Analyzing the Systems Biology Effects of COVID-19 mRNA Vaccines to Assess Their Safety and Putative Side Effects
Abstract
1. Introduction
2. Results
2.1. Deriving Transcriptional Signatures for COVID-19 mRNA Vaccines
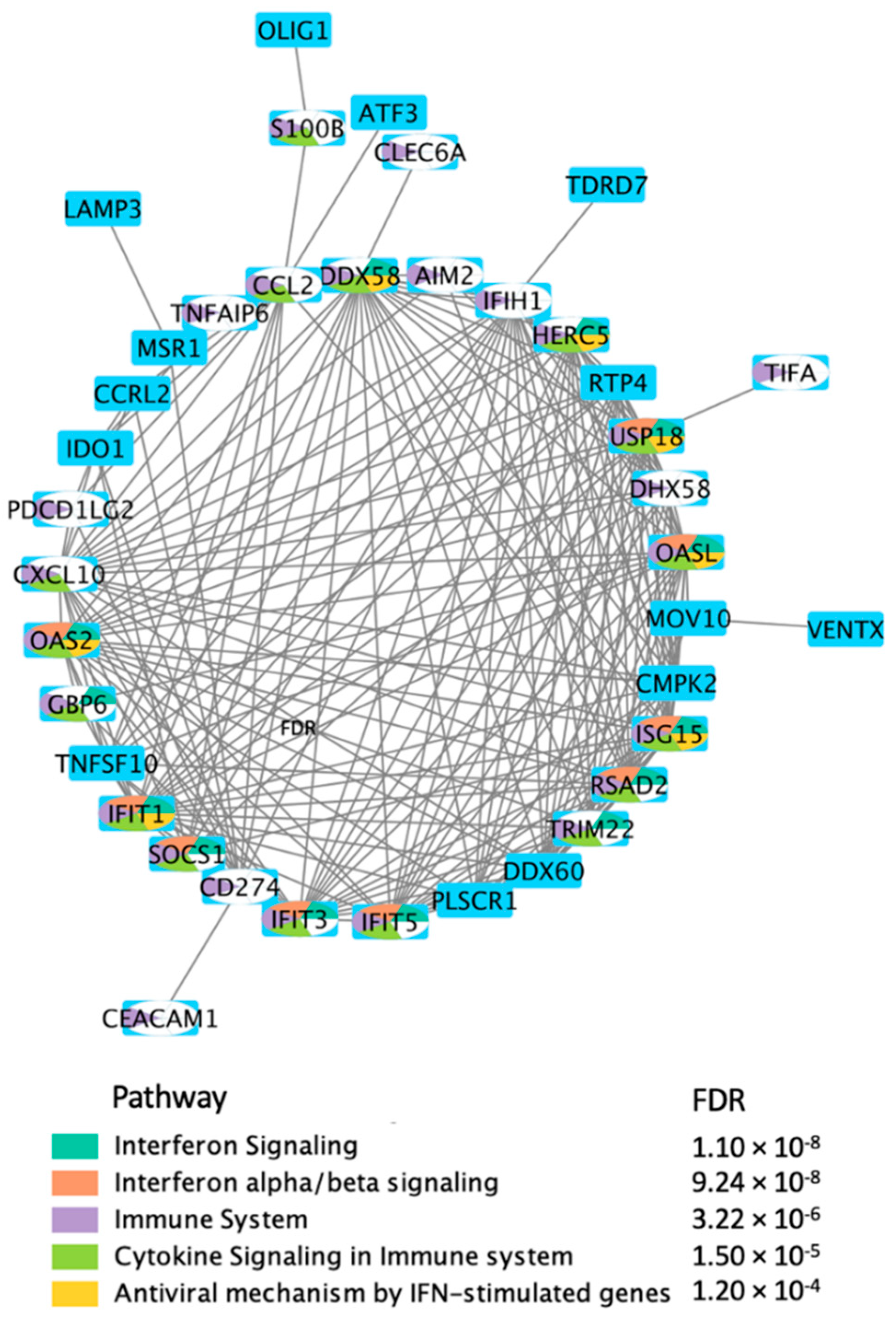
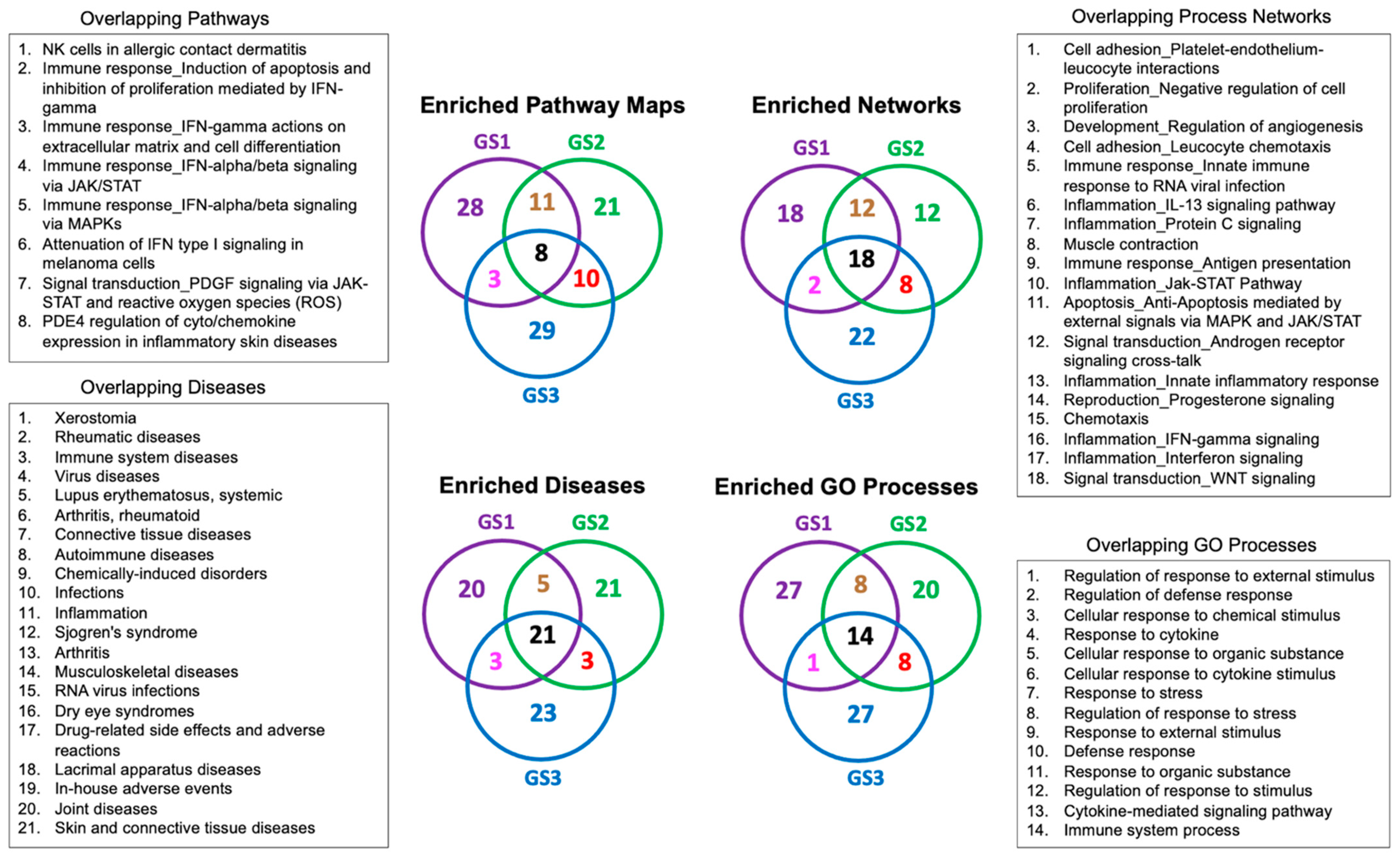
2.2. Enrichment Analysis of the Transcriptional Signatures
2.3. Identifying Transcriptomics Similarities with Small-Molecule Drugs to Aid in Predicting Potential Adverse Events
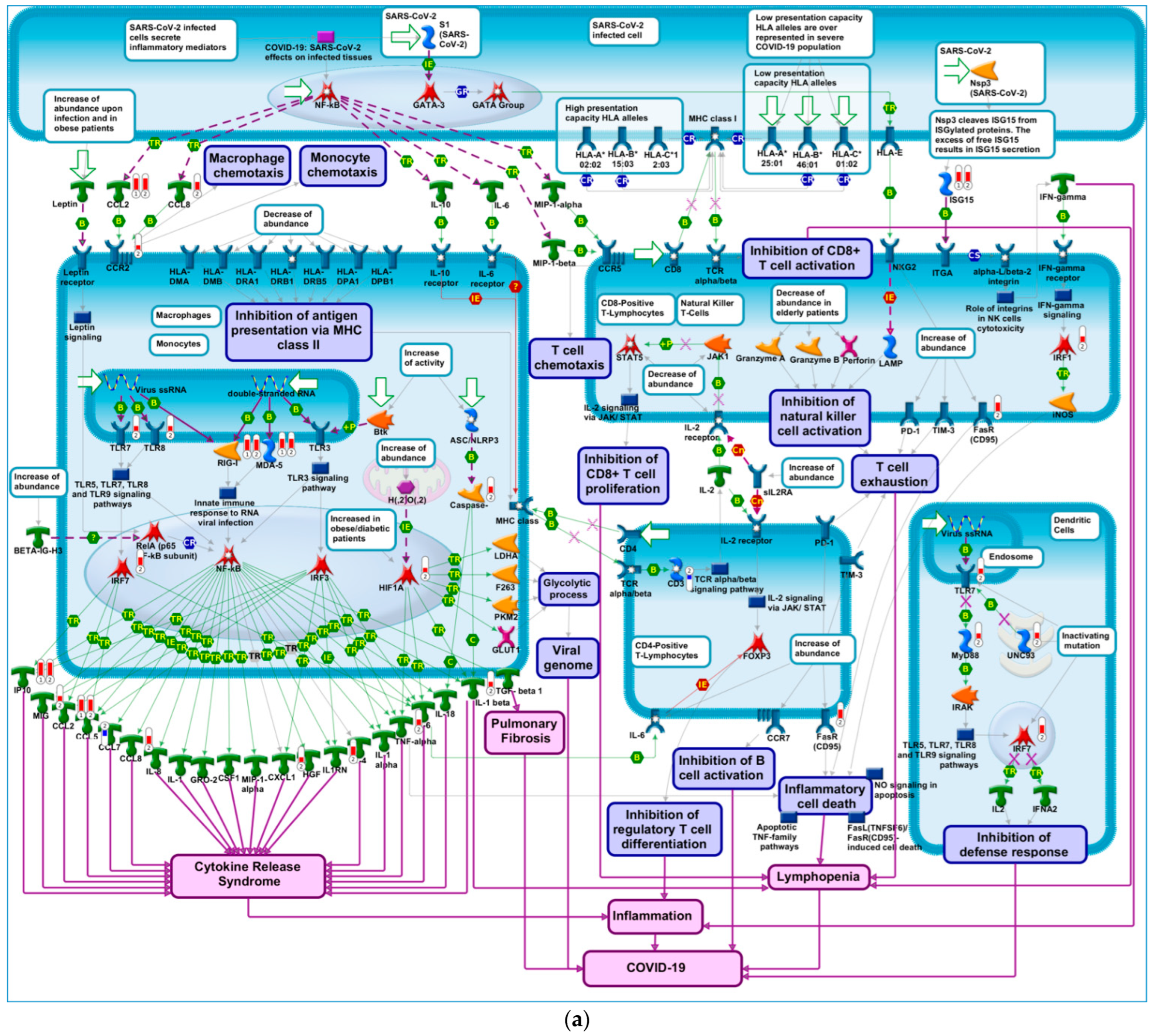
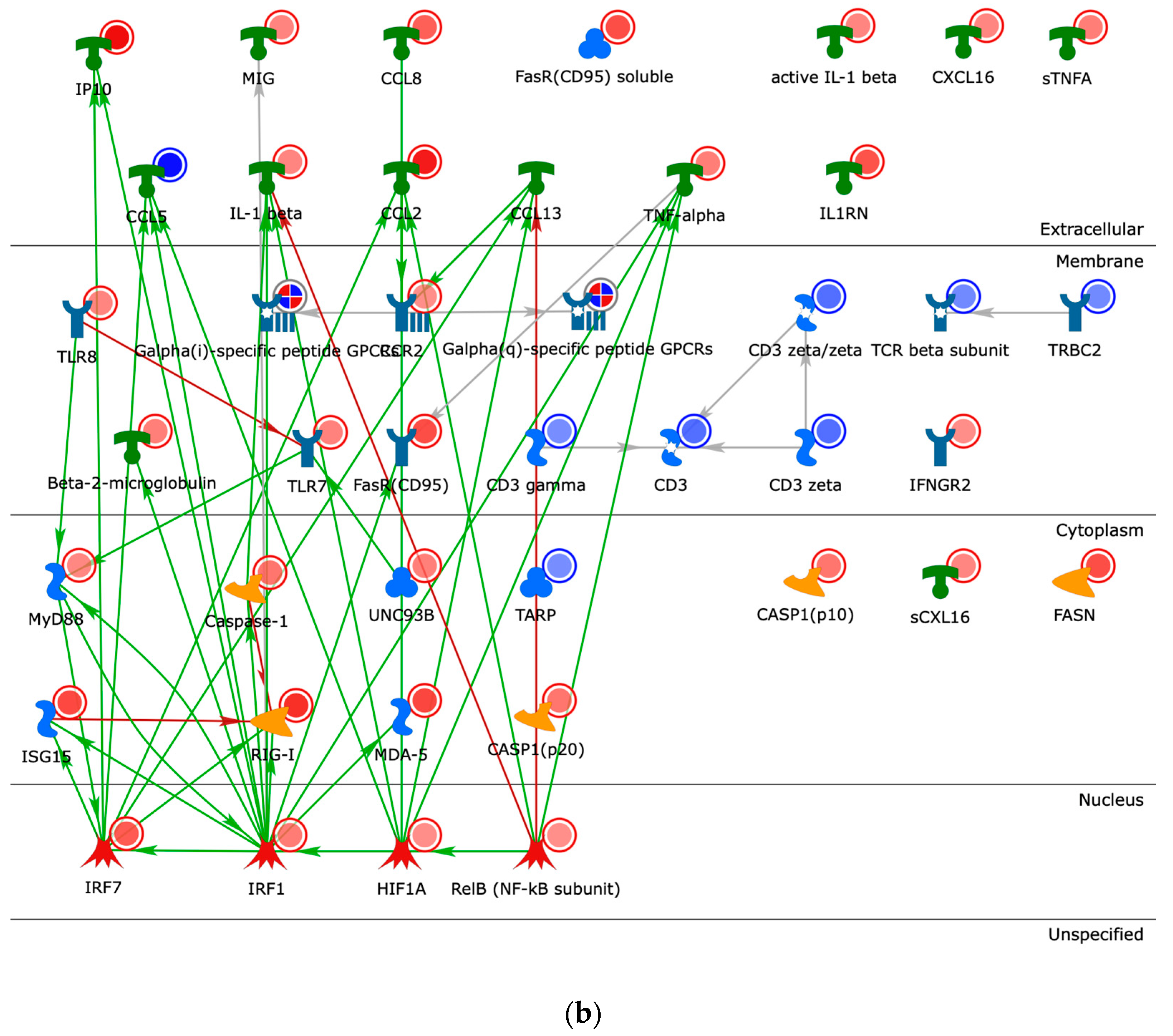
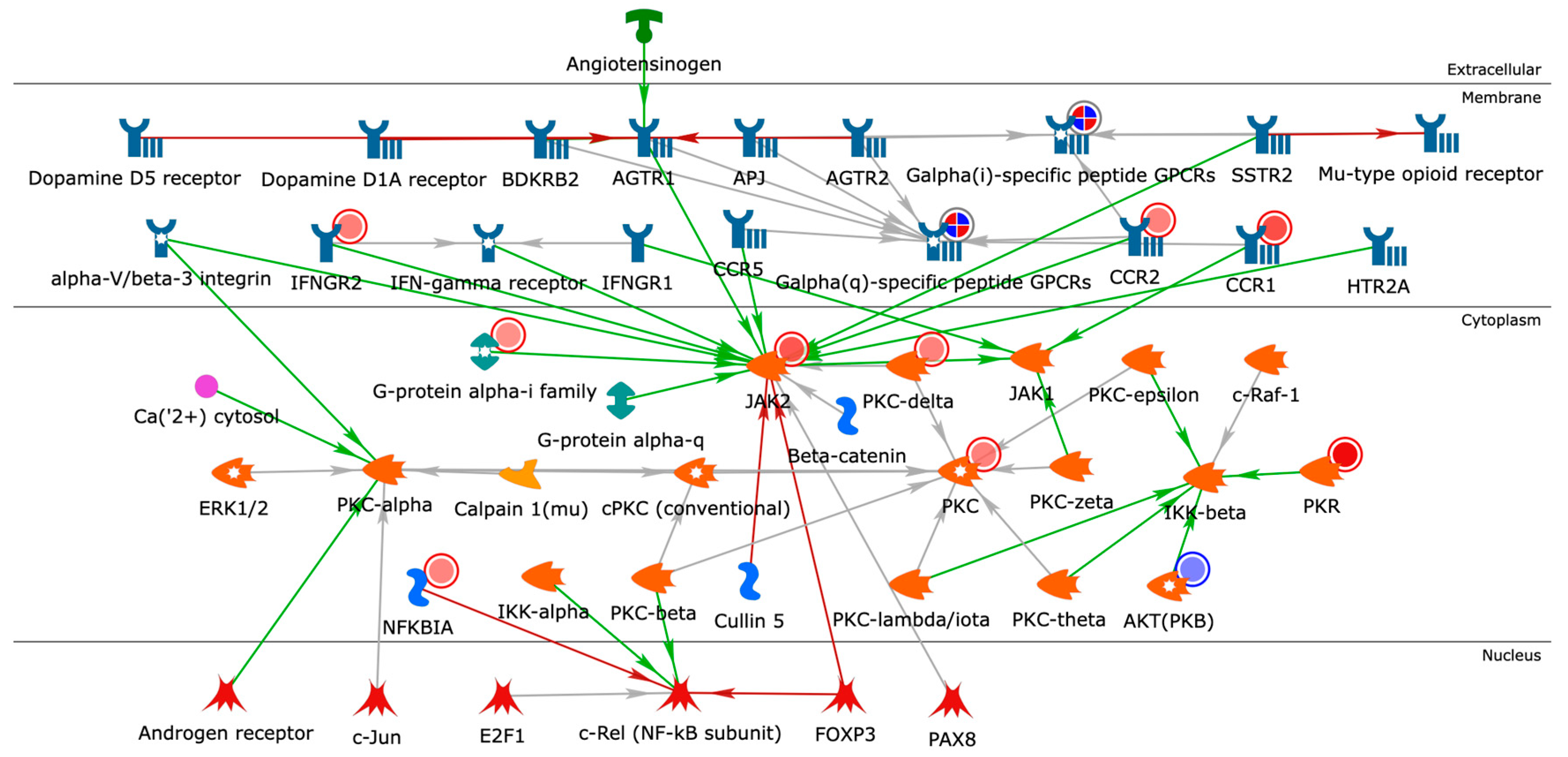
2.4. Building Networks for Drug Targets of CMap Compound Hits to Gain Biological Insights
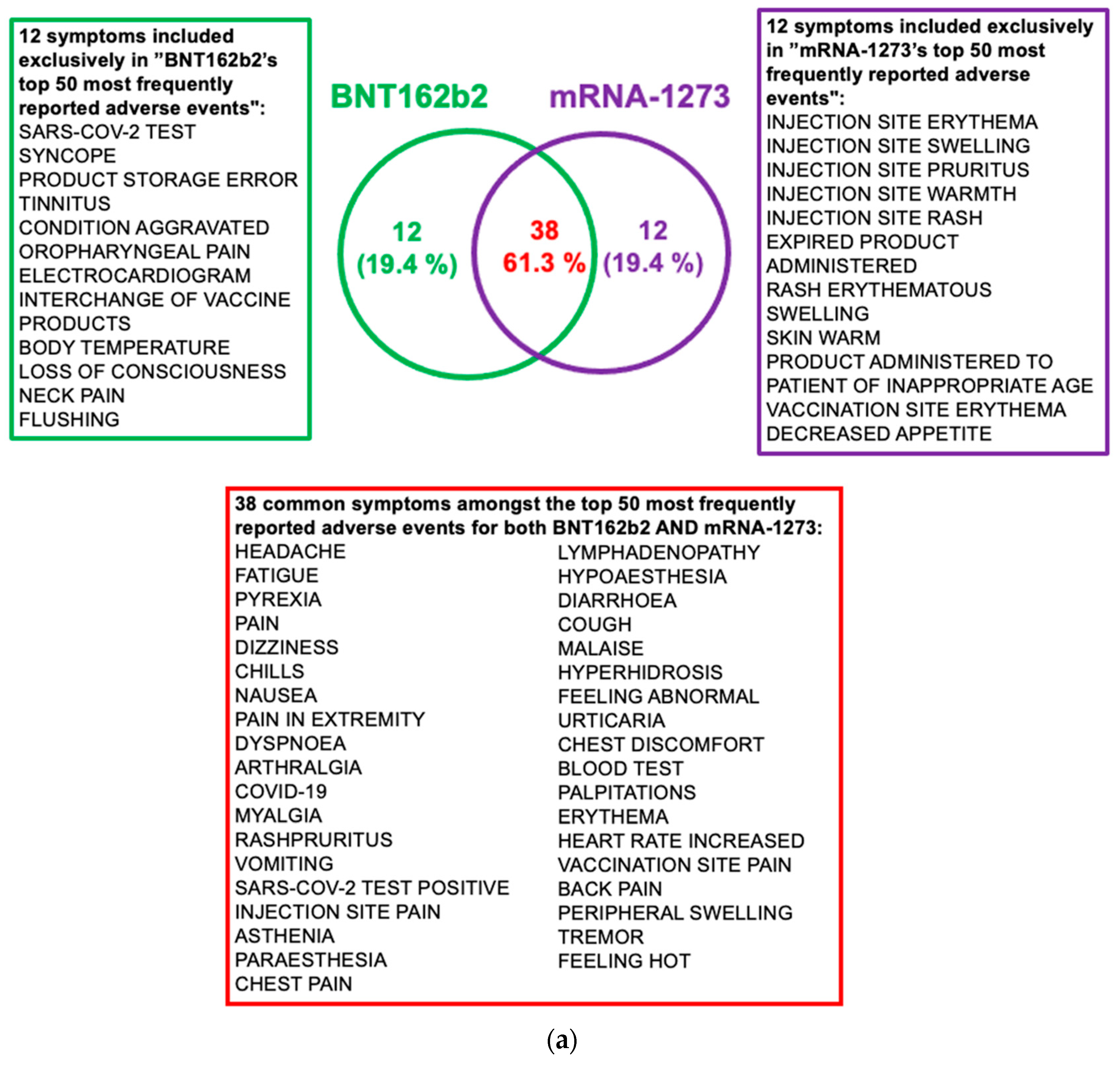
2.5. Post-Vaccine Side Effects Most Frequently Reported in VAERS
3. Discussion
4. Material and Methods
4.1. Integrative Informatics Workflow
4.2. Data Sets
4.2.1. Vaccine Transcriptional Gene Signatures
4.2.2. Vaccine Adverse Events Data Set
4.3. Databases
4.3.1. VAERS
4.3.2. MetaCore™
4.3.3. Comparative Toxicogenomics Database
4.4. Network Building
4.5. OmicSoft Studio
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACE2 | Angiotensin-converting enzyme 2 |
| Ang II | Angiopoietin II |
| BTK | Bruton’s Tyrosine Kinase |
| Calcium | Ca2+ |
| CDC | Centers for Disease Control and Prevention |
| CMap | Connectivity map |
| COVID-19 | Coronavirus disease of 2019 |
| CTD | Comparative toxicogenomics database |
| DEG | Differentially expressed gene |
| FC | Fold change |
| FDA | Food and Drug Administration |
| FDR | False discovery rate |
| GEO | Gene expression omnibus |
| GS | Gene signature |
| INF-gamma | Interferon gamma |
| LNP | Lipid nanoparticle |
| mRNA | Messenger ribonucleic acid |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
| TNF | Tumor necrosis factor |
| VAERS | Vaccine Adverse Event Reporting System |
References
- COVID-19 Vaccines Reduce Hospitalization, Death in People with Prior Infection, Study Finds. Available online: https://med.stanford.edu/news/all-news/2022/03/covid-19-vaccines-prior-infection.html (accessed on 1 May 2022).
