Abstract
Actinobacillus pleuropneumoniae is a major pathogen of swine, which can cause severe pleuropneumonia in pigs, but sometimes the disease can be generalized. Diseases caused by A. pleuropneumoniae are frequent all over the world, resulting in high losses among domestic pigs. However, our knowledge on the occurrence of A. pleuropneumoniae in wild boars and feral pigs is limited. We aimed to examine the carriage of A. pleuropneumoniae by hunted wild boars. The presence of A. pleuropneumoniae was examined in tonsils of 68 hunted wild boars collected at a game processing unit. An in-house designed species-specific PCR test was used to detect the gene of Apx IV toxin, and the samples were inoculated on a modified selective agar. A. pleuropneumoniae was detected in 10 animals (14.7%) by PCR and one A. pleuropneumoniae serotype 12 strain was isolated. The antibiotic resistance pattern of the strain resembled field strains that were isolated from farmed pigs in Hungary. This is the first case for the detection of A. pleuropneumoniae not only using PCR or ELISA, but also its isolation, identification, and serotyping.
1. Introduction
Actinobacillus pleuropneumoniae is a major pathogen of swine. There are different virulence variants of the agent, the highly virulent ones can cause severe fibrino-haemorrhagic, necrotic pneumonia, and fibrinous pleuritis in pigs, but sometimes the disease can be generalized in the infected animals [1,2]. The moderately virulent strains play an important role in the aetiology of porcine respiratory disease complex [3]. The host range of A. pleuropneumoniae is very narrow, it can cause disease only in swine (Sus scrofa), including domestic pigs, wild boars, and feral pigs. However, it was identified once as a causative agent in laying hens, showing clinical signs of infectious coryza [4]. A. pleuropneumoniae is carried in tonsils by infected animals. In addition, it is shed in respiratory discharge, especially when the animals are coughing [2,5,6].
Actinobacillus pleuropneumoniae has two biotypes, biotype I strains can replicate only in the presence of nicotinamide adenine dinucleotide (NAD, V-factor), which is different from the biotype II strains that do not need it. Virulence of A. pleuropneumoniae depends on several virulence factors, including four types of toxins, fimbria, outer membrane proteins, ability of biofilm formation, presence of transporter systems, and different enzymes, that contribute to the virulence of the agent [7,8,9,10]. To date, a total of 19 serovars have been identified on the basis of surface soluble capsular polysaccharide antigens [11]. The frequency of different biotypes and serovars of A. pleuropneumoniae shows great geographical differences [2].
Diseases caused by A. pleuropneumoniae are frequent all over the world, and can cause high losses among domesticated pigs. However, our knowledge on the occurrence of A. pleuropneumoniae in wild boars and feral pigs is limited. A large part (52%) of hunted wild boars proved to be seropositive for A. pleuropneumoniae in Slovenia [12]. More than two thirds (69.7%) of the serum samples were positive in the United States, and the presence of antibodies against different serotypes of A. pleuropneumoniae was identified [13]. In Finland 12.6%, while in Greece 90.5% of the sampled farmed wild boars had antibodies against the agent [14,15]. Moreover, seropositivity to A. pleuropneumoniae was described in Canada. However, the serotypes found in wild boars and domestic pigs were different [16,17]. Polymerase chain reaction (PCR) detected A. pleuropneumoniae in lung and tonsil samples of 38.5% of wild boars in Germany [18], but feral pigs in Australia proved to be free from A. pleuropneumoniae when lungs were examined with PCR [19]. Domestic pigs, wild boars, and feral pigs belong to a single species (Sus scrofa), they can be reservoirs of different viruses and bacteria, and can infect each other [20].
There are several methods to detect A. pleuropneumoniae in tonsils. Isolation of the agent on selective media [21] is rather difficult due to the fact that several other bacteria can overgrow it. Different PCR methods [22,23] or isolation by immunomagnetic separation [24,25] are also available.
The aim of the present examination was an evaluation of the presence of A. pleuropneumoniae in hunted wild boars in Hungary.
2. Results
All of the 38 field bacterial isolates investigated and type strains of A. pleuropneumoniae strains provided a positive reaction in the PCR test that was used to detect the species-specific Apx IV gene of A. pleuropneumoniae, while Streptococcus suis, Staphylococcus aureus subsp. aureus, Escherichia coli, and Proteus mirabilis did not react.
