RAPD-PCR-Based Fingerprinting Method as a Tool for Epidemiological Analysis of Trueperella pyogenes Infections
Abstract
:1. Introduction
2. Results
2.1. Optimization of RAPD-PCR
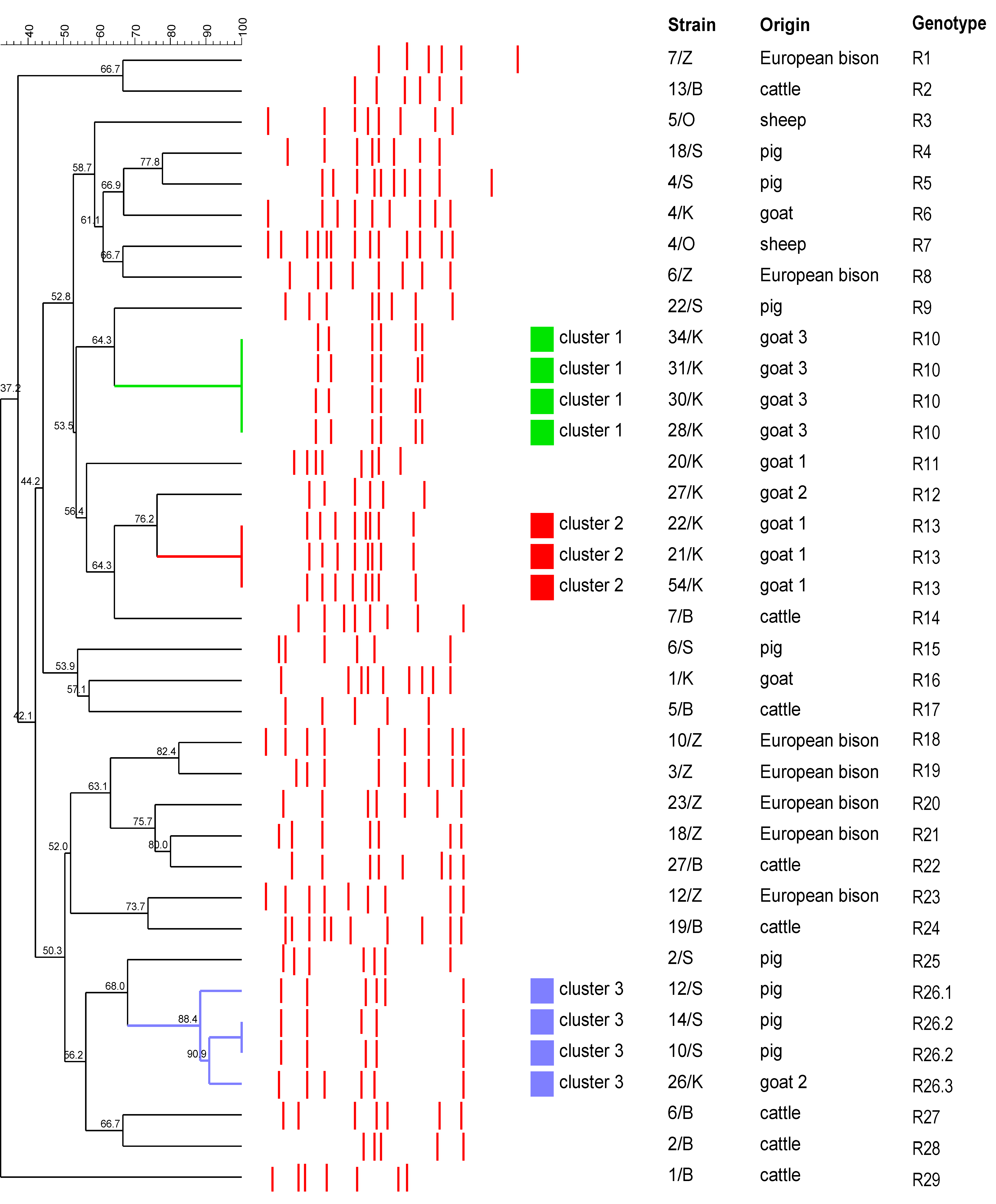
2.2. Typing of T. pyogenes Strains by RAPD-PCR
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. Extraction of DNA
4.3. Optimization of RAPD-PCR Reagents
4.4. RAPD-PCR Conditions
4.5. Gel Electrophoresis
4.6. Analysis of the Amplification Results
4.7. Calculation of a Discriminatory Index
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rzewuska, M.; Kwiecień, E.; Chrobak-Chmiel, D.; Kizerwetter-Świda, M.; Stefańska, I.; Gieryńska, M. Pathogenicity and Virulence of Trueperella pyogenes: A Review. Int. J. Mol. Sci. 2019, 20, 2737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galán-Relaño, Á.; Gómez-Gascón, L.; Luque, I.; Barrero-Domínguez, B.; Casamayor, A.; Cardoso-Toset, F.; Vela, A.I.; Fernández-Garayzábal, J.F.; Tarradas, C. Antimicrobial susceptibility and genetic characterization of Trueperella pyogenes isolates from pigs reared under intensive and extensive farming practices. Vet. Microbiol. 2019, 232, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Galán-Relaño, Á.; Gómez-Gascón, L.; Barrero-Domínguez, B.; Luque, I.; Jurado-Martos, F.; Vela, A.I.; Sanz-Tejero, C.; Tarradas, C. Antimicrobial susceptibility of Trueperella pyogenes isolated from food-producing ruminants. Vet. Microbiol. 2020, 242, 108593. [Google Scholar] [CrossRef] [PubMed]