- Bok, K.; Sitar, S.; Graham, B.S.; Mascola, J.R. Accelerated COVID-19 vaccine development: Milestones, lessons, and prospects. Immunity 2021, 54, 1636–1651. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.Y.; Thone, M.N.; Kwon, Y.J. COVID-19 vaccines: The status and perspectives in delivery points of view. Adv. Drug Deliv. Rev. 2020, 170, 1–25. [Google Scholar] [PubMed]
- Doshi, P. Will COVID-19 vaccines save lives? Current trials aren’t designed to tell us. BMJ 2020, 371, m4037. [Google Scholar] [CrossRef]
- Forni, G.; Mantovani, A. COVID-19 vaccines: Where we stand and challenges ahead. Cell Death Differ. 2021, 28, 626–639. [Google Scholar] [CrossRef] [PubMed]
- Francis, A.I.; Ghany, S.; Gilkes, T.; Umakanthan, S. Review of COVID-19 vaccine subtypes, efficacy and geographical distributions. Postgrad. Med. J. 2022, 98, 389–394. [Google Scholar] [CrossRef]
- Graham, B.S. Rapid COVID-19 vaccine development. Science 2020, 368, 945–946. [Google Scholar] [CrossRef]
- Hotez, P.J.; Nuzhath, T.; Callaghan, T.; Colwell, B. COVID-19 Vaccine Decisions: Considering the Choices and Opportunities. Microb. Infect. 2021, 23, 104811. [Google Scholar] [CrossRef] [PubMed]
- Haynes, B.F.; Corey, L.; Fernandes, P.; Gilbert, P.B.; Hotez, P.J.; Rao, S.; Santos, M.R.; Schuitemaker, H.; Watson, M.; Arvin, A. Prospects for a safe COVID-19 vaccine. Sci. Transl. Med. 2020, 12, eabe0948. [Google Scholar] [CrossRef]
- Pascual-Iglesias, A.; Canton, J.; Ortega-Prieto, A.M.; Jimenez-Guardeño, J.M.; Regla-Nava, J.A. An Overview of Vaccines against SARS-CoV-2 in the COVID-19 Pandemic Era. Pathogens 2021, 10, 1030. [Google Scholar] [CrossRef]
- Feng, Y.; Jiang, H.; Qiu, M.; Liu, L.; Zou, S.; Li, Y.; Guo, Q.; Han, N.; Sun, Y.; Wang, K.; et al. Multi-Epitope Vaccine Design Using an Immunoinformatic Approach for SARS-CoV-2. Pathogens 2021, 10, 737. [Google Scholar] [CrossRef]
- Hohan, R.; Milu, P.; Paraschiv, S.; Casangiu, C.; Tudor, A.; Vlaicu, O.; Banica, L.; Surleac, M.; Florea, D.; Otelea, D. The Predictive Value of Mutation Screening for Anticipating COVID-19 Waves. Pathogens 2021, 10, 1464. [Google Scholar] [CrossRef]
- Al-Qerem, W.A.; Jarab, A.S. COVID-19 vaccination acceptance and its associated factors among a Middle Eastern population. Public Health Front. 2021, 9, 34. [Google Scholar] [CrossRef]
- Pinheiro de Oliveira, F.; Mendes, R.H.; Dobbler, P.T.; Mai, V.; Pylro, V.S.; Waugh, S.G.; Vairo, F.; Refosco, L.F.; Roesch, L.F.; Schwartz, I.V. Phenylketonuria and Gut Microbiota: A Controlled Study Based on Next-Generation Sequencing. PLoS ONE 2016, 11, e0157513. [Google Scholar] [CrossRef]
- Kim, J.; Eygeris, Y.; Gupta, M.; Sahay, G. Self-assembled mRNA vaccines. Adv. Drug Deliv. Rev. 2021, 170, 83–112. [Google Scholar] [CrossRef]
- Kremsner, P.G.; Mann, P.; Kroidl, A.; Leroux-Roels, I.; Schindler, C.; Gabor, J.J.; Schunk, M.; Leroux-Roels, G.; Bosch, J.J.; Fendel, R.; et al. Safety and immunogenicity of an mRNA-lipid nanoparticle vaccine candidate against SARS-CoV-2: A phase 1 randomized clinical trial. Wien. Klin. Wochenschr. 2021, 133, 931–941. [Google Scholar] [CrossRef]
- Witjes, J.; Caris, C.; Mungan, N.; Debruyne, F.; Witjes, W. Results of a randomized phase III trial of sequential intravesical therapy with mitomycin C and bacillus Calmette-Guerin versus mitomycin C alone in patients with superficial bladder cancer. J. Urol. 1998, 160, 1668–1672. [Google Scholar] [CrossRef]
- Pfizer. Phase 1/2/3, Placebo-Controlled, Randomized, Observer-Blind, Dose-Finding Study to Evaluate the Safety, Tolerability, Immunogenicity, and Efficacy of SARS-CoV-2 RNA Vaccine Candidates against COVID-19 in Healthy Individuals. Available online: https://cdn.pfizer.com/pfizercom/2020-11/C4591001_Clinical_Protocol_Nov2020.pdf (accessed on 5 November 2021).
- Chen, T.; Yuan, S.; Wan, X.N.; Zhan, L.; Yu, X.Q.; Zeng, J.H.; Li, H.; Zhang, W.; Hu, X.Y.; Ye, Y.F.; et al. Chinese herb cinobufagin-reduced cancer pain is associated with increased peripheral opioids by invaded CD3/4/8 lymphocytes. Oncotarget 2017, 8, 11425–11441. [Google Scholar] [CrossRef][Green Version]
- Crommelin, D.J.A.; Anchordoquy, T.J.; Volkin, D.B.; Jiskoot, W.; Mastrobattista, E. Addressing the Cold Reality of mRNA Vaccine Stability. J. Pharm. Sci. 2021, 110, 997–1001. [Google Scholar] [CrossRef]
- Schoenmaker, L.; Witzigmann, D.; Kulkarni, J.A.; Verbeke, R.; Kersten, G.; Jiskoot, W.; Crommelin, D.J.A. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int. J. Pharm. 2021, 601, 120586. [Google Scholar] [CrossRef]
- Los, M.; Burek, C.J.; Stroh, C.; Benedyk, K.; Hug, H.; Mackiewicz, A. Anticancer drugs of tomorrow: Apoptotic pathways as targets for drug design. Drug Discov. Today 2003, 8, 67–77. [Google Scholar] [CrossRef]
- Crosti, P.; Malerba, M.; Bianchetti, R. Tunicamycin and Brefeldin A induce in plant cells a programmed cell death showing apoptotic features. Protoplasma 2001, 216, 31–38. [Google Scholar] [CrossRef]
- Dattilo, R.; Mottini, C.; Camera, E.; Lamolinara, A.; Auslander, N.; Doglioni, G.; Muscolini, M.; Tang, W.; Planque, M.; Ercolani, C. Pyrvinium Pamoate Induces Death of Triple-Negative Breast Cancer Stem–Like Cells and Reduces Metastases through Effects on Lipid Anabolism. Cancer Res. 2020, 80, 4087–4102. [Google Scholar] [CrossRef]
- Zhou, X.; Du, J.; Wang, H.; Chen, C.; Jiao, L.; Cheng, X.; Zhou, X.; Chen, S.; Gou, S.; Zhao, W. Repositioning liothyronine for cancer immunotherapy by blocking the interaction of immune checkpoint TIGIT/PVR. Cell Commun. Signal. 2020, 18, 142. [Google Scholar] [CrossRef]
- Von Niessen, A.G.O.; Poleganov, M.A.; Rechner, C.; Plaschke, A.; Kranz, L.M.; Fesser, S.; Diken, M.; Löwer, M.; Vallazza, B.; Beissert, T. Improving mRNA-based therapeutic gene delivery by expression-augmenting 3′ UTRs identified by cellular library screening. Mol. Ther. 2019, 27, 824–836. [Google Scholar] [CrossRef]
- Verbeke, R.; Lentacker, I.; De Smedt, S.C.; Dewitte, H. The dawn of mRNA vaccines: The COVID-19 case. J. Control. Release 2021, 333, 511–520. [Google Scholar] [CrossRef]
- Krantz, M.S.; Liu, Y.; Phillips, E.J.; Stone Jr, C.A. COVID-19 vaccine anaphylaxis: PEG or not? Allergy 2021, 76, 1934. [Google Scholar] [CrossRef]
- Uddin, M.N.; Roni, M.A. Challenges of Storage and Stability of mRNA-Based COVID-19 Vaccines. Vaccines 2021, 9, 1033. [Google Scholar] [CrossRef]
- Comai, S.; Bertazzo, A.; Bailoni, L.; Zancato, M.; Costa, C.V.; Allegri, G. Protein and non-protein (free and protein-bound) tryptophan in legume seeds. Food Chem. 2007, 103, 657–661. [Google Scholar] [CrossRef]
- Oluwagbemigun, K.; Anesi, A.; Ulaszewska, M.; Clarke, G.; Alexy, U.; Schmid, M.; Roden, M.; Herder, C.; Mattivi, F.; Nöthlings, U. Longitudinal relationship of amino acids and indole metabolites with long-term body mass index and cardiometabolic risk markers in young individuals. Sci. Rep. 2020, 10, 6399. [Google Scholar] [CrossRef]
- Konopelski, P.; Mogilnicka, I. Biological Effects of Indole-3-Propionic Acid, a Gut Microbiota-Derived Metabolite, and Its Precursor Tryptophan in Mammals’ Health and Disease. Int. J. Mol. Sci. 2022, 23, 1222. [Google Scholar] [CrossRef]
- Li, Y.; Tenchov, R.; Smoot, J.; Liu, C.; Watkins, S.; Zhou, Q. A comprehensive review of the global efforts on COVID-19 vaccine development. ACS Cent. Sci. 2021, 7, 512–533. [Google Scholar] [CrossRef] [PubMed]
- Rawat, K.; Kumari, P.; Saha, L. COVID-19 vaccine: A recent update in pipeline vaccines, their design and development strategies. Eur. J. Pharmacol. 2020, 892, 173751. [Google Scholar] [CrossRef] [PubMed]
- Samaranayake, L.P.; Seneviratne, C.J.; Fakhruddin, K.S. Coronavirus disease 2019 (COVID-19) vaccines: A concise review. Oral Dis. 2021, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Teo, S.P. Review of COVID-19 mRNA Vaccines: BNT162b2 and mRNA-1273. J. Pharm. Pract. 2021, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Peng, Y.; Xu, H.; Cui, Z.; Williams, R.O. The COVID-19 vaccine race: Challenges and opportunities in vaccine formulation. AAPS PharmSciTech 2020, 21, 225. [Google Scholar] [CrossRef]
- Sangli, S.; Virani, A.; Cheronis, N.; Vannatter, B.; Minich, C.; Noronha, S.; Bhagavatula, R.; Speredelozzi, D.; Sareen, M.; Kaplan, R.B. Thrombosis With Thrombocytopenia After the Messenger RNA–1273 Vaccine. Ann. Intern. Med. 2021, 174, 1480–1482. [Google Scholar] [CrossRef]
- Self, W.H.; Tenforde, M.W.; Rhoads, J.P.; Gaglani, M.; Ginde, A.A.; Douin, D.J.; Olson, S.M.; Talbot, H.K.; Casey, J.D.; Mohr, N.M. Comparative effectiveness of Moderna, Pfizer-BioNTech, and Janssen (Johnson & Johnson) vaccines in preventing COVID-19 hospitalizations among adults without immunocompromising conditions—United States, March–August 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 1337–1343. [Google Scholar]
