Transmission Pathways of the VNN Introduced in Croatian Marine Aquaculture
Abstract
:1. Introduction
2. Results
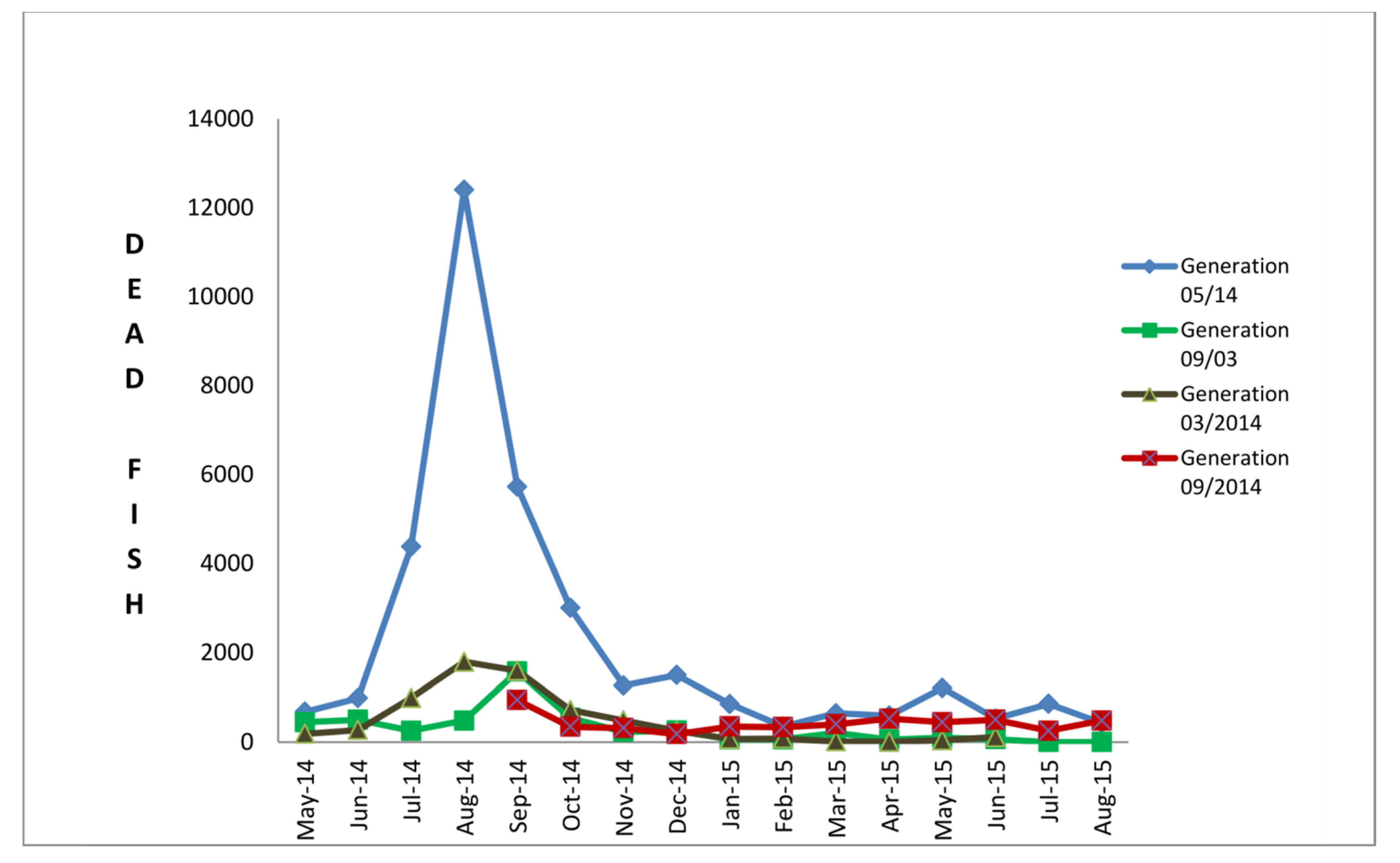
2.1. Necropsy Showed a Typical Pathology Related to VNN
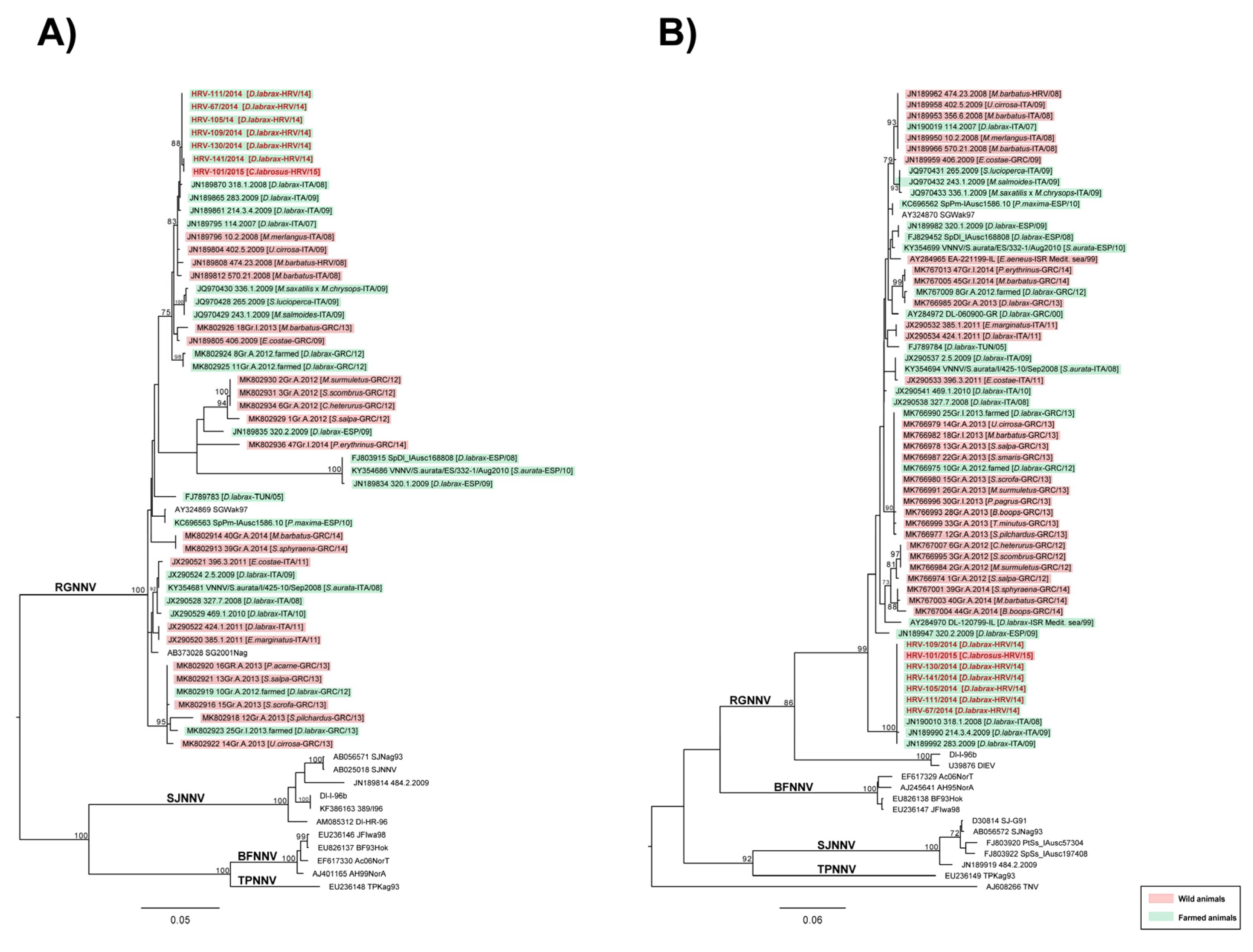
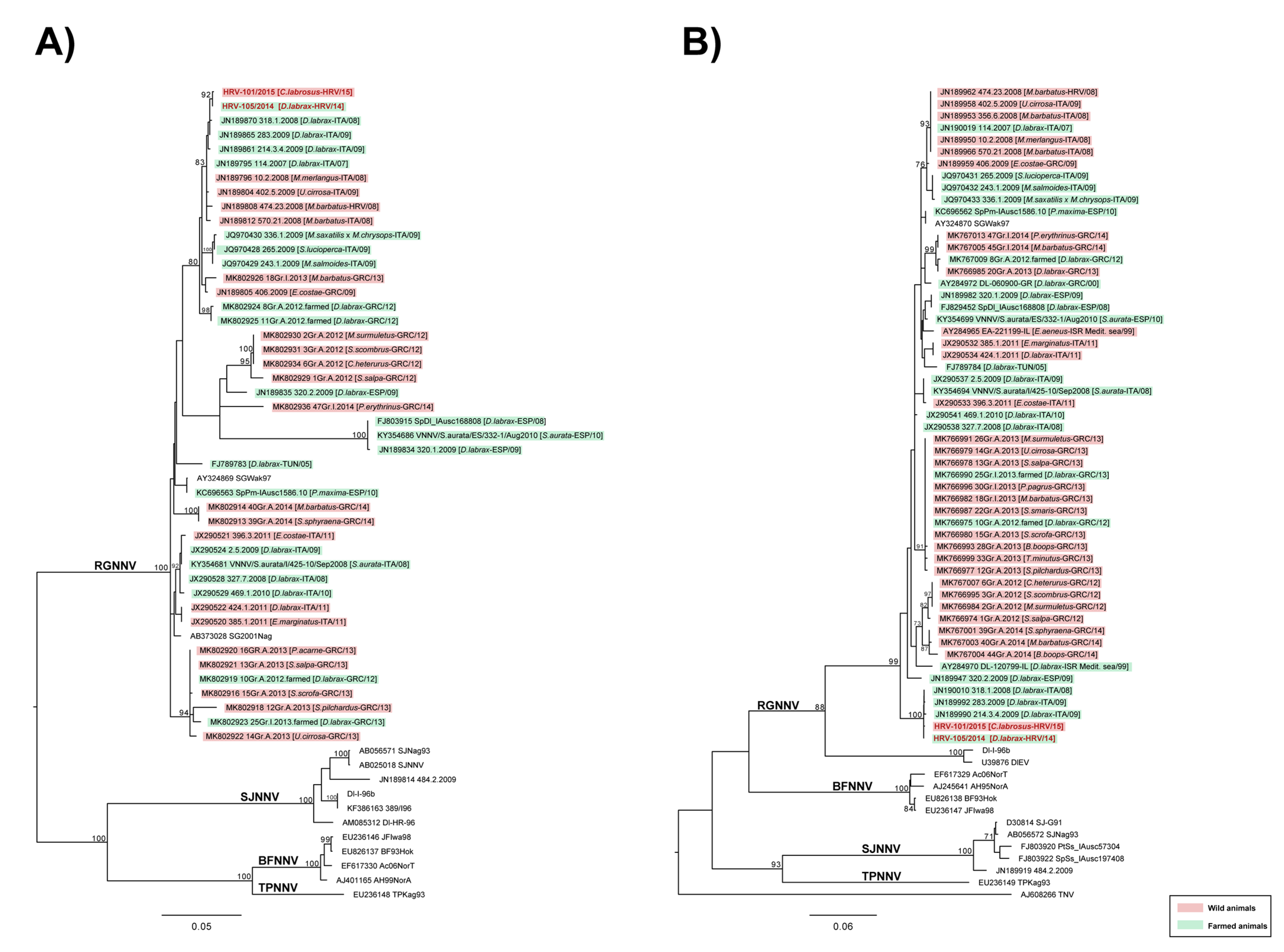
2.2. The Results of Virological and Molecular Investigations
3. Discussion
4. Materials and Methods
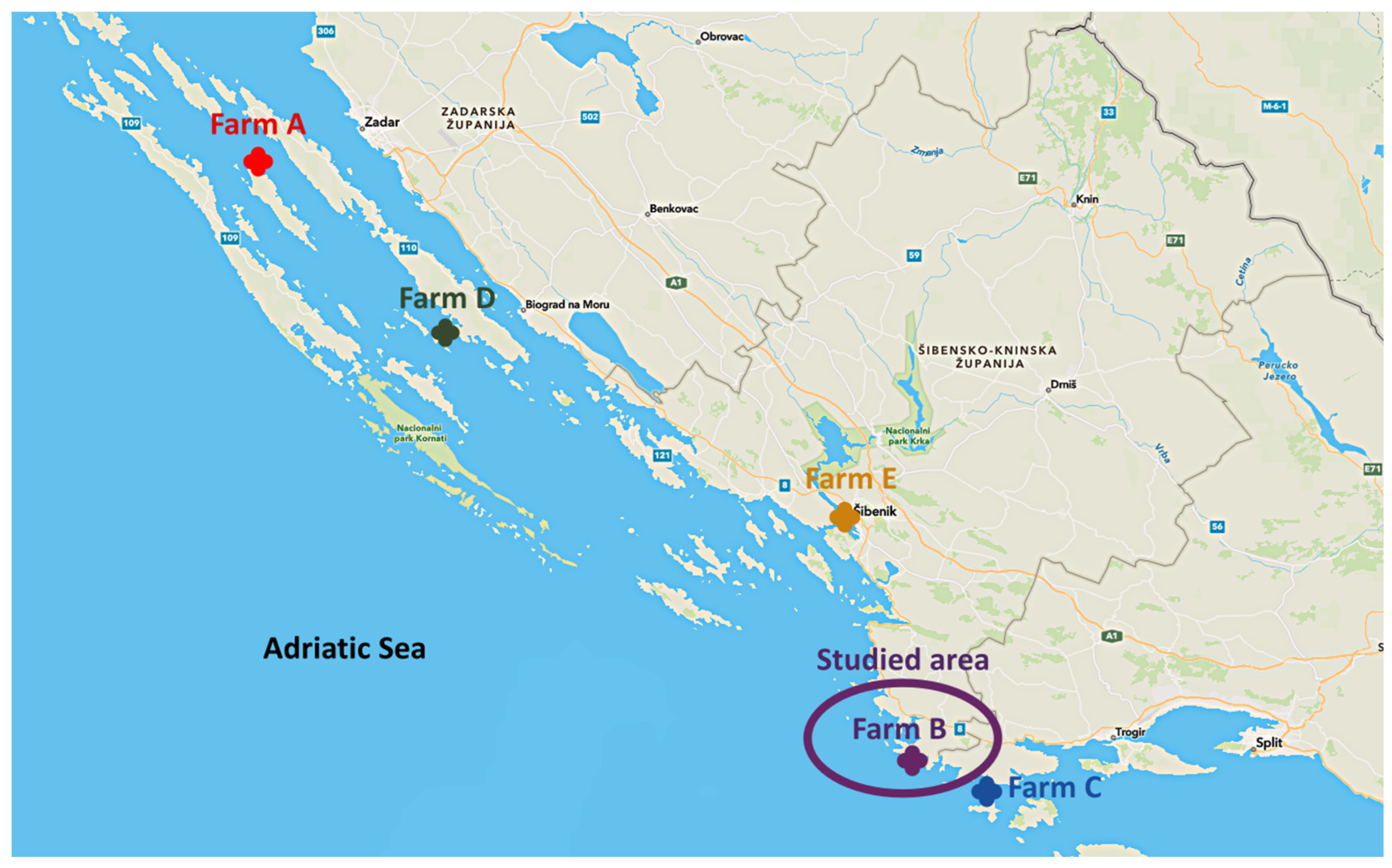
4.1. Description of Disease Outbreaks
4.1.1. Farm A
4.1.2. Farm B
4.1.3. Farm C
4.1.4. Farm D and Farm E
4.1.5. Targeted Surveillance at Farm B
4.2. Necropsy
4.3. Virological Investigations
4.4. Real-Time RT-PCR and Partial Amplification of the RNA1 and RNA2 Genomic Segments
4.5. Whole Genome Sequencing
4.6. Sequencing and Phylogenetic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ministarstvo Poljoprivrede. Ribarstvo. Akvakultura. Uzgoj u Moru. 2022. Available online: https://ribarstvo.mps.hr/default.aspx?id=14 (accessed on 4 January 2022).
- Bondad-Reantaso, M.G.; Lem, A.; Subasinghe, R.P. International Trade in Aquatic Animals and Aquatic Animal Health: What Lessons Have We Learned So Far in Managing the Risks? Fish Pathol. 2009, 44, 107–114. [Google Scholar] [CrossRef]