- Neoh, H.M.; Tan, X.E.; Sapri, H.F.; Tan, T.L. Pulsed-field gel electrophoresis (PFGE): A review of the “gold standard” for bacteria typing and current alternatives. Infect. Genet. Evol. 2019, 74, 103935. [Google Scholar] [CrossRef]
- Sabat, A.J.; Budimir, A.; Nashev, D.; Sá-Leão, R.; van Dijl, J.; Laurent, F.; Grundmann, H.; Friedrich, A.W. ESCMID Study Group of Epidemiological Markers (ESGEM). Overview of molecular typing methods for outbreak detection and epidemiological surveillance. Euro Surveill. 2013, 18, 20380. [Google Scholar] [CrossRef] [Green Version]
- Zhao, K.; Tian, Y.; Yue, B.; Wang, H.; Zhang, X. Virulence determinants and biofilm production among Trueperella pyogenes recovered from abscesses of captive forest musk deer. Arch. Microbiol. 2013, 195, 203–209. [Google Scholar] [CrossRef]
- Nagib, S.; Glaeser, S.P.; Eisenberg, T.; Sammra, O.; Lämmler, C.; Kämpfer, P.; Schauerte, N.; Geiger, C.; Kaim, U.; Prenger-Berninghoff, E.; et al. Fatal infection in three Grey Slender Lorises (Loris lydekkerianus nordicus) caused by clonally related Trueperella pyogenes. BMC Vet. Res. 2017, 13, 273. [Google Scholar] [CrossRef] [Green Version]
- Cardoso-Toset, F.; Gómez-Laguna, J.; Gómez-Gascón, L.; Rodríguez-Gómez, I.M.; Galán-Relaño, A.; Carrasco, L.; Tarradas, C.; Vela, A.I.; Luque, I. Histopathological and microbiological study of porcine lymphadenitis: Contributions to diagnosis and control of the disease. Porc. Health Manag. 2020, 6, 36. [Google Scholar] [CrossRef]
- Taguchi, G.; Wu, Y. Introduction to Off-Line Quality Control; Japan Quality Control Organisation: Negoya, Japan, 1980. [Google Scholar]