- Murray, C.J.; Piot, P. The potential future of the COVID-19 pandemic: Will SARS-CoV-2 become a recurrent seasonal infection? JAMA 2021, 325, 1249–1250. [Google Scholar] [CrossRef]
- Dolgin, E. CureVac COVID vaccine let-down spotlights mRNA design challenges. Nature 2021, 594, 483. [Google Scholar] [CrossRef]
- Cohen, J. What went wrong with CureVac’s mRNA vaccine? Science 2021, 372, 1381. [Google Scholar]
- Jackson, L.A.; Anderson, E.J.; Rouphael, N.G.; Roberts, P.C.; Makhene, M.; Coler, R.N.; McCullough, M.P.; Chappell, J.D.; Denison, M.R.; Stevens, L.J. An mRNA vaccine against SARS-CoV-2—preliminary report. N. Engl. J. Med. 2020, 383, 1920–1931. [Google Scholar] [CrossRef] [PubMed]
- ElBagoury, M.; Tolba, M.M.; Nasser, H.A.; Jabbar, A.; Hutchinson, A. The find of COVID-19 vaccine: Challenges and opportunities. J. Infect. Public Health 2020, 14, 389–416. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.; Wu, N.C.; Mok, C.K. COVID-19 vaccines: Knowing the unknown. Eur. J. Immunol. 2020, 50, 939–943. [Google Scholar] [CrossRef] [PubMed]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
- Sahin, U.; Muik, A.; Derhovanessian, E.; Vogler, I.; Kranz, L.M.; Vormehr, M.; Baum, A.; Pascal, K.; Quandt, J.; Maurus, D. COVID-19 vaccine BNT162b1 elicits human antibody and TH 1 T cell responses. Nature 2020, 586, 594–599. [Google Scholar] [CrossRef] [PubMed]
- Jeyanathan, M.; Afkhami, S.; Smaill, F.; Miller, M.S.; Lichty, B.D.; Xing, Z. Immunological considerations for COVID-19 vaccine strategies. Nat. Rev. Immunol. 2020, 20, 615–632. [Google Scholar] [CrossRef]
- ModernaTX, A. A Study to Evaluate Efficacy, Safety, and Immunogenicity of mRNA-1273 Vaccine in Adults Aged 18 Years and Older to Prevent COVID-19. Available online: https://clinicaltrials.gov/ct2/show/NCT04470427 (accessed on 31 October 2021).
- Zhang, L.; Zlotoff, D.A.; Awadalla, M.; Mahmood, S.S.; Nohria, A.; Hassan, M.Z.; Thuny, F.; Zubiri, L.; Chen, C.L.; Sullivan, R.J. Major adverse cardiovascular events and the timing and dose of corticosteroids in immune checkpoint Inhibitor–Associated myocarditis. Circulation 2020, 141, 2031–2034. [Google Scholar] [CrossRef]
- Karikó, K.; Buckstein, M.; Ni, H.; Weissman, D. Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity 2005, 23, 165–175. [Google Scholar] [CrossRef]
- Lee, G.M. The importance of context in COVID-19 vaccine safety. N. Engl. J. Med. 2021, 385, 1138–1140. [Google Scholar] [CrossRef]
- Moline, H.L.; Whitaker, M.; Deng, L.; Rhodes, J.C.; Milucky, J.; Pham, H.; Patel, K.; Anglin, O.; Reingold, A.; Chai, S.J. Effectiveness of COVID-19 vaccines in preventing hospitalization among adults aged ≥65 years COVID-NET, 13 states, February–April 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 1088–1093. [Google Scholar] [CrossRef]
- Rosenblum, H.G.; Hadler, S.C.; Moulia, D.; Shimabukuro, T.T.; Su, J.R.; Tepper, N.K.; Ess, K.C.; Woo, E.J.; Mba-Jonas, A.; Alimchandani, M. Use of COVID-19 vaccines after reports of adverse events among adult recipients of Janssen (Johnson & Johnson) and mRNA COVID-19 vaccines (Pfizer-BioNTech and Moderna): Update from the Advisory Committee on Immunization Practices—United States, July 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 1094–1099. [Google Scholar] [PubMed]
- MacNeil, J.R.; Su, J.R.; Broder, K.R.; Guh, A.Y.; Gargano, J.W.; Wallace, M.; Hadler, S.C.; Scobie, H.M.; Blain, A.E.; Moulia, D. Updated recommendations from the advisory committee on immunization practices for use of the Janssen (Johnson & Johnson) COVID-19 vaccine after reports of thrombosis with thrombocytopenia syndrome among vaccine recipients—United States, April 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 651. [Google Scholar] [PubMed]
- Pardi, N.; Tuyishime, S.; Muramatsu, H.; Kariko, K.; Mui, B.L.; Tam, Y.K.; Madden, T.D.; Hope, M.J.; Weissman, D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release 2015, 217, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Hajjo, R.; Sabbah, D.A.; Bardaweel, S.K.; Tropsha, A. Shedding the Light on Post-Vaccine Myocarditis and Pericarditis in COVID-19 and Non-COVID-19 Vaccine Recipients. Vaccines 2021, 9, 1186. [Google Scholar] [CrossRef] [PubMed]
- García, J.B.; Ortega, P.P.; Fernández, A.B.; León, A.C.; Burgos, L.R.; Dorta, E.C. Acute myocarditis after administration of the BNT162b2 vaccine against COVID-19. Rev. Esp. Cardiol. 2021, 74, 812–814. [Google Scholar] [CrossRef]
- Coyle, J.; Igbinomwanhia, E.; Sanchez-Nadales, A.; Danciu, S.; Chu, C.; Shah, N. A recovered case of COVID-19 myocarditis and ARDS treated with corticosteroids, tocilizumab, and experimental AT-001. Case Rep. 2020, 2, 1331–1336. [Google Scholar] [CrossRef]
- Calcaterra, G.; Mehta, J.L.; de Gregorio, C.; Butera, G.; Neroni, P.; Fanos, V.; Bassareo, P.P. COVID 19 Vaccine for Adolescents. Concern about Myocarditis and Pericarditis. Pediatr. Rep. 2021, 13, 530–533. [Google Scholar] [CrossRef]
- Turner, J.S.; O’Halloran, J.A.; Kalaidina, E.; Kim, W.; Schmitz, A.J.; Zhou, J.Q.; Lei, T.; Thapa, M.; Chen, R.E.; Case, J.B.; et al. SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature 2021, 596, 109–113. [Google Scholar] [CrossRef]
- Seneff, S.; Nigh, G.; Kyriakopoulos, A.M.; McCullough, P.A. Innate immune suppression by SARS-CoV-2 mRNA vaccinations: The role of G-quadruplexes, exosomes, and MicroRNAs. Food Chem. Toxicol. 2022, 164, 113008. [Google Scholar] [CrossRef]
- Federico, M. Biological and Immune Responses to Current Anti-SARS-CoV-2 mRNA Vaccines beyond Anti-Spike Antibody Production. J. Immunol. Res. 2022, 2022, 4028577. [Google Scholar] [CrossRef]
- Tai, W.; Zhao, G.; Sun, S.; Guo, Y.; Wang, Y.; Tao, X.; Tseng, C.K.; Li, F.; Jiang, S.; Du, L.; et al. A recombinant receptor-binding domain of MERS-CoV in trimeric form protects human dipeptidyl peptidase 4 (hDPP4) transgenic mice from MERS-CoV infection. Virology 2016, 499, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Oliver, S.E. The advisory committee on immunization practices’ interim recommendation for use of moderna COVID-19 vaccine—United States, December 2020. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 1653–1656. [Google Scholar] [CrossRef] [PubMed]
- Ackerman, M.; Henry, D.; Finon, A.; Binois, R.; Esteve, E. Persistent maculopapular rash after the first dose of Pfizer-BioNTech COVID-19 vaccine. J. Eur. Acad. Dermatol. Venereol. 2021, 35, e423–e425. [Google Scholar] [CrossRef]
- Buonaguro, L.; Pulendran, B. Immunogenomics and systems biology of vaccines. Immunol. Rev. 2011, 239, 197–208. [Google Scholar] [CrossRef]
- Ben-Othman, R.; Cai, B.; Liu, A.C.; Varankovich, N.; He, D.; Blimkie, T.M.; Lee, A.H.; Gill, E.E.; Novotny, M.; Aevermann, B.; et al. Systems Biology Methods Applied to Blood and Tissue for a Comprehensive Analysis of Immune Response to Hepatitis B Vaccine in Adults. Front. Immunol. 2020, 11, 580373. [Google Scholar] [CrossRef]
- Six, A.; Bellier, B.; Thomas-Vaslin, V.; Klatzmann, D. Systems biology in vaccine design. Microb. Biotechnol. 2012, 5, 295–304. [Google Scholar] [CrossRef]
- Whitaker, J.A.; Ovsyannikova, I.G.; Poland, G.A. Adversomics: A new paradigm for vaccine safety and design. Expert Rev. Vaccines 2015, 14, 935–947. [Google Scholar] [CrossRef]
- Hajjo, R.; Setola, V.; Roth, B.L.; Tropsha, A. Chemocentric informatics approach to drug discovery: Identification and experimental validation of selective estrogen receptor modulators as ligands of 5-hydroxytryptamine-6 receptors and as potential cognition enhancers. J. Med. Chem. 2012, 55, 5704–5719. [Google Scholar] [CrossRef]
- Jimenez, R.C.; Corpas, M. Bioinformatics workflows and web services in systems biology made easy for experimentalists. Methods Mol. Biol. 2013, 1021, 299–310. [Google Scholar] [CrossRef]
- Hajjo, R.; Tropsha, A. A Systems Biology Workflow for Drug and Vaccine Repurposing: Identifying Small-Molecule BCG Mimics to Reduce or Prevent COVID-19 Mortality. Pharm. Res. 2020, 37, 212. [Google Scholar] [CrossRef] [PubMed]
- Kirwan, G.M.; Johansson, E.; Kleemann, R.; Verheij, E.R.; Wheelock, Å.M.; Goto, S.; Trygg, J.; Wheelock, C.E. Building multivariate systems biology models. Anal. Chem. 2012, 84, 7064–7071. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Matsuoka, Y.; Asai, Y.; Hsin, K.-Y.; Kitano, H. Software for systems biology: From tools to integrated platforms. Nat. Rev. Genet. 2011, 12, 821–832. [Google Scholar] [CrossRef] [PubMed]
- MetaCoreTM Version 20.3 Build 2021. Available online: https://portal.genego.com/ (accessed on 3 March 2022).
