Adaptive Cellular Immunity against African Swine Fever Virus Infections
Abstract
:1. African Swine Fever Virus
2. Antigen-Presenting Cells: Bridging Innate and Adaptive Immunity
2.1. Macrophages
2.2. Dendritic Cells
2.3. Nonconventional APCs
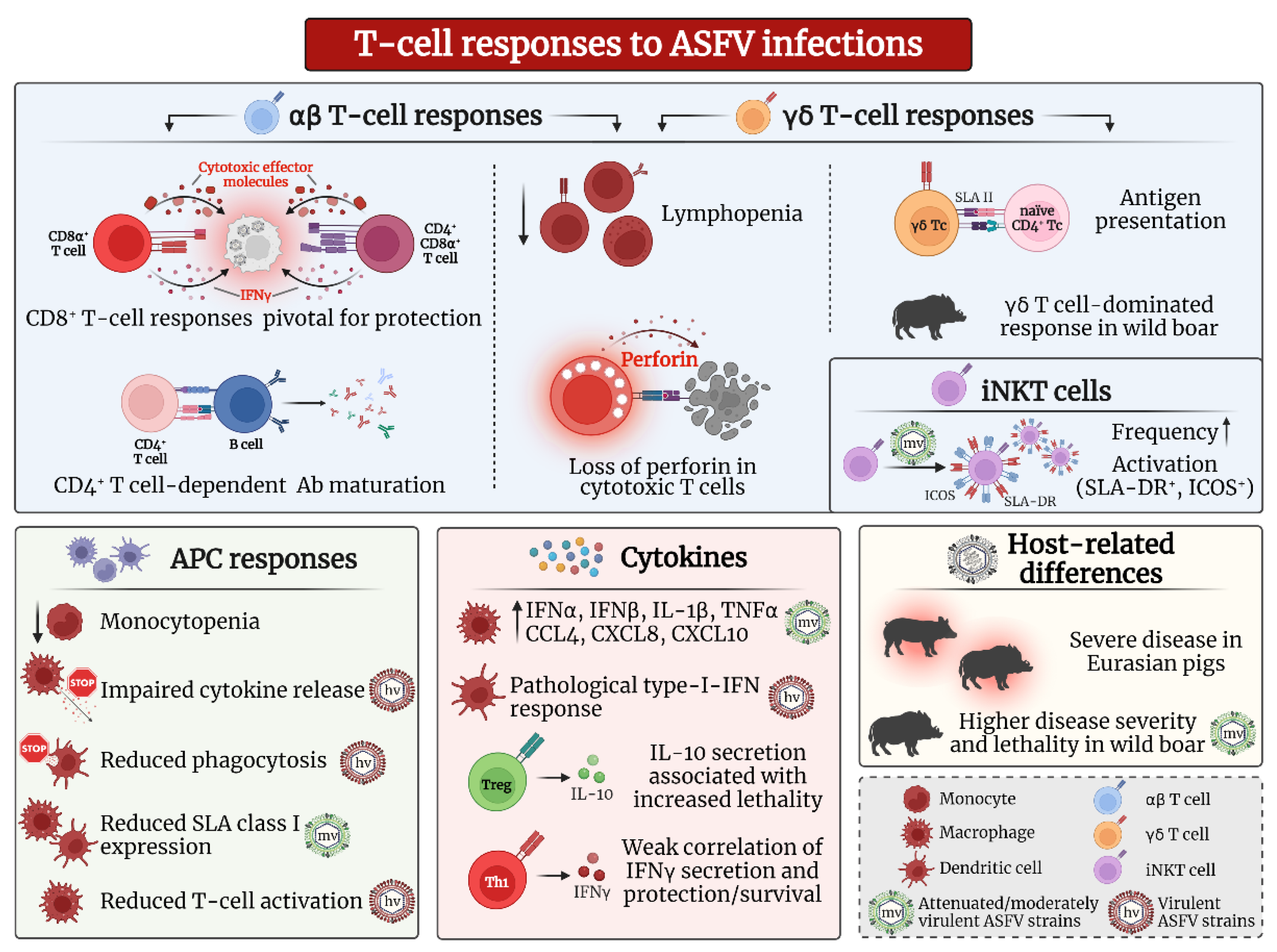
3. T Cell Responses against ASFV Infection
3.1. αβ T Cells
3.1.1. CD8α+ αβ T Cells and Cytotoxic Responses
3.1.2. CD4+ and CD4+/CD8α+ αβ T Cells
3.1.3. Regulatory T Cell Responses
3.2. γδ T Cells
3.3. Nonconventional T Cells
4. Recent Developments and T Cell Epitopes in ASFV Vaccines
4.1. Protection and Induction of IFNγ by Recent ASFV Vaccine Candidates
4.2. CTL Targets
4.3. Recall Targets
5. Concluding Remarks and Open Questions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ward, M.P.; Tian, K.; Nowotny, N. African Swine Fever, the forgotten pandemic. Transbound. Emerg. Dis. 2021, 68, 2637–2639. [Google Scholar] [CrossRef] [PubMed]
- Penrith, M.L.; Vosloo, W.; Jori, F.; Bastos, A.D. African swine fever virus eradication in Africa. Virus Res. 2013, 173, 228–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dixon, L.K.; Stahl, K.; Jori, F.; Vial, L.; Pfeiffer, D.U. African Swine Fever Epidemiology and Control. Annu. Rev. Anim. Biosci. 2020, 8, 221–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alonso, C.; Borca, M.; Dixon, L.; Revilla, Y.; Rodriguez, F.; Escribano, J.M.; Ictv Report, C. ICTV Virus Taxonomy Profile: Asfarviridae. J. Gen. Virol. 2018, 99, 613–614. [Google Scholar] [CrossRef]
- Alejo, A.; Matamoros, T.; Guerra, M.; Andres, G. A Proteomic Atlas of the African Swine Fever Virus Particle. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [Green Version]
- Karger, A.; Perez-Nunez, D.; Urquiza, J.; Hinojar, P.; Alonso, C.; Freitas, F.B.; Revilla, Y.; Le Potier, M.F.; Montoya, M. An Update on African Swine Fever Virology. Viruses 2019, 11, 864. [Google Scholar] [CrossRef] [Green Version]
- Salas, M.L.; Andrés, G. African swine fever virus morphogenesis. Virus Res. 2012, 173, 29–41. [Google Scholar] [CrossRef]