- Cobb, B.D.; Clarkson, J.M. A simple procedure for optimising the polymerase chain reaction (PCR) using modified Taguchi methods. Nucleic Acids Res. 1994, 22, 3801–3805. [Google Scholar] [CrossRef] [Green Version]
- Silva, E.; Gaivão, M.; Leitão, S.; Jost, B.H.; Carneiro, C.; Vilela, C.L.; Lopes da Costa, L.; Mateus, L. Genomic characterization of Arcanobacterium pyogenes isolates recovered from the uterus of dairy cows with normal puerperium or clinical metritis. Vet. Microbiol. 2008, 132, 111–118. [Google Scholar] [CrossRef]
- Tamai, I.A.; Mohammadzadeh, A.; Salehi, T.Z.; Pezhman, M.; Pakbin, B. Investigation of antimicrobial susceptibility and virulence factor genes in Trueperella pyogenes isolated from clinical mastitis cases of dairy cows. Food Sci. Nutr. 2021, 9, 4530–4539. [Google Scholar] [CrossRef]
- Rzewuska, M.; Stefańska, I.; Osińska, B.; Kizerwetter-Świda, M.; Chrobak, D.; Kaba, J.; Bielecki, W. Phenotypic characteristics and virulence genotypes of Trueperella (Arcanobacterium) pyogenes. Vet. Microbiol. 2012, 160, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Rogovskyy, A.S.; Lawhon, S.; Kuczmanski, K.; Gillis, D.C.; Wu, J.; Hurley, H.; Rogovska, Y.V.; Konganti, K.; Yang, C.-Y.; Duncan, K.P. Phenotypic and genotypic characteristics of Trueperella pyogenes isolated from ruminants. J. Vet. Diagn. Investig. 2018, 30, 348–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derakhshandeh, A.; Eraghi, V.; Boroojeni, A.M.; Niaki, M.A.; Zare, S.; Naziri, Z. Virulence factors, antibiotic resistance genes and genetic relatedness of commensal Escherichia coli isolates from dogs and their owners. Microb. Pathog. 2018, 116, 241–245. [Google Scholar] [CrossRef]