- Merad, M.; Martin, J.C. Pathological inflammation in patients with COVID-19: A key role for monocytes and macrophages. Nat. Rev. Immunol. 2020, 20, 355–362. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, J.; Liu, C.; Su, L.; Zhang, D.; Fan, J.; Yang, Y.; Xiao, M.; Xie, J.; Xu, Y.; et al. IP-10 and MCP-1 as biomarkers associated with disease severity of COVID-19. Mol. Med. 2020, 26, 97. [Google Scholar] [CrossRef]
- Keshavarzian, A.; Engen, P.; Bonvegna, S.; Cilia, R. The gut microbiome in Parkinson’s disease: A culprit or a bystander? Prog. Brain Res. 2020, 252, 357–450. [Google Scholar] [CrossRef]
- Birge, R.B.; Ucker, D.S. Innate apoptotic immunity: The calming touch of death. Cell Death Differ. 2008, 15, 1096–1102. [Google Scholar] [CrossRef]
- Azcona-Olivera, J.; Ouyang, Y.-L.; Warner, R.; Linz, J.; Pestka, J. Effects of vomitoxin (deoxynivalenol) and cycloheximide on IL-2, 4, 5 and 6 secretion and mRNA levels in murine CD4+ cells. Food Chem. Toxicol. 1995, 33, 433–441. [Google Scholar] [CrossRef]
- Jiang, Q.; Wei, H.; Tian, Z. Poly I: C enhances cycloheximide-induced apoptosis of tumor cells through TLR3 pathway. BMC Cancer 2008, 8, 12. [Google Scholar] [CrossRef]
- Santiago, B.; Galindo, M.; Palao, G.; Pablos, J.L. Intracellular regulation of Fas-induced apoptosis in human fibroblasts by extracellular factors and cycloheximide. J. Immunol. 2004, 172, 560–566. [Google Scholar] [CrossRef]
- Baskić, D.; Popović, S.; Ristić, P.; Arsenijević, N.N. Analysis of cycloheximide-induced apoptosis in human leukocytes: Fluorescence microscopy using annexin V/propidium iodide versus acridin orange/ethidium bromide. Cell Biol. Int. 2006, 30, 924–932. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, Y.; Turki, T. A new advanced in silico drug discovery method for novel coronavirus (SARS-CoV-2) with tensor decomposition-based unsupervised feature extraction. PLoS ONE 2020, 15, e0238907. [Google Scholar] [CrossRef] [PubMed]
- Blázquez, A.-B.; Saiz, J.-C. Potential for Protein Kinase Pharmacological Regulation in Flaviviridae Infections. Int. J. Mol. Sci. 2020, 21, 9524. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-T.; Chao, T.-L.; Kao, H.-C.; Pang, Y.-H.; Lee, W.-H.; Hsieh, C.-H.; Chang, S.-Y.; Huang, H.-C.; Juan, H.-F. Enhancement of the IFN-β-induced host signature informs repurposed drugs for COVID-19. Heliyon 2020, 6, e05646. [Google Scholar] [CrossRef] [PubMed]
- Weng, T.-Y.; Wu, H.F.; Li, C.-Y.; Hung, Y.-H.; Chang, Y.-W.; Chen, Y.-L.; Hsu, H.-P.; Chen, Y.-H.; Wang, C.-Y.; Chang, J.-Y. Homoharringtonine induced immune alteration for an efficient anti-tumor response in mouse models of non-small cell lung adenocarcinoma expressing Kras mutation. Sci. Rep. 2018, 8, 8216. [Google Scholar] [CrossRef]
- Zhu, M.; Gong, Z.; Wu, Q.; Su, Q.; Yang, T.; Yu, R.; Xu, R.; Zhang, Y. Homoharringtonine suppresses tumor proliferation and migration by regulating EphB4-mediated β-catenin loss in hepatocellular carcinoma. Cell Death Dis. 2020, 11, 632. [Google Scholar] [CrossRef]
- Zheng, Y.; Fang, Z.; Xue, Y.; Zhang, J.; Zhu, J.; Gao, R.; Yao, S.; Ye, Y.; Wang, S.; Lin, C. Specific gut microbiome signature predicts the early-stage lung cancer. Gut Microbes 2020, 11, 1030–1042. [Google Scholar] [CrossRef]
- Schneider, N.F.Z.; Cerella, C.; Simões, C.M.O.; Diederich, M. Anticancer and immunogenic properties of cardiac glycosides. Molecules 2017, 22, 1932. [Google Scholar] [CrossRef]
- Li, Y.; Li, J.; Zhou, K.; He, J.; Cao, J.; An, M.; Chang, Y.-X. A review on phytochemistry and pharmacology of Cortex Periplocae. Molecules 2016, 21, 1702. [Google Scholar] [CrossRef]
- Deng, L.J.; Qi, M.; Li, N.; Lei, Y.H.; Zhang, D.M.; Chen, J.X. Natural products and their derivatives: Promising modulators of tumor immunotherapy. J. Leukoc. Biol. 2020, 108, 493–508. [Google Scholar] [CrossRef]
- Lage, G.; Spratt, J. Antagonism of intravenous digitoxigenin lethality by reserpine pretreatment in the mouse. Proc. Soc. Exp. Biol. Med. 1967, 125, 580–583. [Google Scholar] [CrossRef] [PubMed]
- Möller, M.; Herzer, K.; Wenger, T.; Herr, I.; Wink, M. The alkaloid emetine as a promising agent for the induction and enhancement of drug-induced apoptosis in leukemia cells. Oncol. Rep. 2007, 18, 737–744. [Google Scholar] [CrossRef] [PubMed]
- Csuka, I.; Antoni, F. The effect of emetine on the immune response of mice. Biochem. Pharmacol. 1984, 33, 2061–2063. [Google Scholar] [CrossRef]
- Rodrigues-Mascarenhas, S.; De Oliveira, A.D.S.; Amoedo, N.D.; Affonso-Mitidieri, O.R.; Rumjanek, F.D.; Rumjanek, V.M. Modulation of the immune system by ouabain. Ann. N. Y. Acad. Sci. 2009, 1153, 153–163. [Google Scholar] [CrossRef]
- Cavalcante-Silva, L.H.; Lima, E.D.A.; Carvalho, D.; Sales-Neto, J.M.D.; Alves, A.K.; Galvão, J.G.; Silva, J.S.; Rodrigues-Mascarenhas, S. Much more than a cardiotonic steroid: Modulation of inflammation by ouabain. Front. Physiol. 2017, 8, 895. [Google Scholar] [CrossRef]
- Yu, Y. Repurposing glucocorticoids as adjuvant reagents for immune checkpoint inhibitors in solid cancers. Cancer Biol. Med. 2021, 18, 944. [Google Scholar] [CrossRef]
- Jain, P.; Klotz, J.; Dunavin, N.; Lu, K.; Koklanaris, E.; Draper, D.; Superata, J.; Chinian, F.; Yu, Q.; Keyvanfar, K. Cellular immune profiling after sequential clofarabine and lenalidomide for high risk myelodysplastic syndromes and acute myeloid leukemia. Leuk. Res. Rep. 2017, 7, 40–44. [Google Scholar] [CrossRef]
- Juang, Y.-P.; Liang, P.-H. Biological and pharmacological effects of synthetic saponins. Molecules 2020, 25, 4974. [Google Scholar] [CrossRef]
- Pollard, B.S.; Blanco, J.C.; Pollard, J.R. Classical drug digitoxin inhibits influenza cytokine storm, with implications for COVID-19 therapy. In Vivo 2020, 34, 3723–3730. [Google Scholar] [CrossRef]
- Honda, H.; Nagai, Y.; Matsunaga, T.; Saitoh, S.i.; Akashi-Takamura, S.; Hayashi, H.; Fujii, I.; Miyake, K.; Muraguchi, A.; Takatsu, K. Glycyrrhizin and isoliquiritigenin suppress the LPS sensor Toll-like receptor 4/MD-2 complex signaling in a different manner. J. Leukoc. Biol. 2012, 91, 967–976. [Google Scholar] [CrossRef]
- Hickey, A.R.; Wenger, T.L.; Carpenter, V.P.; Tilson, H.H.; Hlatky, M.A.; Furberg, C.D.; Kirkpatrick, C.H.; Strauss, H.C.; Smith, T.W. Digoxin immune Fab therapy in the management of digitalis intoxication: Safety and efficacy results of an observational surveillance study. J. Am. Coll. Cardiol. 1991, 17, 590–598. [Google Scholar] [CrossRef]
- Hauptman, P.J.; Blume, S.W.; Lewis, E.F.; Ward, S. Digoxin toxicity and use of digoxin immune fab: Insights from a national hospital database. JACC Heart Fail. 2016, 4, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Moore, B.A.; Turler, A.; Pezzone, M.A.; Dyer, K.; Grandis, J.; Bauer, A.J. Tyrphostin AG 126 inhibits development of postoperative ileus induced by surgical manipulation of murine colon. Am. J. Physiol. Gastrointest. Liver Physiol. 2004, 286, G214–G224. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Huang, S.; Wu, Y. UNBS5162 and amonafide inhibits tumor progression in human melanoma by the AKT/mTOR pathway. Cancer Manag. Res. 2019, 11, 2339. [Google Scholar] [CrossRef]
- Tan, Q.; Hu, J.; Zhou, Y.; Wan, Y.; Zhang, C.; Liu, X.; Long, X.; Tan, F.; Zhao, X. Inhibitory Effect of Lactococcus lactis subsp. lactis HFY14 on Diphenoxylate-Induced Constipation in Mice by Regulating the VIP-cAMP-PKA-AQP3 Signaling Pathway. Drug Des. Devel. Ther. 2021, 15, 1971. [Google Scholar] [CrossRef]
- Hughes, B.; Taylor, M.; Sharma, R. Effects of verrucarin A and roridin A, macrocyclic trichothecene mycotoxins, on the murine immune system. Immunopharmacology 1988, 16, 79–87. [Google Scholar] [CrossRef]
- Gambhir, L.; Checker, R.; Sharma, D.; Thoh, M.; Patil, A.; Degani, M.; Gota, V.; Sandur, S.K. Thiol dependent NF-κB suppression and inhibition of T-cell mediated adaptive immune responses by a naturally occurring steroidal lactone Withaferin A. Toxicol. Appl. Pharmacol. 2015, 289, 297–312. [Google Scholar] [CrossRef]