- Doan, Q.K.; Vandeputte, M.; Chatain, B.; Morin, T.; Allal, F. Viral encephalopathy and retinopathy in aquaculture: A review. J. Fish Dis. 2017, 40, 717–742. [Google Scholar] [CrossRef] [Green Version]
- Vendramin, N.; Zrncic, S.; Padrós, F.; Oraic, D.; Le Breton, A.; Zarza, C.; Olesen, N.J. Fish health in Mediterranean Aquaculture, past mistakes and future challenges. Bull. Eur. Assoc. Fish Pathol. 2016, 36, 38–45. [Google Scholar]
- Muniesa, A.; Basurco, B.; Aguilera, C.; Furones, D.; Reverte, C.; Sanjuan Vilaplana, A.; Dverdal Jansen, M.; Brun, E.; Tavornapahich, S. Mapping the knowledge of the main diseases affecting sea bass and sea bream in Mediterranean. Transbound. Emerg. Dis. 2020, 67, 1089–1100. [Google Scholar] [CrossRef] [PubMed]
- Bandín, I.; Souto, S. Betanodavirus and VER Disease: A 30-year Research Review. Pathogens 2020, 9, 106. [Google Scholar] [CrossRef] [Green Version]
- Mori, K.; Nakai, T.; Muroga, K.; Arimoto, M.; Mushiake, K.; Furusawa, I. Properties of a new virus belonging to nodaviridae found in larval striped jack (Pseudocaranx dentex) with nervous necrosis. Virology 1992, 187, 368–371. [Google Scholar] [CrossRef]
- Nishizawa, T.; Furuhashi, M.; Nagai, T.; Nakai, T.; Muroga, K. Genomic classification of fish nodaviruses by molecular phylogenetic analysis of the coat protein gene. Appl. Environ. Microbiol. 1997, 63, 1633–1636. [Google Scholar] [CrossRef] [Green Version]
- Panzarin, V.; Fusaro, A.; Monne, I.; Cappellozza, E.; Patarnello, P.; Bovo, G.; Capua, I.; Holmes, E.C.; Cattoli, G. Molecular epidemiology and evolutionary dynamics of betanodavirus in southern Europe. Infect. Genet. Evol. 2012, 12, 63–70. [Google Scholar] [CrossRef]
- Olveira, J.G.; Souto, S.; Dopazo, C.P.; Thiéry, R.; Barja, J.L.; Bandín, I. Comparative analysis of both genomic segments of betanodaviruses isolated from epizootic outbreaks in farmed fish species provides evidence for genetic reassortment. J. Gen. Virol. 2009, 90, 2940–2951. [Google Scholar] [CrossRef]
- Toffolo, V.; Negrisolo, E.; Maltese, C.; Bovo, G.; Belvedere, P.; Colombo, L.; Valle, L.D. Phylogeny of betanodaviruses and molecular evolution of their RNA polymerase and coat proteins. Mol. Phylogenet. Evol. 2007, 43, 298–308. [Google Scholar] [CrossRef]