- Melo, R.T.; Grazziotin, A.L.; Valadares Júnior, E.C.; Prado, R.R.; Mendonça, E.P.; Monteiro, G.P.; Peres, P.A.B.M.; Rossi, D.A. Evolution of Campylobacter jejuni of poultry origin in Brazil. Food Microbiol. 2019, 82, 489–496. [Google Scholar] [CrossRef]
- Mesini, A.; Mikulska, M.; Giacobbe, D.R.; Del Puente, F.; Gandolfo, N.; Codda, G.; Orsi, A.; Tassinari, F.; Beltramini, S.; Marchese, A.; et al. Changing epidemiology of candidaemia: Increase in fluconazole-resistant Candida parapsilosis. Mycoses 2020, 63, 361–368. [Google Scholar] [CrossRef]
- Ribeiro, L.F.; Sato, R.A.; de Souza Pollo, A.; Marques Rossi, G.A.; do Amaral, L.A. Occurence of methicillin-resistant Staphylococcus spp. on Brazillian dairy farms that produce unpasteurized cheese. Toxins 2020, 12, 779. [Google Scholar] [CrossRef]
- Khalili, Y.; Memar, M.Y.; Farajnia, S.; Khosro, A.; Kafil, H.S.; Ghotaslou, R. Molecular epidemiology and carbapenem resistance of Pseudomonas aeruginosa isolated from patients with burns. J. Wound Care 2021, 30, 135–141. [Google Scholar] [CrossRef]
- Berthold-Pluta, A.; Garbowska, M.; Stefańska, I.; Stasiak-Różańska, L.; Aleksandrzak-Piekarczyk, T.; Pluta, A. Microbiological uality of nuts, dried and candied fruits, including the prevalence of Cronobacter spp. Pathogens 2021, 10, 900. [Google Scholar] [CrossRef]
- Van Belkum, A.; Rassios, P.T.; Dijkshoorn, L.; Haeggman, S.; Cookson, B.; Fry, N.K.; Fussing, V.; Green, J.; Feil, E.; Gerner-Smidt, P.; et al. Guidelines for the validation and applications of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 2007, 3, 1–46. [Google Scholar] [CrossRef]
