Identification of Three Novel Conidiogenesis-Related Genes in the Nematode-Trapping Fungus Arthrobotrys oligospora
Abstract
1. Introduction
2. Results
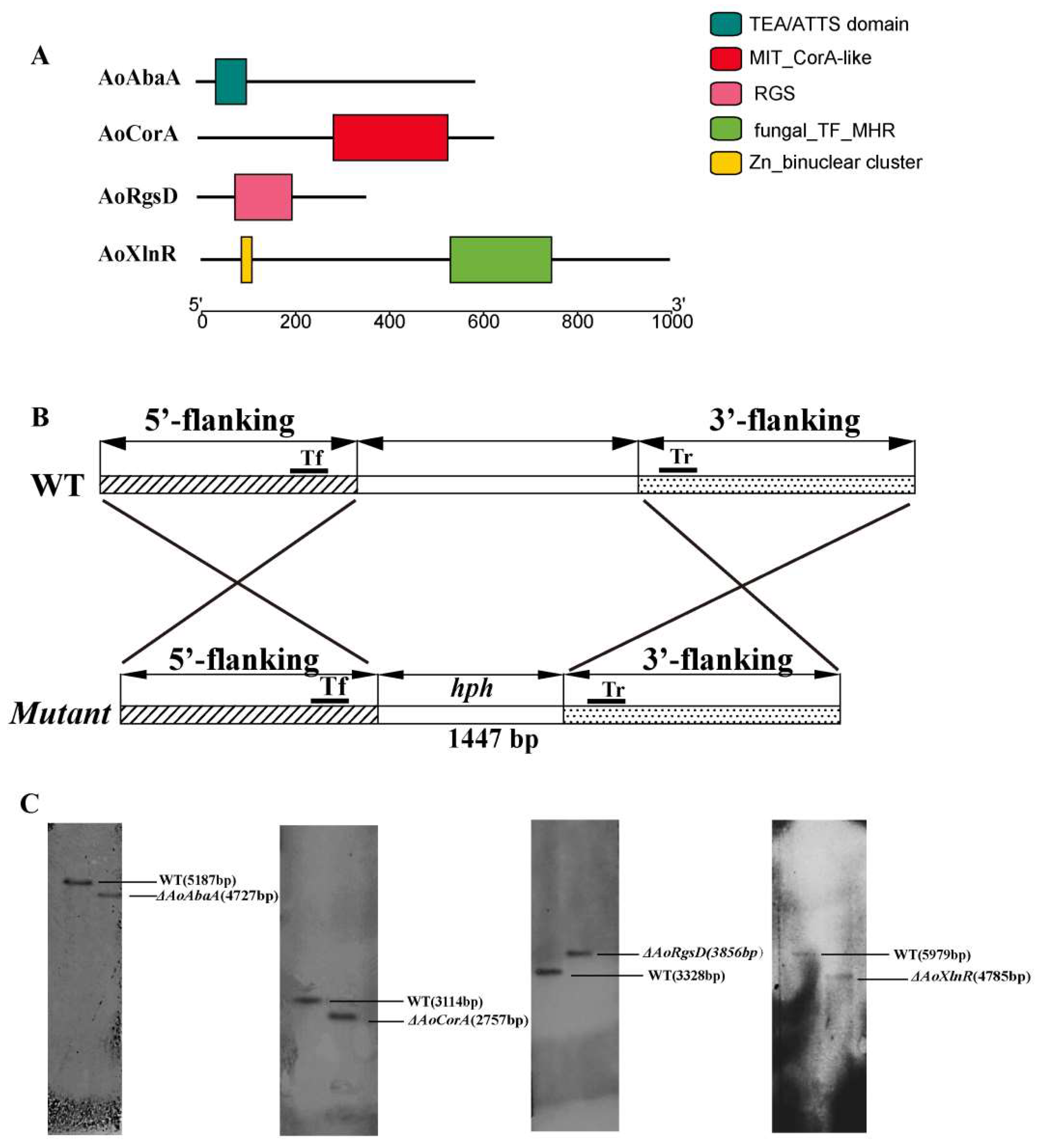
2.1. Identification of AoAbaA, AoXlnR, AoCorA, and AoRgsD in A. oligospora
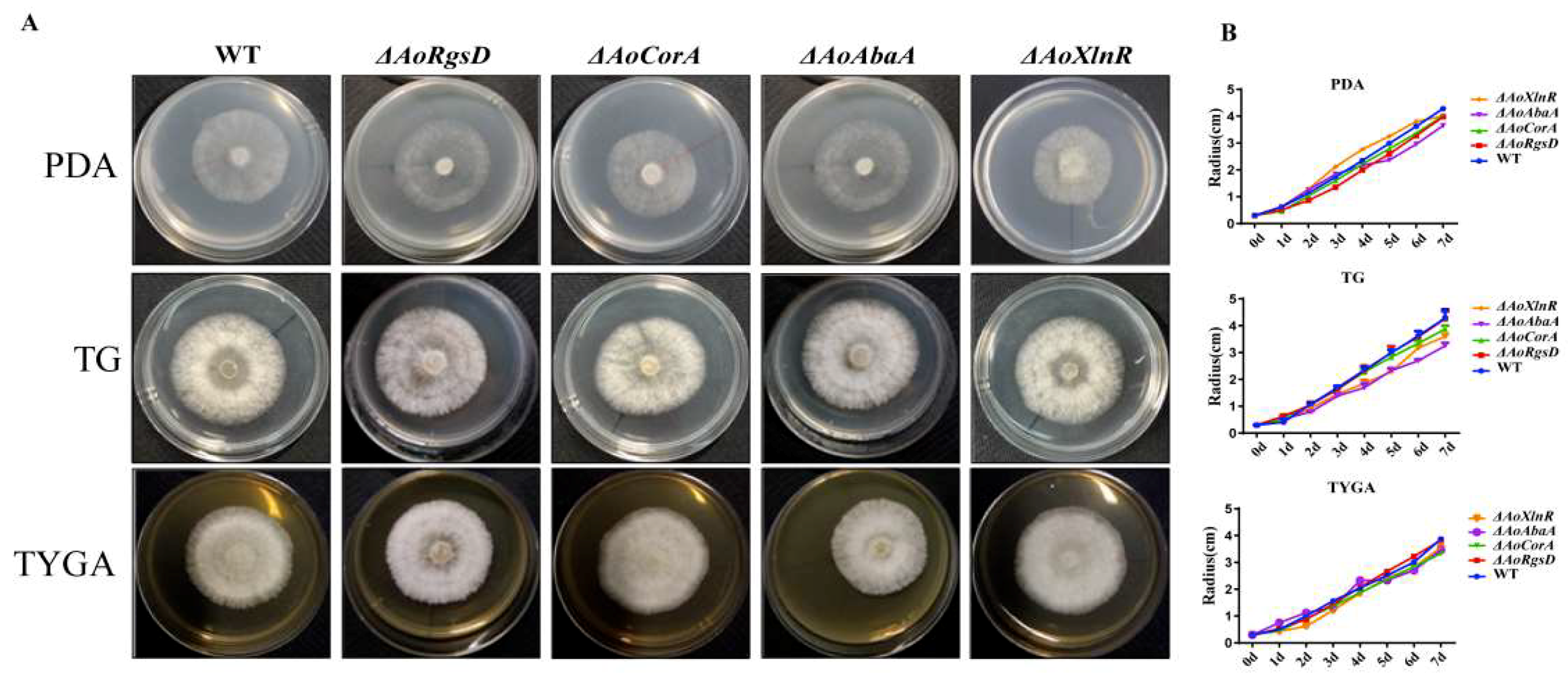
2.2. Phenotypes of AoAbaA, AoXlnR, AoCorA, and AoRgsD Deletion Mutants
2.3. Effects of ΔAoAbaA, ΔAoXlnR, ΔAoCorA, and ΔAoRgsD Deletion Mutations on Hyphal Growth and Responses to Different Stresses
2.4. AoAbaA Is Also Involved in Trap Formation in A. oligospora
2.5. AoAbaA, AoXlnR, AoCorA, and AoRgsD Were Required for Conidiation
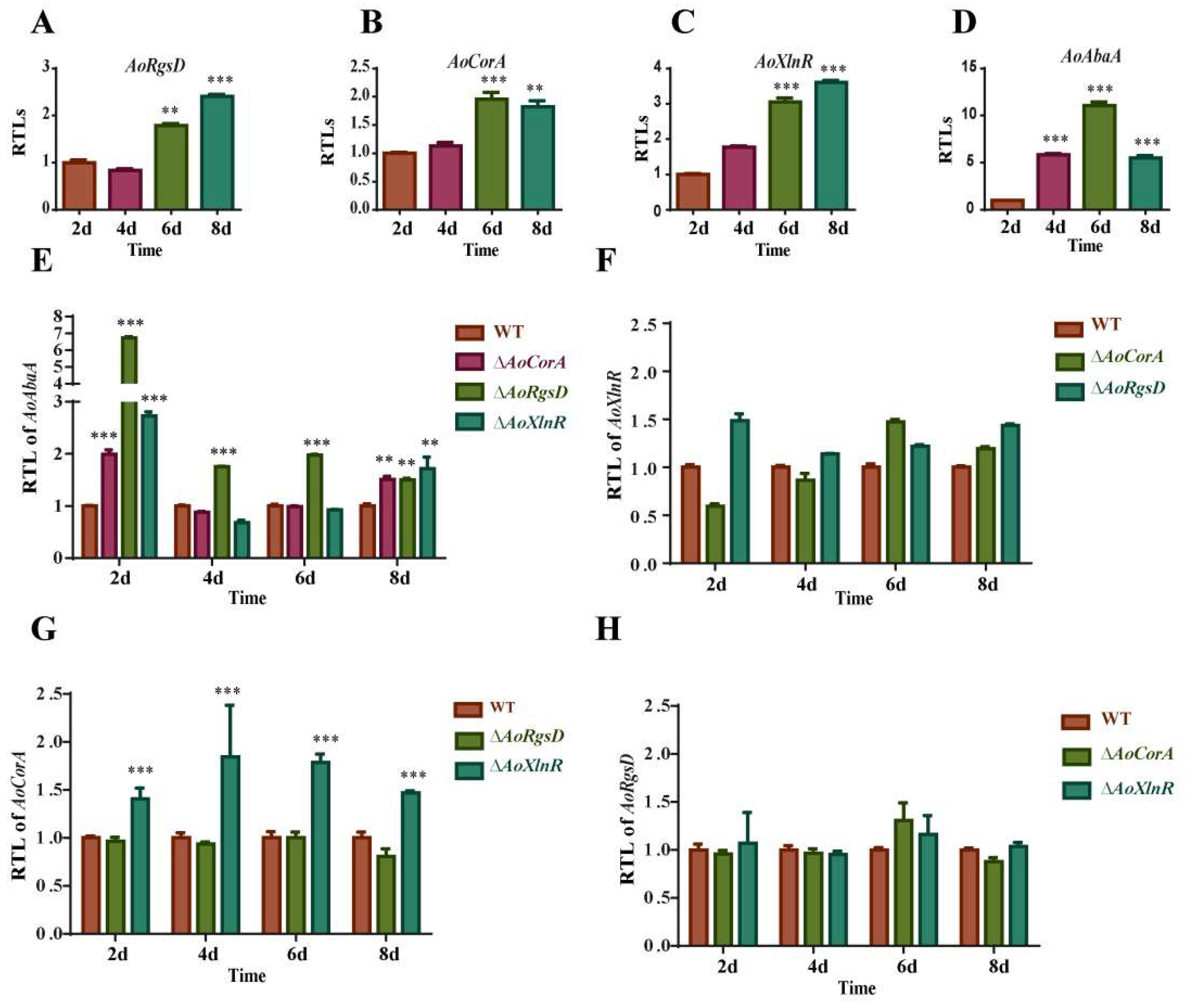
2.6. Relationship between the Expressions of AoAbaA, AoXlnR, AoCorA, and AoRgsD and Conidiation
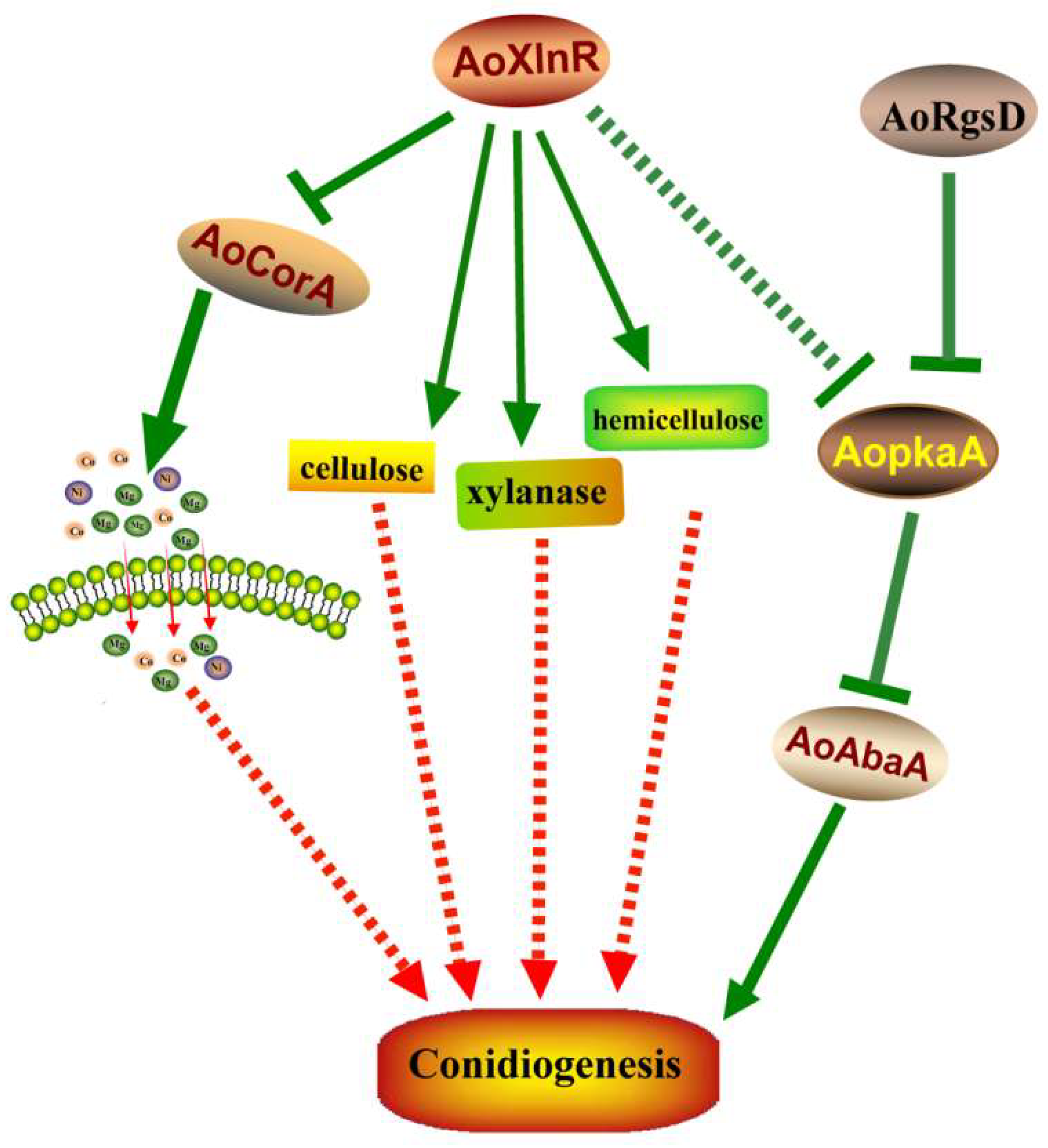
3. Discussion
4. Materials and Methods
4.1. Fungal Strains and Culture Conditions
4.2. Identification of AoAbaA, AoXlnR, AoCorA, and AoRgsD in A. oligospora
4.3. Deletion of the Genes AoAbaA, AoXlnR, AoCorA, and AoRgsD
4.4. Comparison in Growth Rate between Mutants and Wild-Type Strain in Different Conditions
4.5. Comparison of Conidial Production, Morphology, and Germination
4.6. Trap Formation and Pathogenicity Assays
4.7. Quantitative Real-Time PCR (RT-PCR) Analysis
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abad, P.; Gouzy, J.; Aury, J.-M.; Castagnone-Sereno, P.; Danchin, E.G.J.; Deleury, E.; Perfus-Barbeoch, L.; Anthouard, V.; Artiguenave, F.; Blok, V.C.; et al. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat. Biotechnol. 2008, 26, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.-Q.; Hyde, K.D. Nematode-Trapping Fungi; Springer Science & Business: New York, NY, USA, 2014; Volume 23. [Google Scholar]
