberculosis. PLoS Pathog. 2014, 10, e1003953. [Google Scholar] [CrossRef]
- Anes, E.; Portugal, I.; Moniz-Pereira, J. Insertion into the Mycobacterium smegmatis genome of the aph gene through lysogenization with the temperate mycobacteriophage Ms6. FEMS Microbiol. Lett. 1992, 95, 21–25. [Google Scholar] [CrossRef]
- Basra, S.; Anany, H.; Brovko, L.; Kropinski, A.M.; Griffiths, M.W. Isolation and characterization of a novel bacteriophage against Mycobacterium avium subspecies paratuberculosis. Arch. Virol. 2014, 159, 2659–2674. [Google Scholar] [CrossRef] [PubMed]
- Ghazaei, C. Mycobacterium tuberculosis and lipids: Insights into molecular mechanisms from persistence to virulence. J. Res. Med. Sci. 2018, 23, 63. [Google Scholar] [CrossRef]
- Fu, X.; Ding, M.; Zhang, N.; Li, J. Mycobacteriophages: An important tool for the diagnosis of Mycobacterium tuberculosis (Review). Mol. Med. Rep. 2015, 12, 13–19. [Google Scholar] [CrossRef]
- Hatfull, G.F. Mycobacteriophages. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Winthrop, K.L.; Roy, E.E. Mycobacteria and Immunosuppression. In Infections in Systemic Autoimmune Diseases: Risk Factors and Management; Atzeni, F., Galloway, J.B., Gomez-Reino, J.J., Galli, M., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; Volume 16, pp. 83–107. ISBN 1571-5078. [Google Scholar]
- De Lisle, G.W. Mycobacterial infections of animals. Pathology 1991, 23, 12. [Google Scholar] [CrossRef]
- Prasad, R.; Gupta, N.; Banka, A. Multidrug-resistant tuberculosis/rifampicin-resistant tuberculosis: Principles of management. Lung India 2018, 35, 78–81. [Google Scholar] [CrossRef]
- Lange, C.; Chesov, D.; Heyckendorf, J.; Leung, C.C.; Udwadia, Z.; Dheda, K. Drug-resistant tuberculosis: An update on disease burden, diagnosis and treatment. Respirology 2018, 23, 656–673. [Google Scholar] [CrossRef] [PubMed]
- Jing, H.; Wang, H.; Wang, Y.; Deng, Y.; Li, X.; Liu, Z.; Graviss, E.; Ma, X. Prevalence of nontuberculous mycobacteria infection, China, 2004–2009. Emerg. Infect. Dis. J. 2012, 18, 527. [Google Scholar] [CrossRef] [PubMed]
- Gygli, S.M.; Borrell, S.; Trauner, A.; Gagneux, S. Antimicrobial resistance in Mycobacterium tuberculosis: Mechanistic and evolutionary perspectives. FEMS Microbiol. Rev. 2017, 41, 354–373. [Google Scholar] [CrossRef] [PubMed]
- Kalapala, Y.C.; Sharma, P.R.; Agarwal, R. Antimycobacterial potential of mycobacteriophage under disease-mimicking conditions. Front. Microbiol. 2020, 11, 583661. [Google Scholar] [CrossRef] [PubMed]
- Johansen, M.D.; Alcaraz, M.; Dedrick, R.M.; Roquet-Banères, F.; Hamela, C.; Hatfull, G.F.; Kremer, L. Mycobacteriophage-antibiotic therapy promotes enhanced clearance of drug-resistant Mycobacterium abscessus. Dis. Model. Mech. 2021, 14, dmm049159. [Google Scholar] [CrossRef]
- Senhaji-Kacha, A.; Esteban, J.; Garcia-Quintanilla, M. Considerations for phage therapy against Mycobacterium abscessus. Front. Microbiol. 2021, 11, 609017. [Google Scholar] [CrossRef]
- Li, Q.; Zhou, M.; Fan, X.; Yan, J.; Li, W.; Xie, J. Mycobacteriophage SWU1 gp39 can potentiate multiple antibiotics against Mycobacterium via altering the cell wall permeability. Sci. Rep. 2016, 6, 28701. [Google Scholar] [CrossRef]
- Snapper, S.B.; Lugosi, L.; Jekkel, A.; Melton, R.E.; Kieser, T.; Bloom, B.R.; Jacobs, W.R. Lysogeny and transformation in mycobacteria: Stable expression of foreign genes. Proc. Natl. Acad. Sci. USA 1988, 85, 6987–6991. [Google Scholar] [CrossRef]
- Jain, P.; Garing, S.; Verma, D.; Saranathan, R.; Clute-Reinig, N.; Gadwa, J.; Peterson, C.; Hermansky, G.; Astashkina Fernandez, A.; Asare, E.; et al. Nanoluciferase reporter mycobacteriophage for sensitive and rapid detection of Mycobacterium tuberculosis drug susceptibility. J. Bacteriol. 2020, 202, e00411-20. [Google Scholar] [CrossRef]
- Tokunaga, T.; Sellers, M.I. Streptomycin induction of premature lysis of bacteriophage-infected mycobacteria. J. Bacteriol. 1965, 89, 537–538. [Google Scholar] [CrossRef]
- Gupta, A.; Bhakta, S.; Kundu, S.; Gupta, M.; Srivastava, B.S.; Srivastava, R. Fast-growing, non-infectious and intracellularly surviving drug-resistant Mycobacterium aurum: A model for high-throughput antituberculosis drug screening. J. Antimicrob. Chemother. 2009, 64, 774–781. [Google Scholar] [CrossRef]
- David, H.L.; Clavel, S.; Clement, F.; Moniz-Pereira, J. Effects of antituberculosis and antileprosy drugs on mycobacteriophage D29 growth. Antimicrob. Agents Chemother. 1980, 18, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Stanley, E.C.; Mole, R.J.; Smith, R.J.; Glenn, S.M.; Barer, M.R.; McGowan, M.; Rees, C.E.D. Development of a new, combined rapid method using phage and PCR for detection and identification of viable Mycobacterium paratuberculosis bacteria within 48 hours. Appl. Environ. Microbiol. 2007, 73, 1851–1857. [Google Scholar] [CrossRef] [PubMed]
- Grant, I.R. Bacteriophage-Based Methods for Detection of Viable Mycobacterium avium subsp. paratuberculosis and Their Potential for Diagnosis of Johne’s Disease. Front. Vet. Sci. 2021, 8, 205. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kriakov, J.; Vilcheze, C.; Dai, Z.; Hatfull, G.F.; Jacobs, W.R., Jr. R, Jr. Bxz1, a new generalized transducing phage for mycobacteria. FEMS Microbiol. Lett. 2004, 241, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Hatfull, G.F. Phage inactivation by an ethanol-ether extract of Mycobacterium smegmatis. Microbiol. Spectr. 2014, 2, 1–36. [Google Scholar]
- Sundar Raj, C.V.; Ramakrishnan, T. Transduction in Mycobacterium smegmatis. Nature 1970, 228, 280–281. [Google Scholar] [CrossRef]
- Zinder, N.D.; Lederberg, J. Genetic exchange in Salmonella. J. Bacteriol. 1952, 64, 679–699. [Google Scholar] [CrossRef]
- Lennox, E.S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1955, 1, 190–206. [Google Scholar] [CrossRef]
- Tokunaga, T.; Kataoka, T.; Suga, K. Phage Inactivation by an Ethanol-Ether Extract of Mycobacterium smegmatis. Am. Rev. Respir. Dis. 1970, 101, 309–313. [Google Scholar] [CrossRef]
- Tokunaga, T.; Sellers, M.I. Phage Receptor of Mycobacterium smegmatis. In Host-Virus Relationships in Mycobacterium, Nocardia and Actinomyces; Stephen, E.J., Plummer, G., Eds.; Charles C Thomas: Springfield, IL, USA, 1970. [Google Scholar]