Actinobacillus pleuropneumoniae was detected by PCR in tonsils of 10 out of 68 (14.7%) wild boars. The positive animals were shot in different geographical locations in Hungary. Neither the positive nor the negative animals had any postmortem lesions. Moreover, no acute or chronic lesions of A. pleuropneumoniae could be observed.
Only one A. pleuropneumoniae strain could be isolated from tonsils of wild boars (1.47%), the same tonsil was also positive in the PCR test. The isolated A. pleuropneumoniae strain could be assigned to serotype 12. The results are shown on the map (Figure 1).
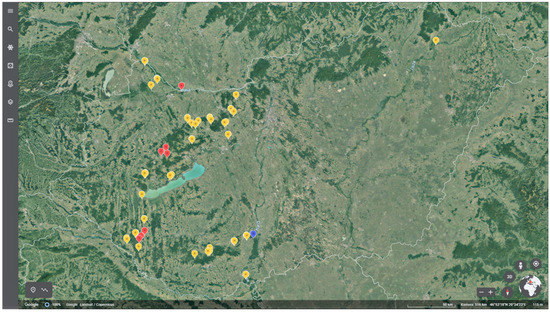
Figure 1.
Detection of A. pleuropneumoniae from wild boars in Hungary (red: PCR positive; blue: PCR and culture positive; yellow: PCR negative).
The minimum inhibitory concentrations of the examined antibiotics are presented in Table 1.

Table 1.
Antibiotic susceptibility of Actinobacillus pleuropneumoniae isolated from wild boars.
3. Discussion
The type specific in-house designed PCR, which was used to detect the gene of Apx IV toxin, proved to be reliable. All of the A. pleuropneumoniae type and field strains provided a positive reaction, while the reactions in the case of other bacterium species remained negative. Detection of the gene of Apx IV toxin is specific, only A. pleuropneumoniae strains carry it, while genes of Apx I-II-III can also be present in other species [10].
Actinobacillus pleuropneumoniae is a widespread bacterium in pig herds. However, there are limited data on the occurrence of the agent in wild boars and feral pigs. In Slovenia, the United States, and Canada, 52–100% of the animals were seropositive for A. pleuropneumoniae [12,13,16]. In addition, 12.6–90% of the farmed boars had antibodies against A. pleuropneumoniae [14,15]. A. pleuropneumoniae was detected in 38.5% of wild boars in Germany [18]. Only 14.7% of the wild boars carried A. pleuropneumoniae in tonsils in Hungary, but we could isolate an A. pleuropneumoniae strain from the tonsil of a wild boar. To the best of our knowledge, this is the first case for the isolation of the agent from wild boars. Isolation of A. pleuropneumoniae using selective media is not a simple task, since other, rapidly growing bacteria can overgrow it [2]. Moreover, isolation of certain serotypes of A. pleuropneumoniae is possible with immunomagnetic separation. However, selective isolation has to be used when the serotype of the targeted strain is not known [24,25].
The isolated strain could be assigned to serotype 12. Serotype 12 strains occur in Hungary, 3.3% of the A. pleuropneumoniae strains belong to this serotype [26].
There are two possible ways of infection among wild boars with A. pleuropneumoniae. Circulation of the agent within wild boar populations is an option. It is assumed in Canada, where wild boars were seropositive for serotype 14, which was not present in domestic pig populations [16]. The other option is infection from domestic pigs in the case of close contact. The A. pleuropneumoniae strain that we isolated was resistant against penicillin, gentamycin, spectinomycin, cefoperazone, tulathromycin, and tilmicosin. The antibiotic resistance pattern of the strain resembled the A. pleuropneumoniae strains isolated from farmed pigs. In addition, it was resistant against those antibiotics that are frequently used in Hungarian pig herds [27]. On the basis of these data, we could hypothesize that the wild boar was most probably infected from a farmed pig. Backyard-raised pigs are usual in rural areas of Hungary, where wild boar populations are also high.
Wild boars can infect farmed pigs with A. pleuropneumoniae. However, the risk is not high in the case of intensive units thanks to the high level of biosecurity, but they can infect backyard-raised pigs, especially during sow heat or estrus. The risk of infection is low in free ranging wild boars since close contact is needed for the infection, but it can be increased among farmed wild boars if the animal density is high.
4. Materials and Methods
4.1. Samples
Tonsils of 68 hunted wild boars were collected at a game processing unit in Hungary. With one exception the wild boars that were shot originated from 8 countries of the western part of the country. They were adult animals of both sexes in average body condition and seemed to be healthy. The tonsil samples were transported on ice to the laboratory immediately after collection, and were examined within 3 h.