- Yang, J.; Wang, L.; Ji, X.; Feng, Y.; Li, X.; Zou, C.; Xu, J.; Ren, Y.; Mi, Q.; Wu, J.; et al. Genomic and proteomic analyses of the fungus Arthrobotrys oligospora provide insights into nematode-trap formation. PLoS Pathog. 2011, 7, e1002179. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Li, X.; Bai, N.; Yang, X.; Zhang, K.Q.; Yang, J. Transcriptomic analysis reveals that Rho GTPases regulate trap development and lifestyle transition of the nematode-trapping fungus Arthrobotrys oligospora. Microbiol. Spectr. 2022, 10, e0175921. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.C.; Li, X.M.; Zhao, N.; Yang, L.; Zhang, K.Q.; Yang, J.K. Regulatory mechanism of trap formation in the nematode-trapping fungi. J. Fungi 2022, 8, 406. [Google Scholar] [CrossRef]
- Etxebeste, O.; Garzia, A.; Espeso, E.A.; Ugalde, U. Aspergillus nidulans asexual development: Making the most of cellular modules. Trends Microbiol. 2010, 18, 569–576. [Google Scholar] [CrossRef]
- Park, H.S.; Yu, J.H. Genetic control of asexual sporulation in filamentous fungi. Curr. Opin. Microbiol. 2012, 15, 669–677. [Google Scholar] [CrossRef]
- Boylan, M.T.; Mirabito, P.M.; Willett, C.E.; Zimmerman, C.R.; Timberlake, W.E. Isolation and physical characterization of three essential conidiation genes from Aspergillus nidulans. Mol. Cell. Biol. 1987, 7, 3113–3118. [Google Scholar] [CrossRef]
- Sewall, T.C.; Mims, C.W.; Timberlake, W.E. Conidium differentiation in Aspergillus nidulans wild-type and wet-white (wetA) mutant strains. Dev. Biol. 1990, 138, 499–508. [Google Scholar] [CrossRef]