- Bisso, G.; Castelnuovo, G.; Nardelli, M.-G.; Orefici, G.; Arancia, G.; Lanéelle, G.; Asselineau, C.; Asselineau, J. A study on the receptor for a mycobacteriophage: Phage phlei. Biochimie 1976, 58, 87–97. [Google Scholar] [CrossRef]
- Goren, M.B.; McClatchy, J.K.; Martens, B.; Brokl, O. Mycosides C: Behavior as receptor site substance for mycobacteriophage D4. J. Virol. 1972, 9, 999–1003. [Google Scholar] [CrossRef] [PubMed]
- Fregnan, G.B.; Smith, D.W.; Randall, H.M. A mutant of a scotochromogenic mycobacterium detected by colony morphology and lipid studies. J. Bacteriol. 1962, 83, 828–836. [Google Scholar] [CrossRef]
- Laneelle, G.; Asselineau, J. Structure of a peptidolipid glycoside isolated from a Mycobacterium. Eur. J. Biochem. 1968, 5, 487–491. [Google Scholar] [CrossRef] [PubMed]
- Lederer, E. Glycolipids of mycobacteria and related microorganisms. Chem. Phys. Lipids 1967, 1, 294–315. [Google Scholar] [CrossRef]
- Donnelly-Wu, M.K.; Jacobs, W.R., Jr.; Hatfull, G.F. Superinfection immunity of mycobacteriophage L5: Applications for genetic transformation of mycobacteria. Mol. Microbiol. 1993, 7, 407–417. [Google Scholar] [CrossRef]
- Chatterjee, S.; Mitra, M.; Das Gupta, S.K. A high yielding mutant of mycobacteriophage L1 and its application as a diagnostic tool. FEMS Microbiol. Lett. 2000, 188, 47–53. [Google Scholar] [CrossRef]
- Dedrick, R.M.; Mavrich, T.N.; Ng, W.L.; Hatfull, G.F. Expression and evolutionary patterns of mycobacteriophage D29 and its temperate close relatives. BMC Microbiol. 2017, 17, 225. [Google Scholar] [CrossRef]
- Halleran, A.; Clamons, S.; Saha, M. Transcriptomic characterization of an infection mycobacteriophage Kampy. PLoS ONE 2015, 10, e0141100. [Google Scholar] [CrossRef]
- Fan, X.; Duan, X.; Tong, Y.; Huang, Q.; Zhou, M.; Wang, H.; Zeng, L.; Young, R.F., 3rd; Xie, J. The global reciprocal reprogramming between mycobacteriophage SWU1 and mycobacterium reveals the molecular strategy of subversion and promotion of phage infection. Front. Microbiol. 2016, 7, 41. [Google Scholar] [CrossRef]
- Hatfull, G.F. Mycobacteriophages: Genes and genomes. Annu. Rev. Microbiol. 2010, 64, 331–356. [Google Scholar] [CrossRef] [PubMed]
- Fullner, K.J.; Hatfull, G.F. Mycobacteriophage L5 infection of Mycobacterium bovis BCG: Implications for phage genetics in the slow-growing mycobacteria. Mol. Microbiol. 1997, 26, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, T.; Nandy, R.; Sau, S. Overexpression of a delayed early gene hlg1 of temperate mycobacteriophage L1 is lethal to both M. smegmatis and E. coli. BMB Rep. 2008, 41, 363–368. [Google Scholar] [CrossRef]
- Hershey, A.D.; Dove, W. Introduction to Lambda. In Lambda II; Hendrix, R.W., Roberts, J.W., Stahl, F.W., Weisberg, R.A., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1983; pp. 3–11. [Google Scholar]
- Sinha, A.; Eniyan, K.; Manohar, P.; Ramesh, N.; Bajpai, U. Characterization and genome analysis of B1 sub-cluster mycobacteriophage PDRPxv. Virus Res. 2020, 279, 197884. [Google Scholar] [CrossRef] [PubMed]
- Eniyan, K.; Sinha, A.; Ahmad, S.; Bajpai, U. Functional characterization of the endolysins derived from mycobacteriophage PDRPxv. World J. Microbiol. Biotechnol. 2020, 36, 83. [Google Scholar] [CrossRef]
- Payne, K.; Sun, Q.; Sacchettini, J.; Hatfull, G.F. Mycobacteriophage Lysin B is a novel mycolylarabinogalactan esterase. Mol. Microbiol. 2009, 73, 367–381. [Google Scholar] [CrossRef]
- Saadhali, S.A.; Hassan, S.; Hanna, L.E.; Ranganathan, U.D.; Kumar, V. Homology modeling, substrate docking, and molecular simulation studies of mycobacteriophage Che12 lysin A. J. Mol. Model. 2016, 22, 180. [Google Scholar] [CrossRef]
- Patton, C.J.; Kotturi, H. Genomic Sequence of Mycobacteriophage OKCentral2016. Genome Announc. 2018, 6, e01208-17. [Google Scholar] [CrossRef]
- Colston, L.E.; Segura-Totten, M.; Shanks, R.A. Characterization and Genome Sequence of the Mycobacteriophage Donny. Microbiol. Resour. Announc. 2020, 9, e00373-20. [Google Scholar] [CrossRef]
- Gigante, A.M.; Olivença, F.; Catalão, M.J.; Leandro, P.; Moniz-Pereira, J.; Filipe, S.R.; Pimentel, M. The mycobacteriophage Ms6 LysB N-terminus displays peptidoglycan binding affinity. Viruses 2021, 13, 1377. [Google Scholar] [CrossRef]
- Catalão, M.J.; Gil, F.; Moniz-Pereira, J.; São-José, C.; Pimentel, M. Diversity in bacterial lysis systems: Bacteriophages show the way. FEMS Microbiol. Rev. 2013, 37, 554–571. [Google Scholar] [CrossRef] [PubMed]
- Catalão, M.J.; Gil, F.; Moniz-Pereira, J.; Pimentel, M. The mycobacteriophage Ms6 encodes a chaperone-like protein involved in the endolysin delivery to the peptidoglycan. Mol. Microbiol. 2010, 77, 672–686. [Google Scholar] [CrossRef]
- Fraga, A.G.; Trigo, G.; Murthy, R.K.; Akhtar, S.; Hebbur, M.; Pacheco, A.R.; Dominguez, J.; Silva-Gomes, R.; Gonçalves, C.M.; Oliveira, H.; et al. Antimicrobial activity of mycobacteriophage D29 Lysin B during Mycobacterium ulcerans infection. PLOS Negl. Trop. Dis. 2019, 13, e0007113. [Google Scholar] [CrossRef] [PubMed]
- Sampson, T.; Broussard, G.W.; Marinelli, L.J.; Jacobs-Sera, D.; Ray, M.; Ko, C.-C.; Russell, D.; Hendrix, R.W.; Hatfull, G.F. Mycobacteriophages BPs, Angel and Halo: Comparative genomics reveals a novel class of ultra-small mobile genetic elements. Microbiology 2009, 155, 2962–2977. [Google Scholar] [CrossRef] [PubMed]
- Delesalle, V.A.; Tanke, N.T.; Vill, A.C.; Krukonis, G.P. Testing hypotheses for the presence of tRNA genes in mycobacteriophage genomes. Bacteriophage 2016, 6, e1219441. [Google Scholar] [CrossRef]
- Dedrick, R.M.; Guerrero Bustamante, C.A.; Garlena, R.A.; Pinches, R.S.; Cornely, K.; Hatfull, G.F. Mycobacteriophage ZoeJ: A broad host-range close relative of mycobacteriophage TM4. Tuberculosis 2019, 115, 14–23. [Google Scholar] [CrossRef]
- Haeseleer, F.; Pollet, J.F.; Bollen, A.; Jacobs, P. Molecular cloning and sequencing of the attachment site and integrase gene of the temperate mycobacteriophage FRAT1. Nucleic Acids Res. 1992, 20, 1420. [Google Scholar] [CrossRef][Green Version]
- Jacobs-Sera, D.; Marinelli, L.J.; Bowman, C.; Broussard, G.W.; Guerrero Bustamante, C.; Boyle, M.M.; Petrova, Z.O.; Dedrick, R.M.; Pope, W.H.; Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES) program; et al. On the nature of mycobacteriophage diversity and host preference. Virology 2012, 434, 187–201. [Google Scholar] [CrossRef]