- Estève, C.; Samson, M.; Guilhem, A.; Nicolas, B.; Leguy-Seguin, V.; Berthier, S.; Bonnotte, B.; Audia, S. Efficacy and safety of dapsone as second line therapy for adult immune thrombocytopenia: A retrospective study of 42 patients. PLoS ONE 2017, 12, e0187296. [Google Scholar] [CrossRef]
- Giaccone, G.; Donadio, M.; Bonardi, G.; Testore, F.; Calciati, A. Teniposide in the treatment of small-cell lung cancer: The influence of prior chemotherapy. Clin. Oncol. 1988, 6, 1264–1270. [Google Scholar] [CrossRef]
- Shepherd, G.M. Hypersensitivity reactions to chemotherapeutic drugs. Clin. Rev. Allergy Immunol. 2003, 24, 253–262. [Google Scholar] [CrossRef]
- Schmidt, A.; Krieg, J.; Clement, H.; Hemmeter, U.; Schulz, E.; Vedder, H.; Heiser, P. Effects of quetiapine, risperidone, 9-hydroxyrisperidone and ziprasidone on the survival of human neuronal and immune cells in vitro. Psychopharmacology 2010, 24, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Mead, J.; Aliya, N.; Wang, L.; Cuconati, A.; Wei, L.; Li, K.; Block, T.M.; Guo, J.-T.; Chang, J. RO 90–7501 Enhances TLR3 and RLR Agonist Induced Antiviral Response. PLoS ONE 2012, 7, e42583. [Google Scholar] [CrossRef] [PubMed]
- Pierson, S.K.; Stonestrom, A.J.; Shilling, D.; Ruth, J.; Nabel, C.S.; Singh, A.; Ren, Y.; Stone, K.; Li, H.; van Rhee, F.; et al. Plasma proteomics identifies a ‘chemokine storm’ in idiopathic multicentric Castleman disease. Am. J. Hematol. 2018, 93, 902–912. [Google Scholar] [CrossRef] [PubMed]
- Ersvaer, E.; Kittang, A.O.; Hampson, P.; Sand, K.; Gjertsen, B.T.; Lord, J.M.; Bruserud, Ø. The protein kinase C agonist PEP005 (ingenol 3-angelate) in the treatment of human cancer: A balance between efficacy and toxicity. Toxins 2010, 2, 174–194. [Google Scholar] [CrossRef]
- Kwaa, A.K.; Goldsborough, K.; Walker-Sperling, V.E.; Pianowski, L.F.; Gama, L.; Blankson, J.N. The effect of Ingenol-B on the suppressive capacity of elite suppressor HIV-specific CD8+ T cells. PLoS ONE 2017, 12, e0174516. [Google Scholar] [CrossRef]
- Kim, M.; Lee, S.-J.; Shin, S.; Park, K.-S.; Park, S.Y.; Lee, C.H. Novel natural killer cell-mediated cancer immunotherapeutic activity of anisomycin against hepatocellular carcinoma cells. Sci. Rep. 2018, 8, 10668. [Google Scholar] [CrossRef]
- Li, R.-Z.; Fan, X.-X.; Duan, F.-G.; Jiang, Z.-B.; Pan, H.-D.; Luo, L.-X.; Zhou, Y.-L.; Li, Y.; Yao, Y.-J.; Yao, X.-J. Proscillaridin A induces apoptosis and suppresses non-small-cell lung cancer tumor growth via calcium-induced DR4 upregulation. Cell Death Dis. 2018, 9, 696. [Google Scholar] [CrossRef]
- Li, H.; Chiappinelli, K.B.; Guzzetta, A.A.; Easwaran, H.; Yen, R.-W.C.; Vatapalli, R.; Topper, M.J.; Luo, J.; Connolly, R.M.; Azad, N.S. Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epithelial cancers. Oncotarget 2014, 5, 587. [Google Scholar] [CrossRef]
- Scofield, R.H.; Kurien, B.T.; Ganick, S.; McClain, M.T.; Pye, Q.; James, J.A.; Schneider, R.I.; Broyles, R.H.; Bachmann, M.; Hensley, K. Modification of lupus-associated 60-kDa Ro protein with the lipid oxidation product 4-hydroxy-2-nonenal increases antigenicity and facilitates epitope spreading. Free Radic. Biol. Med. 2005, 38, 719–728. [Google Scholar] [CrossRef]
- Masci, V.L.; Bernardini, S.; Modesti, L.; Ovidi, E.; Tiezzi, A. Medicinal Plants as a Source of Alkaloids. In Medically Important Plant Biomes: Source of Secondary Metabolites; Springer: New York, NY, USA, 2019; pp. 85–113. [Google Scholar]
- Aniszewski, T. Alkaloids-Secrets of Life:: Aklaloid Chemistry, Biological Significance, Applications and Ecological Role; Elsevier: Amsterdam, The Netherlands, 2007. [Google Scholar]
- House, R.V.; Thomas, P.T.; Kozak, J.T.; Bhargava, H.N. Suppression of immune function by non-peptidic delta opioid receptor antagonists. Neurosci. Lett. 1995, 198, 119–122. [Google Scholar] [CrossRef]
- Zhao, D.; Zhang, L.J.; Huang, T.Q.; Kim, J.; Gu, M.-Y.; Yang, H.O. Narciclasine inhibits LPS-induced neuroinflammation by modulating the Akt/IKK/NF-κB and JNK signaling pathways. Phytomedicine 2021, 85, 153540. [Google Scholar] [CrossRef]
- Yang, Z.; Tao, Y.; Xu, X.; Cai, F.; Yu, Y.; Ma, L. Bufalin inhibits cell proliferation and migration of hepatocellular carcinoma cells via APOBEC3F induced intestinal immune network for IgA production signaling pathway. Biochem. Biophys. Res. Commun. 2018, 503, 2124–2131. [Google Scholar] [CrossRef] [PubMed]
- Baron, S.J.; King, M.S.; Kunji, E.R.; Schirris, T.J. Characterization of drug-induced human mitochondrial ADP/ATP carrier inhibition. Theranostics 2021, 11, 5077. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, Y.; Hartman, S.E.; Kinoshita, E.; Suzuki, H.; Kitaura, J.; Yao, L.; Inagaki, N.; Franco, A.; Hata, D.; Maeda-Yamamoto, M. Terreic acid, a quinone epoxide inhibitor of Bruton’s tyrosine kinase. Proc. Natl. Acad. Sci. USA 1999, 96, 2227–2232. [Google Scholar] [CrossRef] [PubMed]
- Qiang, W.; Dai, Y.; Xing, X.; Sun, X. Identification and validation of a prognostic signature and combination drug therapy for immunotherapy of head and neck squamous cell carcinoma. Comput. Struct. Biotechnol. J. 2021, 19, 1263–1276. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.S.; Huang, P.I.; Chang, Y.L.; Tzao, C.; Chen, Y.W.; Shih, H.C.; Hung, S.C.; Chen, Y.C.; Tseng, L.M.; Chiou, S.H. Cucurbitacin I inhibits tumorigenic ability and enhances radiochemosensitivity in nonsmall cell lung cancer-derived CD133-positive cells. Cancer 2011, 117, 2970–2985. [Google Scholar] [CrossRef] [PubMed]
- Xuan, N.T.; Shumilina, E.; Gulbins, E.; Gu, S.; Götz, F.; Lang, F. Triggering of dendritic cell apoptosis by xanthohumol. Mol. Nutr. Food Res. 2010, 54, S214–S224. [Google Scholar] [CrossRef]
- Lemaire-Gony, S.; Lemaire, P.; Pulsford, A.L. Effects of cadmium and benzo (a) pyrene on the immune system, gill ATPase and EROD activity of European sea bass Dicentrarchus labrax. Aquat. Toxicol. 1995, 31, 297–313. [Google Scholar] [CrossRef]
- Carlson, E.; Li, Y.; Zelikoff, J. Exposure of Japanese medaka (Oryzias latipes) to benzo [a] pyrene suppresses immune function and host resistance against bacterial challenge. Aquat. Toxicol. 2002, 56, 289–301. [Google Scholar] [CrossRef]
- Steinberg, S.F. Cardiac actions of protein kinase C isoforms. Physiology 2012, 27, 130–139. [Google Scholar] [CrossRef]
- Vicencio, J.M.; Ibarra, C.; Estrada, M.; Chiong, M.; Soto, D.; Parra, V.; Diaz-Araya, G.; Jaimovich, E.; Lavandero, S. Testosterone induces an intracellular calcium increase by a nongenomic mechanism in cultured rat cardiac myocytes. Endocrinology 2006, 147, 1386–1395. [Google Scholar] [CrossRef] [PubMed]
- Bednar, F.; Song, C.; Bardi, G.; Cornwell, W.; Rogers, T.J. Cross-desensitization of CCR1, but not CCR2, following activation of the formyl peptide receptor FPR1. J. Immunol. 2014, 192, 5305–5313. [Google Scholar] [CrossRef]
- Golden, K.L.; Marsh, J.D.; Jiang, Y. Testosterone regulates mRNA levels of calcium regulatory proteins in cardiac myocytes. Horm. Metab. Res. 2004, 36, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Ayaz, O.; Howlett, S.E. Testosterone modulates cardiac contraction and calcium homeostasis: Cellular and molecular mechanisms. Biol. Sex Differ. 2015, 6, 9. [Google Scholar] [CrossRef] [PubMed]
- VAERS. Vaccine Adverse Event Reporting System (VAERS) Database. Available online: https://vaers.hhs.gov/ (accessed on 5 June 2021).
- Guo, R.; Chen, L.H.; Xing, C.; Liu, T. Pain regulation by gut microbiota: Molecular mechanisms and therapeutic potential. Br. J. Anaesth. 2019, 123, 637–654. [Google Scholar] [CrossRef] [PubMed]
- COVID-19 Vaccinations in the United States. Available online: https://covid.cdc.gov/covid-data-tracker/#vaccinations_vacc-total-admin-rate-total (accessed on 1 May 2022).