- Marshall, M.A.; Timberlake, W.E. Aspergillus nidulans wetA activates spore-specific gene expression. Mol. Cell. Biol. 1991, 11, 55–62. [Google Scholar] [CrossRef]
- Kim, Y.; Lee, M.W.; Jun, S.C.; Choi, Y.H.; Yu, J.H.; Shin, K.S. RgsD negatively controls development, toxigenesis, stress response, and virulence in Aspergillus fumigatus. Sci. Rep. 2019, 9, 811. [Google Scholar] [CrossRef]
- Kirsten, A.; Herzberg, M.; Voigt, A.; Seravalli, J.; Grass, G.; Scherer, J.; Nies, D.H. Contributions of five secondary metal uptake systems to metal homeostasis of Cupriavidus metallidurans CH34. J. Bacteriol. 2011, 193, 4652–4663. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, Y.; Sano, M.; Kanamaru, K.; Ko, T.; Takeuchi, M.; Kato, M.; Kobayashi, T. Genes regulated by AoXlnR, the xylanolytic and cellulolytic transcriptional regulator, in Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2009, 85, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Marui, J.; Kitamoto, N.; Kato, M.; Kobayashi, T.; Tsukagoshi, N. Transcriptional activator, AoXlnR, mediates cellulose-inductive expression of the xylanolytic and cellulolytic genes in Aspergillus oryzae. FEBS Lett. 2002, 528, 279–282. [Google Scholar] [CrossRef]
- Hasper, A.A.; Visser, J.; de Graaff, L.H. The Aspergillus niger transcriptional activator XlnR, which is involved in the degradation of the polysaccharides xylan and cellulose, also regulates D-xylose reductase gene expression. Mol. Microbiol. 2000, 36, 193–200. [Google Scholar] [CrossRef]