- Hunter, P.R.; Gaston, M.A. Numerical index of the discriminatory ability of typing systems: An application of Simpson’s index of diversity. J. Clin. Microbiol. 1988, 26, 2465–2466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rzewuska, M.; Czopowicz, M.; Gawryś, M.; Markowska-Daniel, I.; Bielecki, W. Relationships between antimicrobial resistance, distribution of virulence factor genes and the origin of Trueperella pyogenes isolated from domestic animals and Eurpean bison (Bison bonasus). Microb. Pathog. 2016, 96, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Kwiecień, E.; Stefańska, I.; Chrobak-Chmiel, D.; Kierwetter-Świda, M.; Moroz, A.; Olech, W.; Spinu, M.; Binek, M.; Rzewuska, M. Trueperella pyogenes isolates from livestock and European Bison (Bison bonasus) as a reservoir of tetracycline resistance determinants. Antibiotics 2021, 10, 380. [Google Scholar] [CrossRef] [PubMed]
- Andrighetto, C.; Zampese, L.; Lombardi, A. RAPD-PCR characterization of lactobacilli isolated from artisanal meat plants and traditional fermented sausages of Vento region (Italy). Lett. Appl. Microbiol. 2001, 33, 26–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, L.J.; Timminis, M.J. Differentiation of Lactobacillus casei, Lactobacillus paracasei and Lactobacillus rhamnosus by polymerase chain reaction. Lett. Appl. Microbiol. 1999, 29, 90–92. [Google Scholar] [CrossRef] [Green Version]
- Nazarowec-White, M.; Farber, J.M. Phenotypic and genotypic typing of food and clinical isolates of Enterobacter sakazakii. J. Med. Microbiol. 1999, 48, 559–567. [Google Scholar] [CrossRef] [Green Version]