- Ford, M.E.; Stenstrom, C.; Hendrix, R.W.; Hatfull, G.F. Mycobacteriophage TM4: Genome structure and gene expression. Tuber. Lung Dis. 1998, 79, 63–73. [Google Scholar] [CrossRef]
- Mayer, O.; Jain, P.; Weisbrod, T.R.; Biro, D.; Ho, L.; Jacobs-Sera, D.; Hatfull, G.F.; Jacobs, W.R., Jr. Fluorescent reporter DS6A mycobacteriophages reveal unique variations in infectibility and phage production in mycobacteria. J. Bacteriol. 2016, 198, 3220–3232. [Google Scholar] [CrossRef]
- Gangadharam, P.R.J.; Stager, C.E. Acid-fastness of Mycobacterium tuberculosis H37Rv following infection by mycobacteriophage DS6A. Tubercle 1976, 57, 203–205. [Google Scholar] [CrossRef]
- Sula, L.; Sulová, J.; Stolcpartová, M. Therapy of experimental tuberculosis in guinea pigs with mycobacterial phages DS-6A, GR-21 T, My-327. Czech. Med. 1981, 4, 209–214. [Google Scholar] [PubMed]
- Zemskova, Z.S.; Dorozhkova, I.R. Pathomorphological assessment of the therapeutic effect of mycobacteriophages in tuberculosis. Probl Tuberk 1991, 11, 63–66. [Google Scholar]
- Pearson, R.E.; Dickson, J.A.; Hamilton, P.T.; Little, M.C.; Beyer, W.F., Jr. Mycobacteriophage DSGA Specific for the Mycobacterium tuberculosis Complex. U.S. Patent No. 5,476,768, 19 December 1995. [Google Scholar]
- Pearson, R.E.; Dickson, J.A.; Hamilton, P.T.; Little, M.C.; Beyer, W.F., Jr. Mycobacteriophage Specific for the Mycobacterium tuberculosis Complex. U.S. Patent No 5,612,182, 18 March 1997. [Google Scholar]
- Pearson, R.E.; Dickson, J.A.; Hamilton, P.T.; Little, M.C.; Beyer, W.F., Jr. Mycobacteriophage Specific for the Mycobacterium tuberculosis Complex. U.S. Patent No 5,582,969, 10 December 1996. [Google Scholar]
- Pearson, R.E.; Dickson, J.A.; Hamilton, P.T.; Little, M.C.; Beyer, W.F., Jr. DNA Polymerase III β-subunit From Mycobacteriophage DS6A. U.S. Patent No. 5,633,159, 27 May 1997. [Google Scholar]
- Foley-Thomas, E.M.; Whipple, D.L.; Bermudez, L.E.; Barletta, R.G. Phage infection, transfection and transformation of Mycobacterium avium complex and Mycobacterium paratuberculosis. Microbiology 1995, 141, 1173–1181. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, W.R.; Tuckman, M.; Bloom, B.R. Introduction of foreign DNA into mycobacteria using a shuttle phasmid. Nature 1987, 327, 532–535. [Google Scholar] [CrossRef] [PubMed]
- Pope, W.H.; Ferreira, C.M.; Jacobs-Sera, D.; Benjamin, R.C.; Davis, A.J.; DeJong, R.J.; Elgin, S.C.R.; Guilfoile, F.R.; Forsyth, M.H.; Harris, A.D.; et al. Cluster K mycobacteriophages: Insights into the evolutionary origins of mycobacteriophage TM4. PLoS ONE 2011, 6, e26750. [Google Scholar] [CrossRef]
- Gan, Y.; Wu, T.; Liu, P.; Guo, S. Characterization and classification of Bo4 as a cluster G mycobacteriophage that can infect and lyse M. tuberculosis. Arch. Microbiol. 2014, 196, 209–218. [Google Scholar] [CrossRef]
- Jones, W.D. Further studies of mycobacteriophage 33D (warsaw) for differentiation of BCG from M. bovis and M. tuberculosis. Tubercle 1979, 60, 55–58. [Google Scholar] [CrossRef]
- Suarez, C.A.; Franceschelli, J.J.; Morbidoni, H.R. Mycobacteriophage CRB2 defines a new subcluster in mycobacteriophage classification. PLoS ONE 2019, 14, e0212365. [Google Scholar] [CrossRef]
- Stella, E.J.; Franceschelli, J.J.; Tasselli, S.E.; Morbidoni, H.R. Analysis of novel mycobacteriophages indicates the existence of different strategies for phage inheritance in mycobacteria. PLoS ONE 2013, 8, e56384. [Google Scholar] [CrossRef]
- Casjens, S.R.; Gilcrease, E.B. Determining DNA packaging strategy by analysis of the termini of the chromosomes in tailed-bacteriophage virions. Methods Mol. Biol. 2009, 502, 91–111. [Google Scholar] [CrossRef] [PubMed]
- Yue, X.; Huang, Y.; Zhang, Y.; Ouyang, H.; Xie, J.; Fu, Z. Mycobacteriophage SWU1-functionalized magnetic particles for facile bioluminescent detection of Mycobacterium smegmatis. Anal. Chim. Acta 2021, 1145, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Yan, J.; Xie, L.; Zeng, L.; Young, R.F., 3rd; Xie, J. Genomic and proteomic features of mycobacteriophage SWU1 isolated from China soil. Gene 2015, 561, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Gomathi, N.S.; Sameer, H.; Kumar, V.; Balaji, S.; Dustackeer, V.N.A.; Narayanan, P.R. In silico analysis of mycobacteriophage Che12 genome: Characterization of genes required to lysogenise Mycobacterium tuberculosis. Comput. Biol. Chem. 2007, 31, 82–91. [Google Scholar] [CrossRef]
- Kumar, V.; Loganathan, P.; Sivaramakrishnan, G.; Kriakov, J.; Dusthakeer, A.; Subramanyam, B.; Chan, J.; Jacobs, W.R., Jr.; Paranji Rama, N. Characterization of temperate phage Che12 and construction of a new tool for diagnosis of tuberculosis. Tuberculosis 2008, 88, 616–623. [Google Scholar] [CrossRef]
- Ford, M.E.; Sarkis, G.J.; Belanger, A.E.; Hendrix, R.W.; Hatfull, G.F. Genome structure of mycobacteriophage D29: Implications for phage evolution11Edited by J. Karn. J. Mol. Biol. 1998, 279, 143–164. [Google Scholar] [CrossRef]
- Foddai, A.; Strain, S.; Whitlock, R.H.; Elliott, C.T.; Grant, I.R. Application of a peptide-mediated magnetic separation-phage assay for detection of viable Mycobacterium avium subsp. paratuberculosis to bovine bulk tank milk and feces samples. J. Clin. Microbiol. 2011, 49, 2017–2019. [Google Scholar] [CrossRef][Green Version]
- Greenstein, R.J.; Su, L.; Grant, I.R.; Foddai, A.C.G.; Turner, A.; Nagati, J.S.; Brown, S.T.; Stabel, J.R. Comparison of a mycobacterial phage assay to detect viable Mycobacterium avium subsp. paratuberculosis with standard diagnostic modalities in cattle with naturally infected Johne disease. Gut Pathog. 2021, 13, 30. [Google Scholar] [CrossRef]
- Beinhauerova, M.; Slana, I. Phage amplification assay for detection of mycobacterial infection: A Review. Microorganisms 2021, 9, 237. [Google Scholar] [CrossRef]
- Foddai, A.C.G.; Grant, I.R. A novel one-day phage-based test for rapid detection and enumeration of viable Mycobacterium avium subsp. paratuberculosis in cows’ milk. Appl. Microbiol. Biotechnol. 2020, 104, 9399–9412. [Google Scholar] [CrossRef]