- Flad, H.-D.; Brandt, E. Platelet-derived chemokines: Pathophysiology and therapeutic aspects. Cell. Mol. Life Sci. 2010, 67, 2363–2386. [Google Scholar] [CrossRef]
- Brandt, E.; Petersen, F.; Ludwig, A.; Ehlert, J.E.; Bock, L.; Flad, H.-D. The β-thromboglobulins and platelet factor 4: Blood platelet-derived CXC chemokines with divergent roles in early neutrophil regulation. J. Leukoc. Biol. 2000, 67, 471–478. [Google Scholar] [CrossRef]
- Harrison, S.L.; Buckley, B.J.R.; Rivera-Caravaca, J.M.; Zhang, J.; Lip, G.Y.H. Cardiovascular risk factors, cardiovascular disease, and COVID-19: An umbrella review of systematic reviews. Eur. Heart J. 2021, 7, 330–339. [Google Scholar] [CrossRef]
- Iba, T.; Levy, J.H. The roles of platelets in COVID-19-associated coagulopathy and vaccine-induced immune thrombotic thrombocytopenia. Trends Cardiovasc. Med. 2021, 21, 1–9. [Google Scholar] [CrossRef]
- Yun, S.-H.; Sim, E.-H.; Goh, R.-Y.; Park, J.-I.; Han, J.-Y. Platelet activation: The mechanisms and potential biomarkers. BioMed Res. Int. 2016, 2016, 9060143. [Google Scholar] [CrossRef]
- Yeaman, M.R. Platelets: At the nexus of antimicrobial defence. Nat. Rev. Microbiol. 2014, 12, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Bautista-Vargas, M.; Bonilla-Abadía, F.; Cañas, C.A. Potential role for tissue factor in the pathogenesis of hypercoagulability associated with in COVID-19. J. Thromb. Thrombolysis 2020, 50, 479–483. [Google Scholar] [CrossRef] [PubMed]
- Mastellos, D.C.; Skendros, P.; Lambris, J.D. Is complement the culprit behind COVID-19 vaccine-related adverse reactions? J. Clin. Investig. 2021, 131, e151092. [Google Scholar] [CrossRef] [PubMed]
- Dias, L.; Soares-dos-Reis, R.; Meira, J.; Ferrão, D.; Soares, P.R.; Pastor, A.; Gama, G.; Fonseca, L.; Fagundes, V.; Carvalho, M. Cerebral Venous Thrombosis after BNT162b2 mRNA SARS-CoV-2 vaccine. J. Stroke Cerebrovasc. Dis. 2021, 30, 105906. [Google Scholar] [CrossRef]
- Ciccone, A.; Zanotti, B. The importance of recognizing cerebral venous thrombosis following anti-COVID-19 vaccination. Eur. J. Intern. Med. 2021, 89, 115–117. [Google Scholar] [CrossRef]
- Greinacher, A.; Thiele, T.; Warkentin, T.E.; Weisser, K.; Kyrle, P.A.; Eichinger, S. Thrombotic Thrombocytopenia after ChAdOx1 nCov-19 Vaccination. N. Engl. J. Med. 2021, 384, 2092–2101. [Google Scholar] [CrossRef]
- Huynh, A.; Kelton, J.G.; Arnold, D.M.; Daka, M.; Nazy, I. Antibody epitopes in vaccine-induced immune thrombotic thrombocytopaenia. Nature 2021, 596, 565–569. [Google Scholar] [CrossRef]
- Tutwiler, V.; Madeeva, D.; Ahn, H.S.; Andrianova, I.; Hayes, V.; Zheng, X.L.; Cines, D.B.; McKenzie, S.E.; Poncz, M.; Rauova, L. Platelet transactivation by monocytes promotes thrombosis in heparin-induced thrombocytopenia. Blood 2016, 127, 464–472. [Google Scholar] [CrossRef]
- Warkentin, T.E.; Greinacher, A. Spontaneous HIT syndrome: Knee replacement, infection, and parallels with vaccine-induced immune thrombotic thrombocytopenia. Thromb. Res. 2021, 204, 40–51. [Google Scholar] [CrossRef]
- McFadyen, J.D.; Stevens, H.; Peter, K. The Emerging Threat of (Micro)Thrombosis in COVID-19 and Its Therapeutic Implications. Circ. Res. 2020, 127, 571–587. [Google Scholar] [CrossRef]
- Martinod, K.; Deppermann, C. Immunothrombosis and thromboinflammation in host defense and disease. Platelets 2021, 32, 314–324. [Google Scholar] [CrossRef] [PubMed]
- De Fabritiis, M.; Angelini, M.L.; Fabbrizio, B.; Cenacchi, G.; Americo, C.; Cristino, S.; Lifrieri, M.F.; Cappuccilli, M.; Spazzoli, A.; Zambianchi, L.; et al. Renal Thrombotic Microangiopathy in Concurrent COVID-19 Vaccination and Infection. Pathogens 2021, 10, 1045. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Yan, H.; Cai, H.; Li, X.; Guan, Q.; Zheng, W.; Chen, R.; Liu, H.; Song, K.; Guo, Z. Statistically controlled identification of differentially expressed genes in one-to-one cell line comparisons of the CMAP database for drug repositioning. J. Transl. Med. 2017, 15, 198. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Polacco, M.; Chen, S.; Schroeder, S.; Hancock, D.; Sanchez, H.; Coe, E. cMap: The comparative genetic map viewer. Bioinformatics 2003, 19, 416–417. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Pepowski, B.; Takahashi, S.; Kron, S.J. A cmap-enabled gene expression signature-matching approach identifies small-molecule inducers of accelerated cell senescence. BMC Genom. 2019, 20, 290. [Google Scholar] [CrossRef]
- Vyleta, M.L.; Wong, J.; Magun, B.E. Suppression of ribosomal function triggers innate immune signaling through activation of the NLRP3 inflammasome. PLoS ONE 2012, 7, e36044. [Google Scholar] [CrossRef]
- Ivanov, K.; Garanina, E.; Rizvanov, A.; Khaiboullina, S. Inflammasomes as Targets for Adjuvants. Pathogens 2020, 9, 252. [Google Scholar] [CrossRef]
- Lonez, C.; Bessodes, M.; Scherman, D.; Vandenbranden, M.; Escriou, V.; Ruysschaert, J.M. Cationic lipid nanocarriers activate Toll-like receptor 2 and NLRP3 inflammasome pathways. Nanomedicine 2014, 10, 775–782. [Google Scholar] [CrossRef]
- Teijaro, J.R.; Farber, D.L. COVID-19 vaccines: Modes of immune activation and future challenges. Nat. Rev. Immunol. 2021, 21, 195–197. [Google Scholar] [CrossRef]
- Davis, A.P.; Grondin, C.J.; Johnson, R.J.; Sciaky, D.; Wiegers, J.; Wiegers, T.C.; Mattingly, C.J. Comparative Toxicogenomics Database (CTD): Update 2021. Nucleic Acids Res. 2020, 49, D1138–D1143. [Google Scholar] [CrossRef]
- Sharaf, A.R.; Narula, J.; Nicol, P.D.; Southern, J.F.; Khaw, B.A. Cardiac sarcoplasmic reticulum calcium ATPase, an autoimmune antigen in experimental cardiomyopathy. Circulation 1994, 89, 1217–1228. [Google Scholar] [CrossRef] [PubMed]
- Kyriakopoulos, A.M.; McCullough, P.A. Synthetic mRNAs; Their Analogue Caps and Contribution to Disease. Diseases 2021, 9, 57. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, F.; Zheng, X.; Su, J.; Yu, Y.; Wang, L.; Zhuang, H. Lipase-catalyzed synthesis of polyhydroxyalkyl furans from unprotected sugars and malononitrile. Process Biochem. 2021, 101, 99–103. [Google Scholar] [CrossRef]
- Fricke-Galindo, I.; Falfán-Valencia, R. Genetics Insight for COVID-19 Susceptibility and Severity: A Review. Front. Immunol. 2021, 12, 622176. [Google Scholar] [CrossRef] [PubMed]
- Sang, E.R.; Tian, Y.; Miller, L.C.; Sang, Y. Epigenetic Evolution of ACE2 and IL-6 Genes: Non-Canonical Interferon-Stimulated Genes Correlate to COVID-19 Susceptibility in Vertebrates. Genes 2021, 12, 154. [Google Scholar] [CrossRef]
- Patone, M.; Mei, X.W.; Handunnetthi, L.; Dixon, S.; Zaccardi, F.; Shankar-Hari, M.; Watkinson, P.; Khunti, K.; Harnden, A.; Coupland, C.A.C.; et al. Risks of myocarditis, pericarditis, and cardiac arrhythmias associated with COVID-19 vaccination or SARS-CoV-2 infection. Nat. Med. 2022, 28, 410–422. [Google Scholar] [CrossRef]
- Agus, A.; Clément, K.; Sokol, H. Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut 2021, 70, 1174–1182. [Google Scholar] [CrossRef]
- Arunachalam, P.S.; Scott, M.K.; Hagan, T.; Li, C.; Feng, Y.; Wimmers, F.; Grigoryan, L.; Trisal, M.; Edara, V.V.; Lai, L. Systems vaccinology of the BNT162b2 mRNA vaccine in humans. Nature 2021, 596, 410–416. [Google Scholar] [CrossRef]
- Tomioka, N.; Kishimoto, C.; Matsumori, A.; Kawai, C. Effects of prednisolone on acute viral myocarditis in mice. J. Am. Coll. Cardiol. 1986, 7, 868–872. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Chatr-aryamontri, A.; Ceol, A.; Palazzi, L.M.; Nardelli, G.; Schneider, M.V.; Castagnoli, L.; Cesareni, G. MINT: The Molecular INTeraction database. Nucleic Acids Res. 2007, 35, D572–D574. [Google Scholar] [CrossRef] [PubMed]
- Wilson, N. Human Protein Reference Database. Nat. Rev. Genet. 2004, 5, 8. [Google Scholar] [CrossRef]
- Liu, T.; Lin, Y.; Wen, X.; Jorissen, R.N.; Gilson, M.K. BindingDB: A web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007, 35, D198–D201. [Google Scholar] [CrossRef] [PubMed]
- Xenarios, I.; Rice, D.W.; Salwinski, L.; Baron, M.K.; Marcotte, E.M.; Eisenberg, D. DIP: The database of interacting proteins. Nucleic Acids Res. 2000, 28, 289–291. [Google Scholar] [CrossRef] [PubMed]
- Stark, C.; Breitkreutz, B.J.; Reguly, T.; Boucher, L.; Breitkreutz, A.; Tyers, M. BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 2006, 34, D535–D539. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Croft, D.; O’Kelly, G.; Wu, G.; Haw, R.; Gillespie, M.; Matthews, L.; Caudy, M.; Garapati, P.; Gopinath, G.; Jassal, B.; et al. Reactome: A database of reactions, pathways and biological processes. Nucleic Acids Res. 2011, 39, D691–D697. [Google Scholar] [CrossRef]
- Karp, P.D.; Weaver, D.; Paley, S.; Fulcher, C.; Kubo, A.; Kothari, A.; Krummenacker, M.; Subhraveti, P.; Weerasinghe, D.; Gama-Castro, S.; et al. The EcoCyc Database. EcoSal Plus 2014, 6, 1–19. [Google Scholar] [CrossRef]
- Schaefer, C.F.; Anthony, K.; Krupa, S.; Buchoff, J.; Day, M.; Hannay, T.; Buetow, K.H. PID: The Pathway Interaction Database. Nucleic Acids Res. 2008, 37, D674–D679. [Google Scholar] [CrossRef]
- Harris, M.A.; Clark, J.; Ireland, A.; Lomax, J.; Ashburner, M.; Foulger, R.; Eilbeck, K.; Lewis, S.; Marshall, B.; Mungall, C.; et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004, 32, D258–D261. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Hu, J.; Newman, M.; Liu, K.; Ge, H. RNA-Seq Analysis Pipeline Based on Oshell Environment. IEEE/ACM Trans. Comput. Biol. Bioinform. 2014, 11, 973–978. [Google Scholar] [CrossRef] [PubMed]