- Toffan, A.; Pascoli, F.; Pretto, T.; Panzarin, V.; Abbadi, M.; Buratin, A.; Quartesan, R.; Gijon, D.; Padros, F. Viral nervous necrosis in gilthead sea bream (Sparus aurata) caused by reassortant betanodavirus RGNNV/SJNNV: An emerging threat for Mediterranean aquaculture. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Volpe, E.; Gustinelli, A.; Caffara, M.; Errani, F.; Quaglio, F.; Fioravanti, M.L.; Ciulli, S. Viral nervous necrosis outbreaks caused by the RGNNV/SJNNV reassortant betanodavirus in gilthead sea bream (Sparus aurata) and European sea bass (Dicentrarchus labrax). Aquaculture 2020, 523, 735155. [Google Scholar] [CrossRef]
- Bitchava, K.; Chassalevris, T.; Lampou, E.; Athanassopoulou, F.; Economou, V.; Dovas, C.I. Occurrence and molecular characterization of betanodaviruses in fish and invertebrates of the Greek territorial waters. J. Fish Dis. 2019, 42, 1773–1783. [Google Scholar] [CrossRef] [PubMed]
- Toffan, A.; Biasini, L.; Pretto, T.; Abbadi, M.; Buratin, A.; Franch, R.; Dalla Rovere, G.; Panzarin, V.M.; Marsella, A.; Bargelloni, L.; et al. Age dependency of RGNNV/SJNNV viral encephalo-retinopathy in Gilthead Sea Bream (Sparus aurata). Aquaculture 2021, 539, 736605. [Google Scholar] [CrossRef]
- Cherif, N.; Thiery, R.; Castric, J.; Biacchesi, S.; Bremont, M.; Thabti, F.; Limem, L.; Hammami, S. Viral encephalopathy and retinopathy of Dicentrarchus labrax and Sparus aurata farmed in Tunisia. Vet. Res. Commun. 2009, 33, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Toffan, A.; Panzarin, V.; Toson, M.; Cecchettin, K.; Pascoli, F. Water temperature affects pathogenicity of different betanodavirus genotypes in experimentally challenged Dicentrarchus labrax. Dis. Aquat. Organ. 2016, 119, 231–238. [Google Scholar] [CrossRef] [Green Version]
- Panzarin, V.M. A Multidisciplinary Approach to the Study of Betanodaviruses in the Mediterranean Basin. Ph.D. Thesis, Università degli Studi di Padova, Dipartimento di Biomedicina Comparata e Alimentazione, Padova, Italy, 2016. Available online: http://paduaresearch.cab.unipd.it/9276/1/panzarin_valentina_tesi.pdf (accessed on 10 February 2022).