- Zhou, D.; Zhu, Y.; Bai, N.; Yang, L.; Xie, M.; Yang, J.; Zhu, M.; Zhang, K.Q.; Yang, J. AoATG5 plays pleiotropic roles in vegetative growth, cell nucleus development, conidiation, and virulence in the nematode-trapping fungus Arthrobotrys oligospora. Sci. China Life Sci. 2022, 65, 412–425. [Google Scholar] [CrossRef]
- Zhou, D.; Zhu, Y.; Bai, N.; Xie, M.; Zhang, K.Q.; Yang, J. Aolatg1 and Aolatg13 Regulate Autophagy and Play Different Roles in Conidiation, Trap Formation, and Pathogenicity in the Nematode-Trapping Fungus Arthrobotrys oligospora. Front. Cell. Infect. Microbiol. 2021, 11, 824407. [Google Scholar] [CrossRef]
- Zhou, D.; Xie, M.; Bai, N.; Yang, L.; Zhang, K.Q.; Yang, J. The autophagy-related gene Aolatg4 regulates hyphal growth, sporulation, autophagosome formation, and pathogenicity in Arthrobotrys oligospora. Front. Microbiol. 2020, 11, 592524. [Google Scholar] [CrossRef]
- Xie, M.; Wang, Y.; Tang, L.; Yang, L.; Zhou, D.; Li, Q.; Niu, X.; Zhang, K.Q.; Yang, J. AoStuA, an APSES transcription factor, regulates the conidiation, trap formation, stress resistance and pathogenicity of the nematode-trapping fungus Arthrobotrys oligospora. Environ. Microbiol. 2019, 21, 4648–4661. [Google Scholar] [CrossRef]
- Zhen, Z.; Xing, X.; Xie, M.; Yang, L.; Yang, X.; Zheng, Y.; Chen, Y.; Ma, N.; Li, Q.; Zhang, K.Q.; et al. MAP kinase Slt2 orthologs play similar roles in conidiation, trap formation, and pathogenicity in two nematode-trapping fungi. Fungal Genet. Biol. 2018, 116, 42–50. [Google Scholar] [CrossRef]
- Xie, M.; Bai, N.; Yang, J.; Jiang, K.; Zhou, D.; Zhao, Y.; Li, D.; Niu, X.; Zhang, K.Q.; Yang, J. Protein kinase Ime2 is required for mycelial growth, conidiation, osmoregulation, and pathogenicity in nematode-trapping fungus Arthrobotrys oligospora. Front. Microbiol. 2019, 10, 3065. [Google Scholar] [CrossRef]