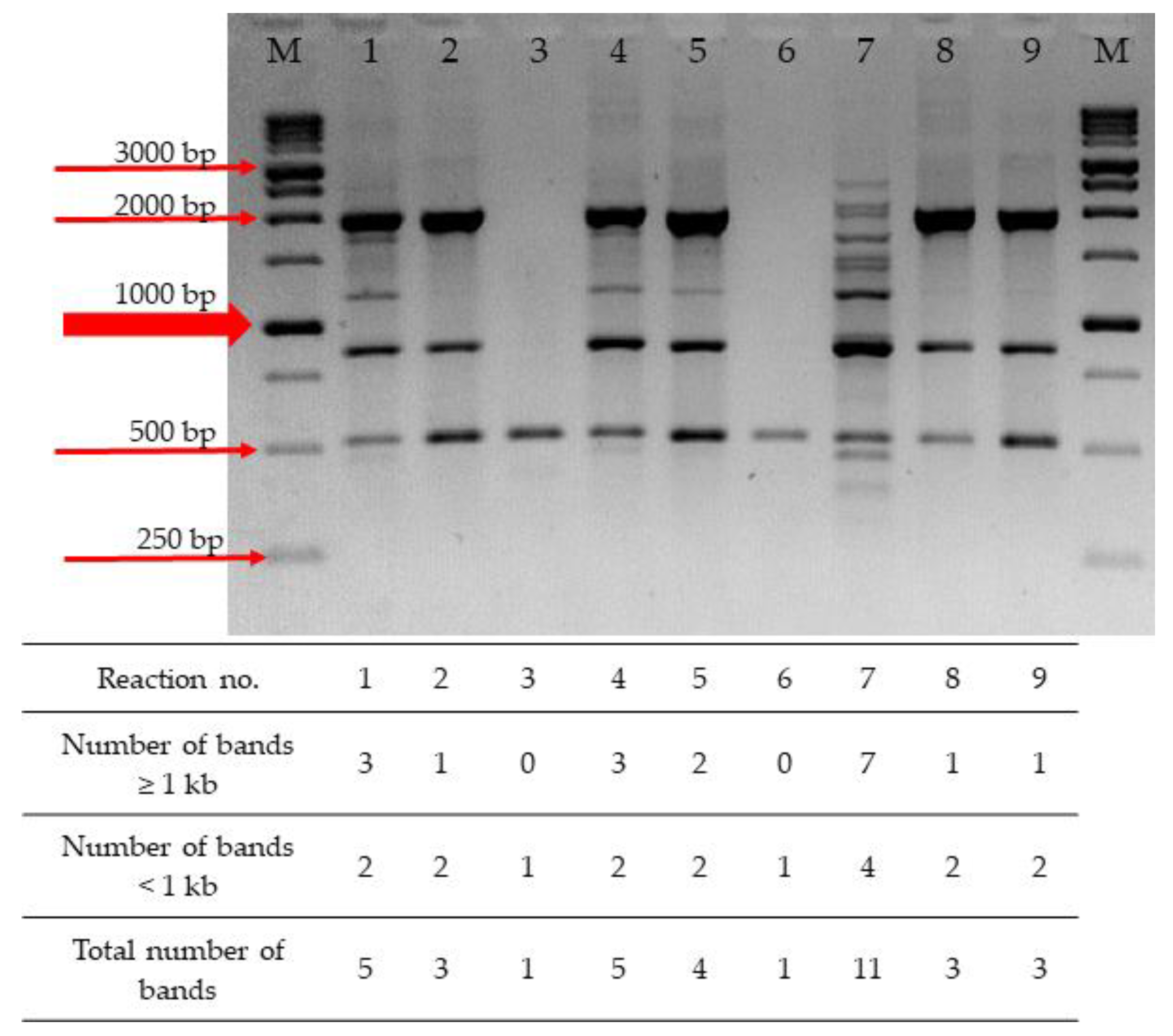
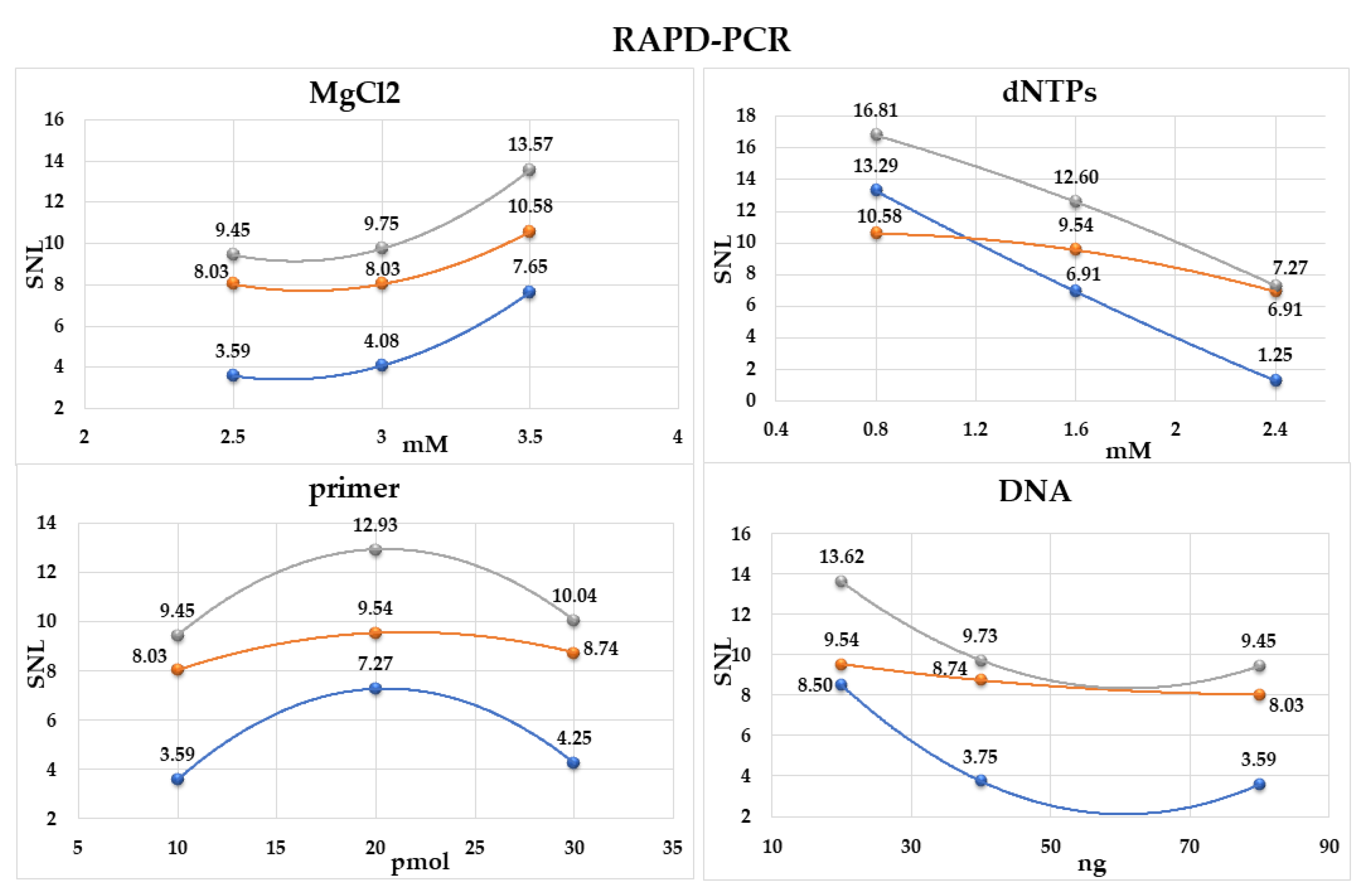
| Reaction No. | MgCl2 (mM) | dNTPs (mM) | Primer (pmol) | DNA (ng) |
|---|---|---|---|---|
| 1. | 2.5 | 0.8 | 10 | 20 |
| 2. | 2.5 | 1.6 | 20 | 40 |
| 3. | 2.5 | 2.4 | 30 | 80 |
| 4. | 3.0 | 0.8 | 20 | 80 |
| 5. | 3.0 | 1.6 | 30 | 20 |
| 6. | 3.0 | 2.4 | 10 | 40 |
| 7. | 3.5 | 0.8 | 30 | 40 |
| 8. | 3.5 | 1.6 | 10 | 80 |
| 9. | 3.5 | 2.4 | 20 | 20 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stefańska, I.; Kwiecień, E.; Górzyńska, M.; Sałamaszyńska-Guz, A.; Rzewuska, M. RAPD-PCR-Based Fingerprinting Method as a Tool for Epidemiological Analysis of Trueperella pyogenes Infections. Pathogens 2022, 11, 562. https://doi.org/10.3390/pathogens11050562
Stefańska I, Kwiecień E, Górzyńska M, Sałamaszyńska-Guz A, Rzewuska M. RAPD-PCR-Based Fingerprinting Method as a Tool for Epidemiological Analysis of Trueperella pyogenes Infections. Pathogens. 2022; 11(5):562. https://doi.org/10.3390/pathogens11050562
Chicago/Turabian StyleStefańska, Ilona, Ewelina Kwiecień, Małgorzata Górzyńska, Agnieszka Sałamaszyńska-Guz, and Magdalena Rzewuska. 2022. "RAPD-PCR-Based Fingerprinting Method as a Tool for Epidemiological Analysis of Trueperella pyogenes Infections" Pathogens 11, no. 5: 562. https://doi.org/10.3390/pathogens11050562
APA StyleStefańska, I., Kwiecień, E., Górzyńska, M., Sałamaszyńska-Guz, A., & Rzewuska, M. (2022). RAPD-PCR-Based Fingerprinting Method as a Tool for Epidemiological Analysis of Trueperella pyogenes Infections. Pathogens, 11(5), 562. https://doi.org/10.3390/pathogens11050562