- Bovo, G.; Nishizawa, T.; Maltese, C.; Borghesan, F.; Mutinelli, F.; Montesi, F.; De Mas, S. Viral encephalopathy and retinopathy of farmed marine fish species in Italy. Virus Res. 1999, 63, 143–146. [Google Scholar] [CrossRef]
- Breuil, G.; Mouchel, O.; Fauvel, C.; Pepin, J.F. Sea bass Dicentrarchus labrax nervous necrosis virus isolates with distinct pathogenicity to sea bass larvae. Di Aqua Org. 2001, 45, 25–31. [Google Scholar] [CrossRef]
- Athanassopoulou, F.; Billinis, C.; Psychas, V.; Karipoglou, K. Viral encephalopathy and retinopathy of Dicentrarchus labrax (L.) farmed in fresh water in Greece. J. Fish Dis. 2003, 26, 361–365. [Google Scholar] [CrossRef]
- Le Breton, A.; Grisez, L.; Sweetman, J.; Ollevier, F. Viral nervous necrosis (VNN) associated with mass mortalities in cage-reared sea bass, Dicentrarchus labrax (L.). J. Fish Dis. 1997, 20, 145–151. [Google Scholar] [CrossRef]
- Munday, B.L.; Kwang, J.; Moody, N. Betanodavirus infections of teleost fish: A review. J. Fish Dis. 2002, 25, 127–142. [Google Scholar] [CrossRef]
- Castric, J.; Thiery, R.; Jeffroy, J.; De Kinkelin, P.; Raymond, J.C. Sea bream Sparus aurata, an asymptomatic contagious fish host for nodavirus. Dis. Aquat. Org. 2001, 47, 33–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korsnes, K.; Karlsbakk, E.; Nylund, A.; Nerland, A.H. Horizontal transmission of nervous necrosis virus between turbot Scophthalmus maximus and Atlantic cod Gadus morhua using cohabitation challenge. Dis. Aquat. Org. 2012, 99, 13–21. [Google Scholar] [CrossRef] [Green Version]
- Frerichs, G.N.; Rodger, H.D.; Peric, Z. Cell culture isolation of piscine neuropathy nodavirus from juvenile seabass, Dicentrarchus labrax. J. Gen. Virol. 1996, 77, 2067–2071. [Google Scholar] [CrossRef] [PubMed]
- Yong, C.Y.; Yeap, S.K.; Omar, A.R.; Tan, W.S. Advances in the study of nodavirus. PeerJ 2017, 5, e3841. [Google Scholar] [CrossRef]
- Faggion, S.; Bertotto, D.; Babbucci, M.; Dalla Rovere, G.; Franch, R.; Bovolenta, M.; Laureau, S.; Pascoli, F.; Toffan, A.; Bargelloni, L.; et al. Resistance to viral nervous necrosis in European sea bass (Dicentrarchus labrax L.): Heritability and relationships with body weight, cortisol concentration, and antibody titer. Genet. Sel. Evol. 2021, 53, 32. [Google Scholar] [CrossRef] [PubMed]
- Fernández Sánchez, J.L.; Le Breton, A.; Brun, E.; Vendramin, N.; Spiliopoulos, G.; Furones, D.; Basurco, B. Assessing the economic impact of diseases in Mediterranean grow-out farms culturing European seabass. Aquaculture 2022, 547, 737530. [Google Scholar] [CrossRef]
- Mushiake, K.; Nishizawa, T.; Nakai, T.; Furusawa, I.; Muroga, K. Control of VNN in striped jack: Selection of spawners based on the detection of SJNNV gene by polymerase chain reaction (PCR). Fish Pathol. 1994, 29, 177–182. [Google Scholar] [CrossRef]
- Watanabe, K.; Nishizawa, T.; Yoshimizu, M. Selection of broodstock candidates of barfin flounder using an ELISA system with recombinant protein of barfin flounder nervous necrosis virus. Dis. Aquat. Org. 2000, 41, 219–223. [Google Scholar] [CrossRef]
- Breuil, G.; Pepin, J.F.; Castric, J.; Fauvel, C.; Thiéry, R. Detection of serum antibodies against nodavirus in wild and farmed adult sea bass: Application to the screening of broodstock in sea bass hatcheries. Bull. Eur. Assoc. Fish Pathol. 2000, 20, 95–100. [Google Scholar]
- Valero, Y.; Arizcun, M.; Esteban, M.A.; Bandin, I.; Olveira, J.G.; Patel, S.; Cuesta, A.; Chaves-Pozo, E. Nodavirus colonizes and replicates in the testis of Gilthead seabream and European sea bass modulating its immune and reproductive functions. PLoS ONE 2015, 10, e0145131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez, D.K.; Mori, K.; Okinaka, Y.; Nakai, T.; Park, S.C. Trash fish can be a source of betanodavirus for cultured marine fish. Aquaculture 2010, 302, 158–163. [Google Scholar] [CrossRef]