- Yang, X.; Ma, N.; Yang, L.; Zheng, Y.; Zhen, Z.; Li, Q.; Xie, M.; Li, J.; Zhang, K.Q.; Yang, J. Two Rab GTPases play different roles in conidiation, trap formation, stress resistance, and virulence in the nematode-trapping fungus Arthrobotrys oligospora. Appl. Microbiol. Biotechnol. 2018, 102, 4601–4613. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.; Zhao, Y.; Wang, Y.; Yang, L.; Li, D.; Yang, J.; Jiang, K.; Zhang, K.Q.; Yang, J. Functional analysis of seven regulators of G protein signaling (RGSs) in the nematode-trapping fungus Arthrobotrys oligospora. Virulence 2021, 12, 1825–1840. [Google Scholar] [CrossRef]
- Lafon, A.; Han, K.H.; Seo, J.A.; Yu, J.H.; d’Enfert, C. G-protein and cAMP-mediated signaling in aspergilli: A genomic perspective. Fungal Genet. Biol. 2006, 43, 490–502. [Google Scholar] [CrossRef] [PubMed]
- Bayram, O.; Braus, G.H. Coordination of secondary metabolism and development in fungi: The velvet family of regulatory proteins. FEMS Microbiol. Rev. 2012, 36, 1–24. [Google Scholar] [CrossRef] [PubMed]
- van Peij, N.N.; Gielkens, M.M.; de Vries, R.P.; Visser, J.; de Graaff, L.H. The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Appl. Environ. Microbiol. 1998, 64, 3615–3619. [Google Scholar] [CrossRef]
- Marui, J.; Tanaka, A.; Mimura, S.; de Graaff, L.H.; Visser, J.; Kitamoto, N.; Kato, M.; Kobayashi, T.; Tsukagoshi, N. A transcriptional activator, AoXlnR, controls the expression of genes encoding xylanolytic enzymes in Aspergillus oryzae. Fungal Genet. Biol. 2002, 35, 157–169. [Google Scholar] [CrossRef]