- Hosseiniporgham, S.; Rebechesu, L.; Pintore, P.; Lollai, S.; Dattena, M.; Russo, S.; Ruiu, A.; Sechi, L.A. A rapid phage assay for detection of viable Mycobacterium avium subsp. paratuberculosis in milk. Sci. Rep. 2022, 12, 475. [Google Scholar] [CrossRef] [PubMed]
- Swift, B.M.C.; Gerrard, Z.E.; Huxley, J.N.; Rees, C.E.D. Factors affecting phage D29 infection: A tool to investigate different growth states of mycobacteria. PLoS ONE 2014, 9, e106690. [Google Scholar] [CrossRef]
- Sassi, M.; Bebeacua, C.; Drancourt, M.; Cambillau, C. The first structure of a mycobacteriophage, the Mycobacterium abscessus subsp. bolletii phage Araucaria. J. Virol. 2013, 87, 8099–8109. [Google Scholar] [CrossRef] [PubMed]
- Ripoll, F.; Pasek, S.; Schenowitz, C.; Dossat, C.; Barbe, V.; Rottman, M.; Macheras, E.; Heym, B.; Herrmann, J.-L.; Daffé, M.; et al. Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS ONE 2009, 4, e5660. [Google Scholar] [CrossRef] [PubMed]
- Sassi, M.; Drancourt, M. Genome analysis reveals three genomospecies in Mycobacterium abscessus. BMC Genom. 2014, 15, 359. [Google Scholar] [CrossRef]
- Sassi, M.; Gouret, P.; Chabrol, O.; Pontarotti, P.; Drancourt, M. Mycobacteriophage-drived diversification of Mycobacterium abscessus. Biol. Direct 2014, 9, 19. [Google Scholar] [CrossRef][Green Version]
- Glickman, C.; Kammlade, S.M.; Hasan, N.A.; Epperson, L.E.; Davidson, R.M.; Strong, M. Characterization of integrated prophages within diverse species of clinical nontuberculous mycobacteria. Virol. J. 2020, 17, 124. [Google Scholar] [CrossRef]
- Amarh, E.D.; Dedrick, R.M.; Garlena, R.A.; Russell, D.A.; Jacobs-Sera, D.; Hatfull, G.F. Genome Sequence of Mycobacterium abscessus Phage phiT46-1. Microbiol. Resour. Announc. 2021, 10, e01421-20. [Google Scholar] [CrossRef]
- Bibb, L.A.; Hatfull, G.F. Integration and excision of the Mycobacterium tuberculosis prophage-like element, φRv1. Mol. Microbiol. 2002, 45, 1515–1526. [Google Scholar] [CrossRef]
- Hatfull, G.F.; Cresawn, S.G.; Hendrix, R.W. Comparative genomics of the mycobacteriophages: Insights into bacteriophage evolution. Res. Microbiol. 2008, 159, 332–339. [Google Scholar] [CrossRef]
- Hatfult, G.F.; Sarkis, G.J. DNA sequence, structure and gene expression of mycobacteriophage L5: A phage system for mycobacterial genetics. Mol. Microbiol. 1993, 7, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.H.; Hatfull, G.F. Mycobacteriophage L5 integrase-mediated site-specific integration in vitro. J. Bacteriol. 1993, 175, 6836–6841. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bajpai, U.; Mehta, A.K.; Eniyan, K.; Sinha, A.; Ray, A.; Virdi, S.; Ahmad, S.; Shah, A.; Arora, D.; Marwaha, D.; et al. Isolation and characterization of bacteriophages from India, with lytic activity against Mycobacterium tuberculosis. Can. J. Microbiol. 2018, 64, 483–491. [Google Scholar] [CrossRef]
- Froman, S.; Will, D.W.; Bogen, E. Bacteriophage active against virulent Mycobacterium tuberculosis. I. Isolation and activity. Am. J. Public Health Nations Health 1954, 44, 1326–1333. [Google Scholar] [CrossRef] [PubMed]
- Clark-Curtiss, J.E. Genome structure of mycobacteria. In Molecular Biology of Mycobacteria; McFadden, J.J., Ed.; Academic Press Ltd.: London, UK, 1990; pp. 77–96. [Google Scholar]
- Pope, W.H.; Jacobs-Sera, D.; Russell, D.A.; Peebles, C.L.; Al-Atrache, Z.; Alcoser, T.A.; Alexander, L.M.; Alfano, M.B.; Alford, S.T.; Amy, N.E.; et al. Expanding the diversity of mycobacteriophages: Insights into genome architecture and evolution. PLoS ONE 2011, 6, e16329. [Google Scholar] [CrossRef] [PubMed]
- Cornely, K.A.; Jancevski, A.V.; Rogers, S.R.; Scola, S.E.; Pinches, R.S.; Perri, C.M.; Brown, M.S.; Cavedon, W.D.; Dubois, H.M.; Fernando, M.A.; et al. Mycobacterium Phage ZoeJ, Complete Genome. Available online: https://www.ncbi.nlm.nih.gov/nuccore/NC_024147.1 (accessed on 17 May 2022).
- Haeseleer, F.; Pollet, J.-F.; Haumont, M.; Bollen, A.; Jacobs, P. Stable integration and expression of the Plasmodium falciparum circumsporozoite protein coding sequence in mycobacteria. Mol. Biochem. Parasitol. 1993, 57, 117–126. [Google Scholar] [CrossRef]
- Curtis, N.; Falkinham, J.O.; Garlena, R.A.; Russell, D.A.; Pope, W.H.; Jacobs-Sera, D.; Hatfull, G.F. Mycobacterium Phage D32, Complete Genome. Available online: https://www.ncbi.nlm.nih.gov/nuccore/1834466630 (accessed on 17 May 2022).
- Sellers, M.I.; Nakamura, R.; Tokunaga, T. The effects of ultraviolet irradiation on mycobacteriophages and their infectious DNAs. J. Gen. Virol. 1970, 7, 233–247. [Google Scholar] [CrossRef]
- Fukai, K.; Sellers, M.I. A morphologic study of mycobacteria infected with D32 phage. Am. Rev. Respir. Dis. 1960, 81, 52–59. [Google Scholar] [CrossRef]
- UniProt Taxonomy. Available online: https://www.uniprot.org/ (accessed on 15 April 2022).
- Fan, X.; Teng, T.; Wang, H.; Xie, J. Biology of a novel mycobacteriophage, SWU1, isolated from Chinese soil as revealed by genomic characteristics. J. Virol. 2012, 86, 10230–10231. [Google Scholar] [CrossRef]
- Hatfull, G.F.; Pedulla, M.L.; Jacobs-Sera, D.; Cichon, P.M.; Foley, A.; Ford, M.E.; Gonda, R.M.; Houtz, J.M.; Hryckowian, A.J.; Kelchner, V.A.; et al. Mycobacterium Phage Che12, Complete Genome. Available online: https://www.ncbi.nlm.nih.gov/nuccore/DQ398043.1 (accessed on 17 May 2022).
- Hatfull, G.F.; Pedulla, M.L.; Jacobs-Sera, D.; Cichon, P.M.; Foley, A.; Ford, M.E.; Gonda, R.M.; Houtz, J.M.; Hryckowian, A.J.; Kelchner, V.A.; et al. Exploring the mycobacteriophage metaproteome: Phage genomics as an educational platform. PLoS Genet. 2006, 2, e92. [Google Scholar] [CrossRef]
- Bowman, B.; Newman, H.; Moritz, J.; Koehler, R. Properties of mycobacteriophage DS6A. II. Lipid composition. Am. Rev. Respir. Dis. 1973, 107, 42–49. [Google Scholar] [PubMed]
- Hatfull, G.F.; the Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science Program; the KwaZulu-Natal Research Institute for Tuberculosis and HIV Mycobacterial Genetics Course Students; the Phage Hunters Integrating Research and Education Program. Complete genome sequences of 138 mycobacteriophages. J. Virol. 2012, 86, 2382–2384. [Google Scholar] [CrossRef]
- Curtis, N.; Falkinham, J.O.; Garlena, R.A.; Russell, D.A.; Pope, W.H.; Jacobs-Sera, D.; Hatfull, G.F. Mycobacterium Phage AN3, Complete Genome. Available online: https://www.ncbi.nlm.nih.gov/nuccore/1834466895 (accessed on 17 May 2022).