4.2. Polymerase Chain Reaction
Polymerase chain reaction was used to detect A. pleuropneumoniae strains in tonsils. DNA was extracted from tonsils with QIAGEN QIAamp® DNA Mini Kit (Qiagen Inc., Germantown, MD, USA), following the instructions of the producer. Our own PCR test was used for the identification of A. pleuropneumoniae. The primers (5′-ACG AACAAC GCG GCT AAT A-3′ and 5′-CTC ACC TAA CGG ACG AGT AAA-3′) were planned to detect the species-specific Apx IV gene of A. pleuropneumoniae. The reaction mixture contained 40.7 μL water, 5 μL buffer, 0.1 μL of 10 mM dNTP, 0.2 μL (5 U/μL) Taq polymerase, 2 μL template, and 1-1 μL of 10 μM primers. After 5 min of initialization at 94 °C, 35 cycles of denaturation at 94 °C, annealing at 55 °C, and extension at 72 °C were carried out, followed by a 7-min-long final elongation step. The amplicon was visualized by gel electrophoresis in 2% agarose gel in Tris–acetate–EDTA buffer (40 mM Tris–acetate and 1 mM EDTA pH 8.3) containing Green Nucleic Acid Gel Stain (Biocenter Ltd., Szeged, Hungary).
The specificity of the test was controlled with a single strain of each of the following: S. suis, S. aureus subsp. aureus, E. coli, and P. mirabilis, which are normally present in the pig, and 38 A. pleuropneumoniae strains including type and field strains. The strains were from our bacterium collection.
4.3. Bacterium Culture, Serotyping, Antibiotic Resistance
Tonsil samples were inoculated onto a selective agar medium, which was described by Jacobsen and Nielsen [21] and that we partially modified. Mueller–Hinton agar (Biolab Ltd., Budapest, Hungary) containing 5% defibrinated sheep blood was used rather than the meat and blood agar. In addition, 50 μg/mL NAD and 1% yeast extract (Biolab Ltd., Budapest, Hungary), as well as 100 μg/mL bacitracin, 1 μg/mL crystal violet, 50 μg/mL nystatin, and 1 μg/mL lincomycin (Sigma-Aldrich, St. Louise, MI, USA) were added. The agar plates were incubated at 37 °C for 72 h and read daily. The isolated bacteria were characterized on the basis of their morphological, cultural, and biochemical features [28,29]. In addition, the identification of the isolated A. pleuropneumoniae strain was confirmed with the above-mentioned PCR. The isolated A. pleuropneumoniae strain was serotyped in the indirect hemagglutination test using hyperimmune sera raised against 1–19 serotype strains of A. pleuropneumoniae, as described [26,30]. The antibiotic resistance of the isolated A. pleuropneumoniae strain was examined by measuring the minimum inhibition concentration in the microdilution method. Briefly, Mueller–Hinton broth (Biolab Ltd., Hungary) with the addition of 10 mg/mL Ca++ (CaCl2 × 2 H2O; Spektrum-3D Ltd., Debrecen, Hungary), 10 mg/mL Mg++ (MgCl2 × 6 H2O; Scharlab Hungary Ltd., Debrecen, Hungary) recommended for fastidious bacteria, and 67 μg/mL NAD medium were used. The tests were carried out in 96-well polystyrene microtiter plates (Kartell, Noviglio, Italy). The breakpoint values of A. pleuropneumoniae were derived, and when they were not available, the values of Mannheimia haemolytica and Actinobacillus sp. were used as reference [31,32].
5. Conclusions
Actinobacillus pleuropneumoniae was detected from tonsils of 14.7% of hunted wild boars, and one serotype 12 strain of A. pleuropneumoniae was isolated. This is the first case for the isolation of A. pleuropneumoniae from wild boars.
Author Contributions
Conceptualization, L.F.; methodology, R.S. and L.M.; validation, L.M. and L.F.; formal analysis, L.M.; investigation, R.S.; resources, L.F.; data curation, L.F.; writing—original draft preparation, R.S.; writing—review and editing, L.M. and L.F.; supervision, L.F.; project administration, L.F.; funding acquisition, L.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Hungarian Scientific Research Fund (OTKA 132833).