- Calero-Nieto, F.; Di Pietro, A.; Roncero, M.I.; Hera, C. Role of the transcriptional activator xlnR of Fusarium oxysporum in regulation of xylanase genes and virulence. Mol. Plant Microbe Interact. 2007, 20, 977–985. [Google Scholar] [CrossRef]
- Schjerling, P.; Holmberg, S. Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators. Nucleic Acids Res. 1996, 24, 4599–4607. [Google Scholar] [CrossRef]
- Kersey, C.M.; Agyemang, P.A.; Dumenyo, C.K. CorA, the magnesium/nickel/cobalt transporter, affects virulence and extracellular enzyme production in the soft rot pathogen Pectobacterium carotovorum. Mol. Plant Pathol. 2012, 13, 58–71. [Google Scholar] [CrossRef]
- Sermon, J.; Wevers, E.M.; Jansen, L.; De Spiegeleer, P.; Vanoirbeek, K.; Aertsen, A.; Michiels, C.W. CorA affects tolerance of Escherichia coli and Salmonella enterica serovar Typhimurium to the lactoperoxidase enzyme system but not to other forms of oxidative stress. Appl. Environ. Microbiol. 2005, 71, 6515–6523. [Google Scholar] [CrossRef]
- Reza, M.H.; Shah, H.; Manjrekar, J.; Chattoo, B.B. Magnesium uptake by CorA transporters is essential for growth, development and infection in the rice blast fungus Magnaporthe oryzae. PLoS ONE 2016, 11, e0159244. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Christianson, T.W.; Sikorski, R.S.; Dante, M.; Shero, J.H.; Hieter, P. Multifunctional yeast high-copy-number shuttle vectors. Gene 1992, 110, 119–122. [Google Scholar] [CrossRef]
- Gray, N. Ecology of nematophagous fungi: Comparison of the soil sprinkling method with the Baermann funnel technique in the isolation of endoparasites. Soil Biol. Biochem. 1984, 16, 81–83. [Google Scholar] [CrossRef]