- Crawford, J.T.; Bates, J.H. Phage Typing of the Mycobacterium-avium intracellulare-scrofulaceum complex. A study of strains of diverse geographic and host origin. Am. Rev. Respir. Dis. 1985, 132, 386–389. [Google Scholar] [CrossRef] [PubMed]
- Curtis, N.; Falkinham, J.O.; Garlena, R.A.; Russell, D.A.; Pope, W.H.; Jacobs-Sera, D.; Hatfull, G.F. Mycobacterium Phage AN9, Complete Genome. Available online: https://www.ncbi.nlm.nih.gov/nuccore/1834466804 (accessed on 17 May 2022).
- Crawford, J.T.; Fitzhugh, J.K.; Bates, J.H. Phage Typing of the Mycobacterium-avium-intracellulare-scrofulaceum complex. Am. Rev. Respir. Dis. 1981, 124, 559–562. [Google Scholar] [CrossRef] [PubMed]
- Curtis, N.; Falkinham, J.O.; Garlena, R.A.; Russell, D.A.; Pope, W.H.; Jacobs-Sera, D.; Hatfull, G.F. Mycobacterium Phage ANI8, Complete Genome. Available online: https://www.ncbi.nlm.nih.gov/nuccore/1834466713 (accessed on 17 May 2022).
- Curtis, N.; Falkinham, J.O.; Garlena, R.A.; Russell, D.A.; Pope, W.H.; Jacobs-Sera, D.; Hatfull, G.F. Mycobacterium Phage VA6, Complete Genome. Available online: https://www.ncbi.nlm.nih.gov/nuccore/1834466119 (accessed on 17 May 2022).
- Curtis, N.; Falkinham, J.O.; Garlena, R.A.; Russell, D.A.; Pope, W.H.; Jacobs-Sera, D.; Hatfull, G.F. Mycobacterium Phage VC3, Complete Genome. Available online: https://www.ncbi.nlm.nih.gov/nuccore/1834466028 (accessed on 17 May 2022).
- Govender, V.S.; Mchunu, L.V.; Naicker, R.N.; Sha, K.I.; Zinyembe, F.; Pillay, B.; Larsen, M.H.; Rubin, E.J.; Kasprowicz, V.O.; Bishai, W.R.; et al. Mycobacterium Phage Muddy, Complete Genome. Available online: https://www.ncbi.nlm.nih.gov/nuccore/KF024728.1 (accessed on 17 May 2022).
- Guerrero-Bustamante, C.A.; Dedrick, R.M.; Garlena, R.A.; Russell, D.A.; Hatfull, G.F. Toward a Phage Cocktail for Tuberculosis: Susceptibility and Tuberculocidal Action of Mycobacteriophages against Diverse Mycobacterium tuberculosis Strains. MBio 2021, 12, e00973-21. [Google Scholar] [CrossRef]
- Amarth, E.D.; Gauthier, C.H.; Dedrick, D.; Garlena, R.A.; Ruissell, D.A.; Jacobs-Sera, D.; Zack, K.M.; Hatfull, G.F. Genome sequence of Mycobacterium abscessus Phage phiT45-1. Microbiol. Resour. Announc. 2021, 10, e00155-21. [Google Scholar] [CrossRef]
- Lamb, D.C.; Follmer, A.H.; Goldstone, J.V.; Nelson, D.R.; Warrilow, A.G.; Price, C.L.; True, M.Y.; Kelly, S.L.; Poulos, T.L.; Stegeman, J.J. On the occurrence of cytochrome P450 in viruses. Proc. Natl. Acad. Sci. USA 2019, 116, 12343–12352. [Google Scholar] [CrossRef]
- Edgington, N.P.; Voshell, S.M.; Ware, V.C.; Akoto, F.F.; Alhout, A.A.; Atwal, G.J.; Balyozian, J.B.; Cadieux, Z.A.; Chop, B.M.; Cresawn, S.G.; et al. Genome sequences of Chancellor, Mitti, and Wintermute, three subcluster K4 phages isolated using Mycobacterium smegmatis mc2155. Genome Announc. 2022, 5, e01070-17. [Google Scholar] [CrossRef]
- Pope, W.H.; Carter, J.T.; Dasher, K.L.; Haynberg, M.C.; Reddi, A.; Shedlock, K.A.; Lapin, J.S.; Prout, A.K.; Grubb, S.R.; Warner, M.H.; et al. Genome sequence of a newly isolated mycobacteriophage, ShedlockHolmes. Genome Announc. 2022, 3, e00597-15. [Google Scholar] [CrossRef]
- Gaballa, J.M.; Keeyon, D.; Rachel, D.; Ryan, N.; Diane, P.; Erin, S.; Yiwei, S.; Boon, T.; Marcia, B.; Olivia, B.; et al. Genome sequences of cluster K mycobacteriophages Deby, LaterM, LilPharaoh, Paola, SgtBeansprout, and Sulley. Microbiol. Resour. Announc. 2022, 8, e01481-18. [Google Scholar] [CrossRef]
- Faith, C.; Josh, K.; Megan, A.; Danielle, B.; Ashleigh, C.; Jessica, D.; Miranda, F.; Leeila, H.; Travis, M.; Jonathan, M.; et al. Complete genome sequence of mycobacteriophage Joy99. Microbiol. Resour. Announc. 2022, 10, e00556-21. [Google Scholar] [CrossRef]
- Franceschelli, J.J.; Suarez, C.A.; Teran, L.; Raya, R.R.; Morbidoni, H.R. Complete genome sequences of Nine mycobacteriophages. Genome Announc. 2014, 2, e00181-14. [Google Scholar] [CrossRef]
- Jacobs-Sera, D.; Catinas, O.; Fernandez-Martinez, M.; Garcia, A.; Garlena, R.A.; Bustamante, C.A.G.; Larsen, M.H.; Medellin, R.H.; Melendez-Ortiz, M.Y.; Melendez-Rivera, C.M.; et al. Genome sequences of mycobacteriophages Kerberos, Pomar16, and StarStuff. Genome Announc. 2017, 5, e00690-17. [Google Scholar] [CrossRef]
- Hatfull, G.F.; Pedulla, M.L.; Ford, M.E.; Houtz, J.M.; Jacobs-Sera, D.; Falbo, J.; Gross, J.; Karthikeyan, T.; Pannunzio, N.; Brucker, W.; et al. Mycobacterium Phage Omega, Complete Genome. Available online: https://www.ncbi.nlm.nih.gov/nuccore/AY129338.1 (accessed on 17 May 2022).
- Della, M.; Palmbos, P.L.; Tseng, H.-M.; Tonkin, L.M.; Daley, J.M.; Topper, L.M.; Pitcher, R.S.; Tomkinson, A.E.; Wilson, T.E.; Doherty, A.J. Mycobacterial Ku and ligase proteins constitute a two-component NHEJ repair machine. Science 2004, 306, 683–685. [Google Scholar] [CrossRef]
- Oftung, F.; Mustafa, A.S.; Wiker, H.G. Extensive sequence homology between the Mycobacterium leprae LSR (12 kDa) antigen and its Mycobacterium tuberculosis counterpart. FEMS Immunol. Med. Microbiol. 2000, 27, 87–89. [Google Scholar] [CrossRef]
- Hatfull, G.F. The Secret Lives of Mycobacteriophages. In Advances in Virous Research, Bacteriophages, Part A; Łobocka, M., Szybalski, W.T., Eds.; Academic Press: Cambridge, MA, USA, 2012; Volume 82, pp. 179–288. ISBN 0065-3527. [Google Scholar]
- Hatfull, G.F.; Pedulla, M.L.; Ford, M.E.; Houtz, J.M.; Jacobs-Sera, D.; Falbo, J.; Gross, J.; Karthikeyan, T.; Pannunzio, N.R.; Brucker, W.; et al. Mycobacterium Phage Cjw1, Complete Genome. Available online: https://www.ncbi.nlm.nih.gov/nuccore/AY129331.1 (accessed on 17 May 2022).