- Nakai, T.; Mori, K.; Sugaya, T.; Nishioka, T.; Mushiake, K.; Yamashita, H. Current Knowledge on Viral Nervous Necrosis (VNN) and its Causative Betanodaviruses. Isr. J. Aquac. 2009, 61, 198–207. [Google Scholar]
- Gardner, I.; Saksida, S.; Dixon, B.; McKenzie, P.; Johnson, S. Pathogen exchange between wild and farmed finfish: Evidence to assess pathogen source and factors associated with clinical disease occurrence. Bull. Aquacult. Assoc. Can. 2014, 111, 4–35. [Google Scholar]
- Volpe, E.; Grodzki, M.; Panzarin, V.; Guercio, A.; Purpari, G.; Serratore, P.; Ciulli, S. Detection and molecular characterization of betanodaviruses retrieved from bivalve molluscs. J. Fish Dis. 2018, 41, 603–611. [Google Scholar] [CrossRef]
- Errani, F.; Ponti, M.; Volpe, E.; Ciulli, S. Spatial and seasonal variability of human and fish viruses in mussels inside and offshore of Ravenna’s harbor (Adriatic Sea, Italy). J. Appl. Microbiol. 2021, 130, 994–1008. [Google Scholar] [CrossRef]
- Gaicopello, C.; Foti, M.; Bottari, T.; Fisichella, V.; Barbera, G. Detection of viral encephalopathy and retinopathy virus (VERV) in wild marine fish species of the South Thyrrenian Sea (Central Mediterranean). J. Fish Dis. 2013, 36, 819–821. [Google Scholar] [CrossRef]
- Ciulli, S.; Galletti, E.; Grodzki, M.; Alessi, A.; Battilani, M.; Prosperi, S. Isolation and genetic characterization of Betanodavirus from wild marine fish from the Adriatic Sea. Vet. Res. Commun. 2007, 31, 221–224. [Google Scholar] [CrossRef]
- Vendramin, N.; Patarnello, P.; Toffan, A.; Panzarin, V.; Cappellozza, E.; Tedesco, P.; Terlizzi, P.; Terregino, C.; Cattoli, G. Viral Encephalopathy and Retinopathy in groupers (Epinephelus spp.) in southern Italy: A threat for wild endangered species? BMC Vet. Res. 2013, 9, 20. [Google Scholar] [CrossRef] [Green Version]
- Panzarin, V.; Patarnello, P.; Mori, A.; Rampazzo, E.; Cappellozza, E.; Bovo, G.; Cattoli, G. Development and validation of a real-time TaqMan PCR assay for the detection of betanodavirus in clinical specimens. Arch. Virol. 2010, 155, 1193–1203. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating max-imum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Farm | Date of Fry Import | Sea Temperature °C | Sampling Date | ANALYSIS CODE | Fish Weight (g) | Sea Temperature °C | rRTPCR (Cq) | SSN1 Cell Culture |
|---|---|---|---|---|---|---|---|---|
| A | 07/06/2014 | 20 | 13/06/2014 | 67/14 | 8.0 | 21 | 11.63 | CPE* |
| B | 13/05/2014 | 18.5 | 23/06/2014 | 71/14 | 12.5 | 21 | 31.6 | No CPE |
| B | 13/05/2014 | 18 | 03/08/2014 | 105/14 | 19.5 | 23 | 12.57 | CPE |
| C | 18/05/2014 | 16 | 04/09/2014 | 111/14 | 17.5 | 24 | 19.2 | CPE |
| D | 06/06/2014 | 20 | 10/10/2014 | 130/14 | 50.8 | 22 | 17.57 | CPE |
| E | 06/06/2014 | 20 | 24/10/2014 | 141/14 | 84.3 | 21 | 19.8 | CPE |
| Sampling Date | Fish Species—Cage Label | Origin | Date of Import | Analysis Code | Fish Weight (g) | Fish Age | Sea Temperature °C | rRT-PCR (Cq) |
|---|---|---|---|---|---|---|---|---|
| 23/06/2014 | ESB–C | Hatchery XL | 05/2014 | 71/14 | 12.5 | Fry | 21 | 31.6 |
| 03/08/2014 | ESB–M1 | Hatchery XL | 05/2014 | 105/14 | 19.5 | Fry | 23 | 12.57 |
| 03/08/2014 | ESB–A | Hatchery FZ | 09/2013 | 105/14 | 29.39 | Year | 23 | 26.71 |
| 18/08/2014 | ESB–17 | Hatchery FZ | 03/2014 | 109/14 | 44.6 | Fry | 25 | 15.28 |
| 18/08/2014 | ESB–OK6 | Hatchery FZ | 09/2013 | 109/14 | 108.91 | Year | 25 | 34.09 |
| 18/08/2014 | ESB–OK5 | Hatchery XL | 05/2013 | 109/14 | 193.73 | Year | 25 | 33.45 |
| 18/08/2014 | GSB–OK3 | Hatchery FZ | 03/2014 | 109/14 | 47.42 | Fry | 25 | Neg. |