Institutional Review Board Statement
Ethical review and approval were waived for this study, due to the fact that the samples were collected from carcasses of hunted wild boars with consent of the owner.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors thank L. Hetényi D.V.M. for support in the collection of tonsils.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hoeltig, D.; Rohde, J.; Frase, R.; Nietfeld, F.; Waldmann, K.-H.; Valentin-Weigand, P.; Meens, J. Multi-organ spreading of Actinobacillus pleuropneumoniae serovar 7 in weaned pigs during the first week after experimental infection. Vet. Res. 2018, 49, 97. [Google Scholar] [CrossRef] [PubMed]
- Gottschalk, M.; Broes, A. Actinobacillosis. In Diseases of Swine, 11th ed.; Zimmerman, J.J., Karriker, L.A., Ramirez, A., Schwartz, K.J., Stevenson, G.W., Zhang, J., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2019; pp. 749–766. [Google Scholar]
- Hansen, M.S.; Pors, S.E.; Jensen, H.E.; Bille-Hansen, V.; Bisgaard, M.; Flachs, E.M.; Nielsen, O.L. An investigation of the pathology and pathogens associated with porcine respiratory disease complex in Denmark. J. Com. Pathol. 2010, 143, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Márquez, V.M.P.; Ochoa, J.L.; Cruz, C.V.; Alonso, P.S.; Olmedo-Alvarez, G.; Vaca, S.P.; Abascal, E.N. Isolation of Actinobacillus pleuropneumoniae from layer hens showing clinical signs of infectious coryza. Avian Dis. 2014, 58, 638–641. [Google Scholar] [CrossRef] [PubMed]
- Bossé, J.T.; Janson, H.; Sheehan, B.J.; Beddek, A.J.; Rycroft, A.N.; Kroll, J.S.; Langford, P.R. Actinobacillus pleuropneumoniae: Pathobiology and pathogenesis of infection. Microbes Infect. 2002, 4, 225–235. [Google Scholar] [CrossRef]
- Sassu, E.L.; Frömbling, J.; Duvigneau, J.C.; Miller, I.; Müllebner, A.; Gutiérrez, A.M.; Grunert, T.; Patzl, M.; Saalmüller, A.; von Altrock, A.; et al. Host-pathogen interplay at primary infection sites in pigs challenged with Actinobacillus pleuropneumoniae. BMC Vet. Res. 2017, 13, 64. [Google Scholar] [CrossRef][Green Version]
- Chiers, K.; de Waele, T.; Pasmans, F.; Ducatelle, R.; Haesebrouck, F. Virulence factors of Actinobacillus pleuropneumoniae involved in colonisation, persistence and induction of lesions in its porcine host. Vet. Res. 2010, 41, 65. [Google Scholar] [CrossRef]
- Grasteau, A.; Tremblay, Y.D.N.; Labrie, J.; Jacques, M. Novel genes associated with biofilm formation of Actinobacillus pleuropneumoniae. Vet. Microbiol. 2011, 153, 134–143. [Google Scholar] [CrossRef]
- Sassu, E.L.; Bossé, J.T.; Tobias, T.J.; Gottschalk, M.; Langford, P.R.; Hennig-Pauka, I. Update on Actinobacillus pleuropneumoniae. Knowledge, gaps and challenges. Tranbound. Emerg. Dis. 2018, 65 (Suppl. 1), 72–90. [Google Scholar] [CrossRef]
- Frey, J. RTX toxins of animal pathogens and their role as antigens in vaccines and diagnostics. Toxins 2019, 11, 719. [Google Scholar] [CrossRef]
- Stringer, O.W.; Bossé, J.T.; Lacouture, S.; Gottschalk, M.; Fodor, L.; Angen, Ø.; Velazquez, E.; Penny, P.; Lei, L.; Langford, P.R.; et al. Proposal of Actinobacillus pleuropneumoniae serovar 19, and reformulation of previous multiplex PCRs for capsule-specific typing of all known serovars. Vet. Microbiol. 2021, 255, 109021. [Google Scholar] [CrossRef]
- Vengust, G.; Valencak, Z.; Bidovec, A.A. serological survey of selected pathogens in wild boar in Slovenia. J. Vet. Med. B 2006, 53, 24–27. [Google Scholar] [CrossRef] [PubMed]
- Baroch, J.A.; Gagnon, C.A.; Lacouture, S.; Gottschalk, M. Exposure of feral swine (Sus scrofa) in the United States to selected pathogens. Canad. J. Vet. Res. 2015, 79, 74–78. [Google Scholar]