- Laal, S.; Sharma, Y.D.; Prasad, H.K.; Murtaza, A.; Singh, S.; Tangri, S.; Misra, R.S.; Nath, I. Recombinant fusion protein identified by lepromatous sera mimics native Mycobacterium leprae in T-cell responses across the leprosy spectrum. Proc. Natl. Acad. Sci. USA 1991, 88, 1054–1058. [Google Scholar] [CrossRef]
- Dao, V.L.; Xia, L.; Bancroft, C.T. Genome sequences of Mycobacterium smegmatis phages Fefferhead and ShamWow. Microbiol. Resour. Announc. 2022, 10, e00973-21. [Google Scholar] [CrossRef]
- Pedulla, M.L.; Ford, M.E.; Houtz, J.M.; Karthikeyan, T.; Wadsworth, C.; Lewis, J.A.; Jacobs-Sera, D.; Falbo, J.; Gross, J.; Pannunzio, N.R.; et al. Origins of highly mosaic mycobacteriophage genomes. Cell 2003, 113, 171–182. [Google Scholar] [CrossRef]
- Sellers, M.I.; Runnals, H.R. Mycobacteriophage. I. Physicochemical characterization. J. Bacteriol. 1961, 81, 442–447. [Google Scholar] [CrossRef]
- Penso, G.; Ortali, V. Studies and research on mycobacteria. Note II. The phages of mycobacteria. Rend. Ist. Sup. Sanita 1949, 12, 903–918. [Google Scholar]
- Hnatko, S.I. The isolation of bacteriophages for mycobacteria with reference to phage typing of the genus. Can. J. Med. Sci. 1953, 31, 462–473. [Google Scholar] [CrossRef]
- Sørensen, P.E.; Van Den Broeck, W.; Kiil, K.; Jasinskyte, D.; Moodley, A.; Garmyn, A.; Ingmer, H.; Butaye, P. New insights into the biodiversity of coliphages in the intestine of poultry. Sci. Rep. 2020, 10, 15220. [Google Scholar] [CrossRef]
- Hendrix, R.W.; Casjens, S.R. The Double Stranded DNA Viruses. In Virus Taxonomy; Fauquet, C.M., Mayo, M.A., Maniloff, J., Desselberger, U., Ball, L.A., Eds.; Academic Press: San Diego, CA, USA, 2005; pp. 33–276. ISBN 978-0-12-249951-7. [Google Scholar]
- Schäfer, R.; Huber, U.; Franklin, R.M. Chemical and physical properties of mycobacteriophage D29. Eur. J. Biochem. 1977, 73, 239–246. [Google Scholar] [CrossRef]
- Timme, T.L.; Brennan, P.J. Induction of bacteriophage from members of the Mycobacterium avium, Mycobacterium intracellulare, Mycobacterium scrofulaceum Serocomplex. Microbiology 1984, 130, 2059–2066. [Google Scholar] [CrossRef]
- Nieto-Fernandez, F.E.; Noutsos, C.; Nissen, J.; Abdelsalam, Y.; Ackloo, J.; Banger, N.; Chan, H.; Chittineedi, T.; Duplessy, I.; Dyce, M.; et al. Complete genome sequence of Rahel, a C1 Cluster mycobacteriophage. Microbiol. Resour. Announc. 2020, 9, e01071-20. [Google Scholar] [CrossRef]
- Lavigne, R.; Ceyssens, P.J. Myoviridae. In Virus Taxonomy; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Elsevier: San Diego, CA, USA, 2012; pp. 46–62. ISBN 978-0-12-384684-6. [Google Scholar]
- Hendrix, R.W.; Casjens, S.R.; Lavigne, R. Siphoviridae. In Virus Taxonomy; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Elsevier: San Diego, CA, USA, 2012; pp. 86–98. ISBN 978-0-12-384684-6. [Google Scholar]
- Hatfull, G.F.; Jacobs-Sera, D.; Lawrence, J.G.; Pope, W.H.; Russell, D.A.; Ko, C.-C.; Weber, R.J.; Patel, M.C.; Germane, K.L.; Edgar, R.H.; et al. Comparative genomic analysis of 60 Mycobacteriophage genomes: Genome clustering, gene acquisition, and gene size. J. Mol. Biol. 2010, 397, 119–143. [Google Scholar] [CrossRef]
- Pope, W.H.; Mavrich, T.N.; Garlena, R.A.; Guerrero-Bustamante, C.A.; Jacobs-Sera, D.; Montgomery, M.T.; Russell, D.A.; Warner, M.H.; Hatfull, G.F.; Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES). Bacteriophages of Gordonia spp. Display a spectrum ofdiversity and genetic relationships. MBio 2017, 8, e01069-17. [Google Scholar] [CrossRef]
- Byrum, C.A.; Korey, C.A.; Jordan, Z.; Zhou, Y.; Taylor, S.; Alfajardo, J.; Delesalle, V.A. Complete genome sequence of the cluster F1 mycobacteriophage KingMidas. Microbiol. Resour. Announc. 2020, 9, e01557-19. [Google Scholar] [CrossRef]
- Dedrick, R.M.; Marinelli, L.J.; Newton, G.L.; Pogliano, K.; Pogliano, J.; Hatfull, G.F. Functional requirements for bacteriophage growth: Gene essentiality and expression in mycobacteriophage Giles. Mol. Microbiol. 2013, 88, 577–589. [Google Scholar] [CrossRef]
- Ko, C.-C.; Hatfull, G.F. Mycobacteriophage Fruitloop gp52 inactivates Wag31 (DivIVA) to prevent heterotypic superinfection. Mol. Microbiol. 2018, 108, 443–460. [Google Scholar] [CrossRef]
- Dedrick, R.M.; Jacobs-Sera, D.; Bustamante, C.A.G.; Garlena, R.A.; Mavrich, T.N.; Pope, W.H.; Reyes, J.C.C.; Russell, D.A.; Adair, T.; Alvey, R.; et al. Prophage-mediated defence against viral attack and viral counter-defence. Nat. Microbiol. 2017, 2, 16251. [Google Scholar] [CrossRef]
- Dedrick, R.M.; Mavrich, T.N.; Ng, W.L.; Cervantes Reyes, J.C.; Olm, M.R.; Rush, R.E.; Jacobs-Sera, D.; Russell, D.A.; Hatfull, G.F. Function, expression, specificity, diversity and incompatibility of actinobacteriophage parABS systems. Mol. Microbiol. 2016, 101, 625–644. [Google Scholar] [CrossRef]
- Beste, D.J.V.; Espasa, M.; Bonde, B.; Kierzek, A.M.; Stewart, G.R.; McFadden, J. The genetic requirements for fast and slow growth in mycobacteria. PLoS ONE 2009, 4, e5349. [Google Scholar] [CrossRef]
- Smith, I. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin. Microbiol. Rev. 2003, 16, 463–496. [Google Scholar] [CrossRef]
- Hosseiniporgham, S.; Biet, F.; Ganneau, C.; Bannantine, J.P.; Bay, S.; Sechi, L.A. A Comparative Study on the Efficiency of Two Mycobacterium avium subsp. paratuberculosis (MAP)-Derived Lipopeptides of L3P and L5P as Capture Antigens in an In-House Milk ELISA Test. Vaccines 2021, 9, 997. [Google Scholar] [CrossRef]
- Hosseiniporgham, S.; Cubeddu, T.; Rocca, S.; Sechi, L.A. Identification of Mycobacterium avium subsp. paratuberculosis (map) in sheep milk, a zoonotic problem. Microorganisms 2020, 8, 1264. [Google Scholar] [CrossRef]
- Gutierrez, C.; Somoskovi, A. Human Pathogenic Mycobacteria. In Reference Module in Biomedical Sciences; Elsevier: Amsterdam, The Netherlands, 2014; ISBN 978-0-12-801238-3. [Google Scholar]