| 18/08/2014 | GSB–14 | Hatchery FZ | 04/2014 | 109/14 | 38.36 | Fry | 25 | Neg. |
| 18/08/2014 | GSB–15 | Hatchery FZ | 04/2014 | 109/14 | 39.57 | Fry | 25 | Neg. |
| 05/09/2014 | ESB * | Hatchery FZ | 09/2014 | 115/14 | 7.8 | Fry | 24 | Neg. |
| 05/09/2014 | Bogue (Boops boops) | Caught close to the farm | n/a | 115/14 | 139 | n/a | 24 | Neg. |
| 05/09/2014 | Thicklip grey mullet (Chelon labrosus) | Caught close to the farm | n/a | 115/14 | 447 | n/a | 24 | Neg. |
| 05/09/2014 | Salema (Sarpa salpa) | Caught close to the farm | n/a | 115/14 | 210 | n/a | 24 | Neg. |
| 05/09/2014 | Mussel (Mytilus galloprovincialis) | Collected from the cages | n/a | 114/14 | n/a | n/a | 24 | Neg. |
| 22/09/2014 | ESB–C | Hatchery FZ | 09/2014 | 125/14 | 8.3 | Fry | 23 | 37.22 |
| 03/12/2014 | ESB–M1 | Hatchery XL | 05/2014 | 162/14 | 73.29 | Fry | 17.2 | 26.56 |
| 03/12/2014 | ESB–B | Hatchery XL | 05/2014 | 162/14 | 72.43 | Fry | 17.2 | 25.73 |
| 25/02/2015 | ESB–M1 | Hatchery XL | 05/2014 | 12/15 | 85 | Year | 12.5 | 34.56 |
| 25/02/2015 | ESB–M2 | Hatchery XL | 05/2014 | 12/15 | 74.31 | Year | 12.5 | 33.27 |
| 02/04/2015 | ESB–M1 | Hatchery XL | 05/2014 | 23/15 | 61.7 | Year | 14.5 | 30.1 |
| 30/04/2015 | ESB * | Hatchery FZ | 04/2015 | 36/15 | 3.28 | Fry | 16 | Neg. |
| 27/05/2015 | ESB–C1 | Hatchery FZ | 04/2015 | 56/15 | 5.86 | Fry | 19 | Neg. |
| 27/05/2015 | ESB–C2 | Hatchery FZ | 04/2015 | 56/15 | 4.9 | Fry | 19 | Neg. |
| 17/06/2015 | ESB–C1 | Hatchery FZ | 04/2015 | 67/15 | 8.53 | Fry | 23 | 35.85 |
| 17/06/2015 | ESB–C2 | Hatchery FZ | 04/2015 | 67/15 | 7.96 | Fry | 23 | 34.92 |
| 02/07/2015 | ESB–C1+C2 | Hatchery FZ | 04/2015 | 72/15 | 9.04 | Fry | 23 | Neg. |
| 02/07/2015 | ESB–OK1 | Hatchery FZ | 09/2014 | 72/15 | 81.23 | Year | 23 | 28.94 |
| 02/07/2015 | ESB–OK4 | Hatchery XL | 05/2014 | 72/15 | 159.5 | Year | 23 | 26.67 |
| 12/08/2015 | ESB–OK1 | Hatchery FZ | 09/2014 | 101/15 | 151 | Year | 26 | Neg. |
| 12/08/2015 | ESB–OK6 | Hatchery FZ | 04/2014 | 101/15 | 240.5 | Year | 26 | 30.82 |
| 12/08/2015 | ESB–OK2 | Hatchery XL | 05/2014 | 101/15 | 192.5 | Year | 26 | 33.44 |
| 12/08/2015 | ESB–OK4 | Hatchery XL | 05/2014 | 101/15 | 189 | Year | 26 | 31.25 |
| 12/08/2015 | Bogue (Boops boops) | Caught close to the farm | n/a | 101/15 | 248 | n/a | 26 | Neg. |
| 12/08/2015 | Salema (Sarpa salpa) | Caught close to the farm | n/a | 101/15 | 485 | n/a | 24 | Neg. |
| 12/08/2015 | Thicklip grey mullet (Chelon labrosus) | Caught close to the farm | n/a | 101/15 | 280 | n/a | 24 | 31.30 |
| 12/08/2015 | Annular seabream (Diplodus annularis) | Caught close to the farm | n/a | 101/15 | 97 | n/a | 24 | Neg. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zrnčić, S.; Brnić, D.; Panzarin, V.; Abbadi, M.; Lojkić, I.; Zupičić, I.G.; Oraić, D. Transmission Pathways of the VNN Introduced in Croatian Marine Aquaculture. Pathogens 2022, 11, 418. https://doi.org/10.3390/pathogens11040418
Zrnčić S, Brnić D, Panzarin V, Abbadi M, Lojkić I, Zupičić IG, Oraić D. Transmission Pathways of the VNN Introduced in Croatian Marine Aquaculture. Pathogens. 2022; 11(4):418. https://doi.org/10.3390/pathogens11040418
Chicago/Turabian StyleZrnčić, Snježana, Dragan Brnić, Valentina Panzarin, Miriam Abbadi, Ivana Lojkić, Ivana Giovanna Zupičić, and Dražen Oraić. 2022. "Transmission Pathways of the VNN Introduced in Croatian Marine Aquaculture" Pathogens 11, no. 4: 418. https://doi.org/10.3390/pathogens11040418
APA StyleZrnčić, S., Brnić, D., Panzarin, V., Abbadi, M., Lojkić, I., Zupičić, I. G., & Oraić, D. (2022). Transmission Pathways of the VNN Introduced in Croatian Marine Aquaculture. Pathogens, 11(4), 418. https://doi.org/10.3390/pathogens11040418