- Hälli, O.; Ala-Kurikka, E.; Wallgren, P.; Heinonen, M. Actinobacillus pleuropneumoniae seroprevalence in farmed wild boars in Finland. J. Zoo Wildlife Med. 2014, 45, 813–818. [Google Scholar] [CrossRef] [PubMed]
- Marinou, K.A.; Papatsiros, V.G.; Gkotsopoulos, E.K.; Odatzoglu, P.K.; Athanasiou, L.V. Exposure of extensively farmed wild boars (Sus scrofa scrofa) to selected pig pathogens in Greece. Vet. Quart. 2015, 35, 97–101. [Google Scholar] [CrossRef]
- McGregor, G.F.; Gottschalk, M.; Godson, D.L.; Wilkins, W.; Bollinger, T.K. Disease risks associated with free-ranging wild boar in Saskatchewan. Can. Vet. J. 2015, 56, 839–844. [Google Scholar]
- Lacouture, S.; Gottschalk, M. Distribution of Actinobacillus pleuropneumoniae (from 2015 to June 2020) and Glaesserella parasuis (from 2017 to June 2020) serotypes isolated from diseased pigs in Quebec. Can. Vet. J. 2020, 61, 1261–1263. [Google Scholar]
- Reiner, G.; Fresen, C.; Bronnert, S.; Haack, I.; Willems, H. Prevalence of Actinobacillus pleuropneumoniae infection in hunted wild boars (Sus scrofa) in Germany. J. Wildlife Dis. 2010, 46, 551–555. [Google Scholar] [CrossRef][Green Version]
- Pearson, H.E.; Toribio, J.-A.L.M.L.; Hernandez-Jover, M.; Marshall, D.; Lapidge, S.J. Pathogen presence in feral pigs and their movement around two commercial piggeries in Queensland, Australia. Vet. Rec. 2014, 174, 325. [Google Scholar] [CrossRef]
- Ruiz-Fons, F.; Segalés, J.; Gortázar, C. A review of viral diseases of the European wild boar: Effects of population dynamics and reservoir role. Vet. J. 2008, 176, 158–169. [Google Scholar] [CrossRef]
- Jacobsen, M.J.; Nielsen, J.P. Development and evaluation of a selective and indicative medium for isolation of Actinobacillus pleuropneumoniae from tonsils. Vet. Microbiol. 1995, 47, 191–197. [Google Scholar] [CrossRef]
- Frey, J.; Beck, M.; van den Bosch, J.F.; Segers, R.P.A.M.; Nicolet, J. Development of an efficient PCR method for toxin typing of Actinobacillus pleuropneumoniae strains. Mol. Cell. Probes 1995, 9, 277–282. [Google Scholar] [CrossRef]
- Gottschalk, M. The challenge of detecting herds sub-clinically infected with Actinobacillus pleuropneumoniae. Vet. J. 2015, 206, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Gagné, A.; Lacouture, S.; Broes, A.; D’Allaire, S.; Gottschalk, M. Development of an immunomagnetic method for selective isolation of Actinobacillus pleuropneumoniae serotype 1 from tonsils. J. Clin. Microbiol. 1998, 36, 251–254. [Google Scholar] [CrossRef] [PubMed]
- Angen, O.; Heegaard, P.M.H.; Lavritsen, D.T.; Sorensen, V. Isolation of Actinobacillus pleuropneumoniae serotype 2 by immunomagnetic separation. Vet. Microbiol. 2001, 79, 19–29. [Google Scholar] [CrossRef]
- Sárközi, R.; Makrai, L.; Fodor, L. Actinobacillus pleuropneumoniae serotypes in Hungary. Acta Vet. Hung. 2018, 66, 343–349. [Google Scholar] [CrossRef]
- Sárközi, R.; Makrai, L.; Tóth, A.; Fodor, L. Hazai Actinobacillus pleuropneumoniae törzsek antibiotikum-érzékenysége. Magy. Állatorvosok. 2014, 136, 643–649. (In Hungarian) [Google Scholar]
- Barrow, G.I.; Feltham, R.K.A. Cowan and Steel’s Manual for the Identification of Medical Bacteria, 3rd ed.; Cambridge University Press: Cambridge, UK, 2003. [Google Scholar]
- Markey, B.; Leonard, F.; Archambault, M.; Cullinane, A.; Maguire, D. Clinical Veterinary Microbiology, 2nd ed.; Mosby-Elsevier: Edinburgh, UK, 2013. [Google Scholar]
- Sárközi, R.; Makrai, L.; Fodor, L. Identification of a proposed new serovar of Actinobacillus pleuropneumoniae: Serovar 16. Acta Vet. Hung. 2015, 63, 444–450. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 5th ed.; CLSI standard VET01; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 4th ed.; CLSI supplement VET08; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).