- Bardarov, S.; Bardarov, S.; Pavelka, M.S.; Sambandamurthy, V.; Larsen, M.; Tufariello, J.; Chan, J.; Hatfull, G.; Jacobs, W.R. Specialized transduction: An efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 2002, 148, 3007–3017. [Google Scholar] [CrossRef]
- Piuri, M.; Hatfull, G.F. Fluoromycobacteriophages for drug susceptibility testing (DST) of mycobacteria. Methods Mol. Biol. 2019, 1898, 27–36. [Google Scholar] [CrossRef]
- McNerney, R.; Kambashi, B.S.; Kinkese, J.; Tembwe, R.; Godfrey-Faussett, P. Development of a bacteriophage phage replication assay for diagnosis of pulmonary tuberculosis. J. Clin. Microbiol. 2004, 42, 2115–2120. [Google Scholar] [CrossRef]
- Foddai, A.C.G.; Grant, I.R. Sensitive and specific detection of viable Mycobacterium avium subsp. paratuberculosis in raw milk by the peptide-mediated magnetic separation-phage assay. J. Appl. Microbiol. 2017, 122, 1357–1367. [Google Scholar] [CrossRef]
- Arutyunov, D.; Singh, U.; El-Hawiet, A.; Seckler, H.D.S.; Nikjah, S.; Joe, M.; Bai, Y.; Lowary, T.L.; Klassen, J.S.; Evoy, S.; et al. Mycobacteriophage cell binding proteins for the capture of mycobacteria. Bacteriophage 2014, 4, e960346. [Google Scholar] [CrossRef]
- Jacobs, M.; Wnendt, S.; Stahl, U. High-efficiency electro-transformation of Escherichia coli with DNA from ligation mixtures. Nucleic Acids Res. 1990, 18, 1653. [Google Scholar] [CrossRef][Green Version]
- Jagadeesh, G.; Nataraja, K.N.; Udayakumar, M. Shock waves can enhance bacterial transformation with plasmid DNA. Curr. Sci. 2004, 87, 734–735. [Google Scholar]
- Jarlier, V.; Nikaido, H. Mycobacterial cell wall: Structure and role in natural resistance to antibiotics. FEMS Microbiol. Lett. 1994, 123, 11–18. [Google Scholar] [CrossRef]
- Kieser, K.J.; Rubin, E.J. How sisters grow apart: Mycobacterial growth and division. Nat. Rev. Microbiol. 2014, 12, 550–562. [Google Scholar] [CrossRef]
- Datey, A.; Subburaj, J.; Gopalan, J.; Chakravortty, D. Mechanism of transformation in Mycobacteria using a novel shockwave assisted technique driven by in-situ generated oxyhydrogen. Sci. Rep. 2017, 7, 8645. [Google Scholar] [CrossRef]
- Jacobs, W.R.; Barletta, R.G.; Udani, R.; Chan, J.; Kalkut, G.; Sosne, G.; Kieser, T.; Sarkis, G.J.; Hatfull, G.F.; Bloom, B.R. Rapid assessment of drug susceptibilities of Mycobacterium tuberculosis by means of luciferase reporter phages. Pap. Vet. Biomed. Sci. 1993, 260, 819–822. [Google Scholar] [CrossRef]
- Piuri, M.; Jacobs, W.R.; Hatfull, G.F. Fluoromycobacteriophages for rapid, specific, and sensitive antibiotic susceptibility testing of Mycobacterium tuberculosis. PLoS ONE 2009, 4, e4870. [Google Scholar] [CrossRef]
- Bardarov, S.; Kriakov, J.; Carriere, C.; Yu, S.; Vaamonde, C.; McAdam, R.A.; Bloom, B.R.; Hatfull, G.F.; Jacobs, W.R. Conditionally replicating mycobacteriophages: A system for transposon delivery to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 1997, 94, 10961–10966. [Google Scholar] [CrossRef]
- Sassetti, C.M.; Boyd, D.H.; Rubin, E.J. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 2003, 48, 77–84. [Google Scholar] [CrossRef]
- Mouton, J.M.; Heunis, T.; Dippenaar, A.; Gallant, J.L.; Kleynhans, L.; Sampson, S.L. Comprehensive characterization of the attenuated double auxotroph Mycobacterium tuberculosis ΔleuDΔpanCD as an alternative to H37Rv. Front. Microbiol. 2019, 10, 1922. [Google Scholar] [CrossRef] [PubMed]
- Franch, O.; Han, X.; Marcussen, L.B.; Givskov, A.; Andersen, M.B.; Godbole, A.A.; Harmsen, C.; Nørskov-Lauritsen, N.; Thomsen, J.; Pedersen, F.S.; et al. A new DNA sensor system for specific and quantitative detection of mycobacteria. Nanoscale 2019, 11, 587–597. [Google Scholar] [CrossRef]
- Tokunaga, T.; Sellers, M.I. Infection of Mycobacterium smegmatis with D29 phage DNA. J. Exp. Med. 1964, 119, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Foddai, A.; Elliott, C.T.; Grant, I.R. Optimization of a phage amplification assay to permit accurate enumeration of viable Mycobacterium avium subsp. paratuberculosis cells. Appl. Environ. Microbiol. 2009, 75, 3896–3902. [Google Scholar] [CrossRef][Green Version]
- Kalantri, S.; Pai, M.; Pascopella, L.; Riley, L.; Reingold, A. Bacteriophage-based tests for the detection of Mycobacterium tuberculosis in clinical specimens: A systematic review and meta-analysis. BMC Infect. Dis. 2005, 5, 59. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Swift, B.M.C.; Denton, E.J.; Mahendran, S.A.; Huxley, J.N.; Rees, C.E.D. Development of a rapid phage-based method for the detection of viable Mycobacterium avium subsp. paratuberculosis in blood within 48 h. J. Microbiol. Methods 2013, 94, 175–179. [Google Scholar] [CrossRef]
- Swift, B.M.C.; Meade, N.; Barron, E.S.; Bennett, M.; Perehenic, T.; Hughes, V.; Stevenson, K.; Rees, C.E.D. The development and use of Actiphage to detect viable mycobacteria from bovine tuberculosis and Johne’s disease-infected animals. Microb. Biotechnol. 2020, 13, 738–746. [Google Scholar] [CrossRef]
- Foddai, A.; Elliott, C.T.; Grant, I.R. Maximizing capture efficiency and specificity of magnetic separation for Mycobacterium avium subsp. paratuberculosis cells. Appl. Environ. Microbiol. 2010, 76, 7550–7558. [Google Scholar] [CrossRef]
- Foddai, A.C.G.; Grant, I.R. An optimised milk testing protocol to ensure accurate enumeration of viable Mycobacterium avium subsp. paratuberculosis by the PMS-phage assay. Int. Dairy J. 2015, 51, 16–23. [Google Scholar] [CrossRef]
- Swift, B.M.C.; Convery, T.W.; Rees, C.E.D. Evidence of Mycobacterium tuberculosis complex bacteraemia in intradermal skin test positive cattle detected using phage-RPA. Virulence 2016, 7, 779–788. [Google Scholar] [CrossRef]
- Zeller, S.; Ferneini, E.M. Tuberculosis and Mycobacterial Infections of the Head and Neck. In Head, Neck, and Orofacial Infections: A Multidisciplinary Approach: Hupp, J.R., Ferneini, E.M., Eds.; Elsevier: St. Louis, MO, USA, 2016; pp. 416–421. ISBN 978-0-323-28945-0. [Google Scholar]