- Baldini, F.; Hertel, J.; Sandt, E.; Thinnes, C.C.; Neuberger-Castillo, L.; Pavelka, L.; Betsou, F.; Krüger, R.; Thiele, I. Parkinson’s disease-associated alterations of the gut microbiome predict disease-relevant changes in metabolic functions. BMC Biol. 2020, 18, 62. [Google Scholar] [CrossRef] [PubMed]


| Description | Pfizer–BioNTech | Moderna | CureVac |
|---|---|---|---|
| Country | Pfizer (Pfizer, New York, NY, USA)–BioNTech (BioNTech, Mainz, Germany) | Moderna, Cambridge, MA, USA | CureVac, Tübingen, Germany |
| Vaccine platform [17,18,19,20,21,22,23,24,25] | mRNA: BNT162a1, BNT162b1, BNT162b2, and BNT162c2 | mRNA: mRNA-1273 | mRNA: CVnCoV |
| Vaccine genetic material composition (mRNA) [20,26,27,28] |
|
|
|
| LNPs j composition [4,19,20,21,27,29] |
|
|
|
| Molar lipid ratios (%) ionizable cationic lipid: neutral lipid: cholesterol: PEG-ylated lipid [21,27] | 46.3:9.4:42.7:1.6 | 50:10:38.5:1.5 | 50:10:38.5:1.5 |
| Molar N/P ratios N = nitrogen (ionizable group cationic lipid) P = phosphate (nucleotide group) [21,27] | Vaccine makers evaluated 6 different formulations | Vaccine makers evaluated 6 different formulations | Vaccine makers evaluated 6 different formulations |
| Buffer [4,19,20,21,29] | Phosphate (PO4−2) (KH2PO4, Na2HPO4·2H2O) | Tris (Tromethamine) | NA l |
| Extra excipients [4,19,20,21,29] | KCl, NaCl, Sucrose, and H2O for vaccination | CH3COONa, Sucrose, and H2O for vaccination | Saline |
| Dose, dosing regimen, and route of administration [17,18,19,20,21,22,23,24,25] | 30 μg (0.3 mL), day 1-day21 | 100 μg (0.5 mL), day 1-day29 | 12 μg (NA mL), day 1-day29 |
| Stability condition [18,20,30,31,32] | (−80–60 °C), Up to 6 months | −20 °C, Up to 6 months | ≤−60 °C, Up to 3 months |
| Temperature range (−20–−80 °C) | |||
| Temperature range (−2–−8 °C) | Up to 5 days | Up to 30 days | Up to 3 months |
| Room temperature | Up to 2 h (mixing with 1.8 mL NaCl expands the span till 6 h) | Up to 12 h | Up to 24 h |
| Clinical information [2,33,34,35,36,37,38] | BNT162b1 (2417899–75-1), BNT162b2 (2417899-77-3) | mRNA-1273 (2457298-05-2) | CVnCoV n (2541470-90-8) |
| CAS m registry number (RN) | |||
| Clinical trial registration number | NCT04368728; NCT04760132 | NCT04470427; NCT04649151; NCT04760132 | NCT04449276; ISRCTN73765130 NCT04515147, PER-054-20; NCT04652102, EUCTR2020-003998-22; NCT04652102, EUCTR2020-003998-22; EUCTR2020-004066-19, NCT04674189; NCT04838847; NCT04848467; NCT04860258 |
| Clinical stage | Phase 4 | Phase 4 | Phase 3 |
| Target protein | Prefusion stabilized (S-2P) o transmembrane attached whole sequence spike protein | Prefusion stabilized (S-2P) transmembrane attached whole sequence spike protein | Prefusion stabilized (S-2P) transmembrane attached spike protein |
| Furin cleavage site | Native | Native | Entire S1/S2 p cleavage domain and transmembrane domain |
| Real world vaccine effectiveness against original SARS-CoV-2 strain of Wuhan [2,8,39,40,41,42] | 64–99% | 68–99% | 47% |
| Real world vaccine effectiveness against SARS-CoV-2 variants [2,8,39,40,41,42] | α (B.1.1.7) 65–100% q (84–100%) r | α (B.1.1.7) 79–100% q (90–96%) r | α (B.1.1.7) NA l |
| β (B.1.351) 75–88% q (95–100%)r | β (B.1.351) 88–96% q (96–100%) r | β (B.1.351) NA | |
| γ (P.1) 79–88% q (95–100%) r | γ (P.1) 79–88% q (95–100%) r | γ (P.1) NA | |
| δ (B.1.617.2) 79–88% q (96%) r | δ (B.1.617.2) NA | δ (B.1.617.2) NA | |
| o (B.1.1529) NA | o (B.1.1529) NA | o (B.1.1529) NA |
| # | Pathway Map a | FDR b | #Map Objects c | #Overlapping Objects d | Overlapping Objects |
|---|---|---|---|---|---|
| 1 | Immune response_IFN-alpha/beta signaling via JAK/STAT | 2.05 × 10−1 | 62 | 9 | USP18, IP10, CCL2, Apo-2L(TNFSF10), ERAP140, RIG-G, PNPase(old-35), RSAD2, ISG15 |
| 2 | Immune response_IFN-alpha/beta signaling via MAPKs | 9.20 × 10−7 | 73 | 7 | IP10, PL scramblase 1, GCH1, Apo-2L(TNFSF10), RIG-G, RSAD2, ISG15 |
| 3 | Glomerular injury in Lupus Nephritis | 1.37 × 10−3 | 92 | 5 | MDA-5, RIG-I, IP10, CCL2, IFI56 |
| 4 | Macrophage-induced immunosuppression in the tumor microenvironment | 1.37 × 10−3 | 97 | 5 | MSR1, PD-L1, CCL2, PD-L2, IDO1 |
| 5 | COVID-19: immune dysregulation | 1.37 × 10−3 | 100 | 5 | MDA-5, RIG-I, IP10, CCL2, ISG15 |
| 6 | Macrophage and dendritic cell phenotype shift in cancer | 1.37 × 10−3 | 100 | 5 | MSR1, IP10, Apo-2L(TNFSF10), SOCS1, IDO1 |
| 7 | Immune response_IFN-gamma actions on extracellular matrix and cell differentiation | 1.73 × 10−3 | 54 | 4 | OAS2, IP10, GCH1, 2′-5′-oligoadenylate synthetase |
| 8 | Vascular endothelial cell damage in SLE | 2.79 × 10−3 | 63 | 4 | MSR1, PD-L1, CCL2, PD-L2 |
| 9 | Immune response_Innate immune response to RNA viral infection | 4.07 × 10−3 | 28 | 3 | MDA-5, RIG-I, LGP2 |
| 10 | Immune response_IFN-gamma actions on blood cells | 4.07 × 10−3 | 28 | 3 | PD-L1, PD-L2, SOCS1 |
| # | Compound | CMap Score a | Description | Confidence b | Immune Effects |
|---|---|---|---|---|---|
| 1 | Cycloheximide | 98.31 | Protein synthesis inhibitor | High | [82,83,84,85] |
| 2 | QL-XII-47 | 96.48 | BTK inhibitor | High | [86,87,88] |
| 3 | Homoharringtonine | 94.71 | Protein synthesis inhibitor | High | [89,90,91] |
| 4 | Periplocymarin | 94.38 | Apoptosis stimulant | High | [92,93] |
| 5 | Digitoxigenin | 94.11 | ATPase inhibitor | High | [94,95] |
| 6 | Emetine | 94.05 | Protein synthesis inhibitor | High | [96,97] |
| 7 | Ouabain | 93.62 | ATPase inhibitor | High | [98,99] |
| 8 | Cephaeline | 92.26 | Protein synthesis inhibitor | High | [100] |
| 9 | Clofarabine | 92.11 | Ribonucleoside reductase inhibitor | High | [101] |
| 10 | Sarmentogenin | 91.66 | ATPase inhibitor | High | [102] |
| 11 | Digitoxin | 91.34 | ATPase inhibitor | High | [103] |
| 12 | Isoliquiritigenin | 90.85 | Guanylate cyclase activator | High | [104] |
| 13 | Digoxin | 90.38 | ATPase inhibitor | High | [105,106] |
| 14 | Tyrphostin-AG-126 | 98.84 | ERK1/2 phosphorylation inhibitor | Intermediate | [107] |
| 15 | Amonafide | 98.73 | Topoisomerase inhibitor | Intermediate | [108] |
| 16 | Diphenoxylate | 98.50 | Opioid receptor agonist | Intermediate | [109] |
| 17 | Verrucarin-a | 98.38 | Protein synthesis inhibitor | Intermediate | [110] |
| 18 | Withaferin-a | 97.96 | IKK inhibitor | Intermediate | [111] |
| 19 | Dapsone | 96.32 | Bacterial antifolate | Intermediate | [112] |
| 20 | Teniposide | 95.99 | Topoisomerase inhibitor | Intermediate | [113,114] |
| 21 | Ziprasidone | 95.86 | Dopamine receptor antagonist | Intermediate | [115] |
| 22 | RO-90-7501 | 95.80 | Beta amyloid inhibitor | Intermediate | [116] |
| 23 | XMD-1150 | 95.38 | Leucine rich repeat kinase inhibitor | Intermediate | [117] |
| 24 | Ingenol | 94.47 | PKC activator | Intermediate | [118,119] |
| 25 | XMD-892 | 93.85 | MAP kinase inhibitor | Intermediate | [117] |
| 26 | Anisomycin | 93.69 | DNA synthesis inhibitor | Intermediate | [120] |
| 27 | Proscillaridin | 91.37 | ATPase inhibitor | Intermediate | [121] |
| 28 | Azacitidine | 91.29 | DNA methyltransferase inhibitor | Intermediate | [122] |
| 29 | 4-hydroxy-2-nonenal | 90.47 | Cytotoxic lipid peroxidation product | Intermediate | [123] |
| 30 | Dubinidine | 90.37 | Anti-epileptic | Intermediate | [124,125] |
| 31 | BNTX | 89.95 | Opioid receptor antagonist | Intermediate | [126] |
| 32 | Narciclasine | 89.85 | Coflilin signaling pathway activator | Intermediate | [127] |
| 33 | Mitomycin-c | 88.45 | DNA alkylating agent | Intermediate | [17] |
| 34 | Bufalin | 87.17 | ATPase inhibitor | Intermediate * | [128] |
| 35 | Cinobufagin | 86.79 | ATPase inhibitor | Intermediate * | [19] |
| 36 | Brefeldin-a | 86.54 | Protein synthesis inhibitor | Intermediate * | [23] |
| 37 | Pyrvinium-pamoate | 78.80 | AKT inhibitor | Low | [24] |
| 38 | Liothyronine | 72.93 | Thyroid hormone stimulant | Low | [25] |
| 39 | CD-437 | 70.27 | Retinoid receptor agonist | Low | [22,129] |
| 40 | Terreic-acid | 66.04 | BTK inhibitor | Low * | [130] |
| 41 | Minaprine | 63.46 | Serotonin reuptake inhibitor | Low | [131] |
| 42 | Cucurbitacin-i | 61.47 | JAK inhibitor | Low | [132] |
| 43 | Xanthohumol | 60.13 | ATPase inhibitor | Low * | [133] |
| 44 | Benzo(a)pyrene | 54.66 | Carcinogen | Low | [134,135] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hajjo, R.; Sabbah, D.A.; Tropsha, A. Analyzing the Systems Biology Effects of COVID-19 mRNA Vaccines to Assess Their Safety and Putative Side Effects. Pathogens 2022, 11, 743. https://doi.org/10.3390/pathogens11070743
Hajjo R, Sabbah DA, Tropsha A. Analyzing the Systems Biology Effects of COVID-19 mRNA Vaccines to Assess Their Safety and Putative Side Effects. Pathogens. 2022; 11(7):743. https://doi.org/10.3390/pathogens11070743
Chicago/Turabian StyleHajjo, Rima, Dima A. Sabbah, and Alexander Tropsha. 2022. "Analyzing the Systems Biology Effects of COVID-19 mRNA Vaccines to Assess Their Safety and Putative Side Effects" Pathogens 11, no. 7: 743. https://doi.org/10.3390/pathogens11070743
APA StyleHajjo, R., Sabbah, D. A., & Tropsha, A. (2022). Analyzing the Systems Biology Effects of COVID-19 mRNA Vaccines to Assess Their Safety and Putative Side Effects. Pathogens, 11(7), 743. https://doi.org/10.3390/pathogens11070743