- Colot, H.V.; Park, G.; Turner, G.E.; Ringelberg, C.; Crew, C.M.; Litvinkova, L.; Weiss, R.L.; Borkovich, K.A.; Dunlap, J.C. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 2006, 103, 10352. [Google Scholar] [CrossRef]
- Li, J.; Wu, R.; Wang, M.; Borneman, J.; Yang, J.; Zhang, K.Q. The pH sensing receptor AopalH plays important roles in the nematophagous fungus Arthrobotrys oligospora. Fungal Biol. 2019, 123, 547–554. [Google Scholar] [CrossRef]
- Xie, X.Q.; Li, F.; Ying, S.H.; Feng, M.G. Additive contributions of two manganese-cored superoxide dismutases (MnSODs) to antioxidation, UV tolerance and virulence of Beauveria bassiana. PLoS ONE 2012, 7, e30298. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Miao, Q.; Zhou, Z.; Lu, S.; Li, J. Identification of Three Novel Conidiogenesis-Related Genes in the Nematode-Trapping Fungus Arthrobotrys oligospora. Pathogens 2022, 11, 717. https://doi.org/10.3390/pathogens11070717
Liu X, Miao Q, Zhou Z, Lu S, Li J. Identification of Three Novel Conidiogenesis-Related Genes in the Nematode-Trapping Fungus Arthrobotrys oligospora. Pathogens. 2022; 11(7):717. https://doi.org/10.3390/pathogens11070717
Chicago/Turabian StyleLiu, Xiaoying, Qiao Miao, Zong Zhou, Siyi Lu, and Juan Li. 2022. "Identification of Three Novel Conidiogenesis-Related Genes in the Nematode-Trapping Fungus Arthrobotrys oligospora" Pathogens 11, no. 7: 717. https://doi.org/10.3390/pathogens11070717
APA StyleLiu, X., Miao, Q., Zhou, Z., Lu, S., & Li, J. (2022). Identification of Three Novel Conidiogenesis-Related Genes in the Nematode-Trapping Fungus Arthrobotrys oligospora. Pathogens, 11(7), 717. https://doi.org/10.3390/pathogens11070717