- Meacci, F.; Orrù, G.; Iona, E.; Giannoni, F.; Piersimoni, C.; Pozzi, G.; Fattorini, L.; Oggioni, M.R. Drug resistance evolution of a Mycobacterium tuberculosis strain from a noncompliant patient. J. Clin. Microbiol. 2005, 43, 3114–3120. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.-R.; Sheng, W.-H.; Hung, C.-C.; Yu, C.-J.; Lee, L.-N.; Hsueh, P.-R. Mycobacterium abscessus Complex Infections in Humans. Emerg. Infect. Dis. 2015, 21, 1638–1646. [Google Scholar] [CrossRef] [PubMed]
- Palella, F.J.; Delaney, K.M.; Moorman, A.C.; Loveless, M.O.; Fuhrer, J.; Satten, G.A.; Aschman, D.J.; Holmberg, S.D. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N. Engl. J. Med. 1998, 338, 853–860. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, J.E.; Hanson, D.; Dworkin, M.S.; Frederick, T.; Bertolli, J.; Lindegren, M.L.; Holmberg, S.; Jones, J.L. Epidemiology of human immunodeficiency virus-associated opportunistic infections in the United States in the era of highly active antiretroviral therapy. Clin. Infect. Dis. 2000, 30, S5–S14. [Google Scholar] [CrossRef]
- Azimi, T.; Mosadegh, M.; Nasiri, M.J.; Sabour, S.; Karimaei, S.; Nasser, A. Phage therapy as a renewed therapeutic approach to mycobacterial infections: A comprehensive review. Infect. Drug Resist. 2019, 12, 2943–2959. [Google Scholar] [CrossRef]
- Aslam, S.; Lampley, E.; Wooten, D.; Karris, M.; Benson, C.; Strathdee, S.; Schooley, R.T. Lessons learned from the first 10 consecutive cases of intravenous bacteriophage therapy to treat multidrug-resistant bacterial infections at a single center in the United States. Open Forum Infect. Dis. 2020, 7, ofaa389. [Google Scholar] [CrossRef]
- Broxmeyer, L.; Sosnowska, D.; Miltner, E.; Chacón, O.; Wagner, D.; McGarvey, J.; Barletta, R.G.; Bermudez, L.E. Killing of Mycobacterium avium and Mycobacterium tuberculosis by a mycobacteriophage delivered by a nonvirulent mycobacterium: A model for phage therapy of intracellular bacterial pathogens. J. Infect. Dis. 2002, 186, 1155–1160. [Google Scholar] [CrossRef]
- Arora, G.; Chaudhary, D.; Kidwai, S.; Sharma, D.; Singh, R. CitE enzymes are essential for Mycobacterium tuberculosis to establish infection in macrophages and guinea pigs. Front. Cell. Infect. Microbiol. 2018, 8, 385. [Google Scholar] [CrossRef]
- Papet, I.; Rémond, D.; Dardevet, D.; Mosoni, L.; Polakof, S.; Peyron, M.A.; Savary-Auzeloux, I. Sulfur Amino Acids and Skeletal Muscle. In Nutrition and Skeletal Muscle; Walrand, S., Ed.; Academic Press: Cambridge, MA, USA, 2019; pp. 335–363. [Google Scholar] [CrossRef]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef]
- Chakraborty, P.; Kumar, A. The extracellular matrix of mycobacterial biofilms: Could we shorten the treatment of mycobacterial infections? Microb. Cell 2019, 6, 105–122. [Google Scholar] [CrossRef]
- Jiang, Z.; Wei, J.; Liang, Y.; Peng, N.; Li, Y. Aminoglycoside antibiotics inhibit mycobacteriophage infection. Antibiotics 2020, 9, 714. [Google Scholar] [CrossRef]
- Malik, D.J.; Sokolov, I.J.; Vinner, G.K.; Mancuso, F.; Cinquerrui, S.; Vladisavljevic, G.T.; Clokie, M.R.J.; Garton, N.J.; Stapley, A.G.F.; Kirpichnikova, A. Formulation, stabilisation and encapsulation of bacteriophage for phage therapy. Adv. Colloid Interface Sci. 2017, 249, 100–133. [Google Scholar] [CrossRef] [PubMed]
- Colom, J.; Cano-Sarabia, M.; Otero, J.; Cortés, P.; Maspoch, D.; Llagostera, M. Liposome-encapsulated bacteriophages for enhanced oral phage therapy against Salmonella spp. Appl. Environ. Microbiol. 2015, 81, 4841–4849. [Google Scholar] [CrossRef] [PubMed]
- Olivier, K.N.; Griffith, D.E.; Eagle, G.; McGinnis, J.P.; Micioni, L.; Liu, K.; Daley, C.L.; Winthrop, K.L.; Ruoss, S.; Addrizzo-Harris, D.J.; et al. Randomized trial of liposomal amikacin for inhalation in nontuberculous mycobacterial lung disease. Am. J. Respir. Crit. Care Med. 2017, 195, 814–823. [Google Scholar] [CrossRef] [PubMed]
- Carrigy, N.B.; Chang, R.Y.; Leung, S.S.Y.; Harrison, M.; Petrova, Z.; Pope, W.H.; Hatfull, G.F.; Britton, W.J.; Chan, H.-K.; Sauvageau, D.; et al. Antituberculosis bacteriophage D29 delivery with a vibrating mesh nebulizer, jet nebulizer, and soft mist inhaler. Pharm. Res. 2017, 34, 2084–2096. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.P.; Costa, A.R.; Pinto, G.; Meneses, L.; Azeredo, J. Current challenges and future opportunities of phage therapy. FEMS Microbiol. Rev. 2020, 44, 684–700. [Google Scholar] [CrossRef] [PubMed]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef]
- Mangalea, M.R.; Duerkop, B.A. Fitness trade-offs resulting from bacteriophage resistance potentiate synergistic antibacterial strategies. Infect. Immun. 2020, 88, e00926-19. [Google Scholar] [CrossRef]
- Dedrick, R.M.; Smith, B.E.; Garlena, R.A.; Russell, D.A.; Aull, H.G.; Mahalingam, V.; Divens, A.M.; Guerrero-Bustamante, C.A.; Zack, K.M.; Abad, L.; et al. Mycobacterium abscessus strain morphotype determines phage susceptibility, the repertoire of therapeutically useful phages, and phage resistance. MBio 2021, 12, e03431-20. [Google Scholar] [CrossRef]
- Dedrick, R.M.; Aull, H.G.; Jacobs-Sera, D.; Garlena, R.A.; Russell, D.A.; Smith, B.E.; Mahalingam, V.; Abad, L.; Gauthier, C.H.; Hatfull, G.F. The prophage and plasmid mobilome as a likely driver of Mycobacterium abscessus diversity. MBio 2021, 12, e03441-20. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hosseiniporgham, S.; Sechi, L.A. A Review on Mycobacteriophages: From Classification to Applications. Pathogens 2022, 11, 777. https://doi.org/10.3390/pathogens11070777
Hosseiniporgham S, Sechi LA. A Review on Mycobacteriophages: From Classification to Applications. Pathogens. 2022; 11(7):777. https://doi.org/10.3390/pathogens11070777
Chicago/Turabian StyleHosseiniporgham, Sepideh, and Leonardo A. Sechi. 2022. "A Review on Mycobacteriophages: From Classification to Applications" Pathogens 11, no. 7: 777. https://doi.org/10.3390/pathogens11070777
APA StyleHosseiniporgham, S., & Sechi, L. A. (2022). A Review on Mycobacteriophages: From Classification to Applications. Pathogens, 11(7), 777. https://doi.org/10.3390/pathogens11070777