- Andres, G.; Charro, D.; Matamoros, T.; Dillard, R.S.; Abrescia, N.G.A. The cryo-EM structure of African swine fever virus unravels a unique architecture comprising two icosahedral protein capsids and two lipoprotein membranes. J. Biol. Chem. 2020, 295, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Carrasco, L.; de Lara, F.C.; Gomez-Villamandos, J.C.; Bautista, M.J.; Villeda, C.J.; Wilkinson, P.J.; Sierra, M.A. The pathogenic role of pulmonary intravascular macrophages in acute African swine fever. Res. Vet. Sci. 1996, 61, 193–198. [Google Scholar] [CrossRef]
- Carrasco, L.; de Lara, F.C.; Martin de las Mulas, J.; Gomez-Villamandos, J.C.; Hervas, J.; Wilkinson, P.J.; Sierra, M.A. Virus association with lymphocytes in acute African swine fever. Vet. Res. 1996, 27, 305–312. [Google Scholar]
- Martins, C.L.; Scholl, T.; Mebus, C.A.; Fisch, H.; Lawman, M.J. Modulation of porcine peripheral blood-derived macrophage functions by in vitro infection with African swine fever virus (ASFV) isolates of different virulence. Viral Immunol. 1987, 1, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Mebus, C.A. African swine fever. Adv. Virus Res. 1988, 35, 251–269. [Google Scholar] [PubMed]
- Ramiro-Ibanez, F.; Escribano, J.M.; Alonso, C. Application of a monoclonal antibody recognizing protein p30 to detect African swine fever virus-infected cells in peripheral blood. J. Virol. Methods 1995, 55, 339–345. [Google Scholar] [CrossRef]
- Perez, J.; Rodriguez, F.; Fernandez, A.; Martin de las Mulas, J.; Gomez-Villamandos, J.C.; Sierra, M.A. Detection of African swine fever virus protein VP73 in tissues of experimentally and naturally infected pigs. J. Vet. Diagn. Investig. 1994, 6, 360–365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blome, S.; Gabriel, C.; Beer, M. Pathogenesis of African swine fever in domestic pigs and European wild boar. Virus Res. 2013, 173, 122–130. [Google Scholar] [CrossRef]
- Gomez-Villamandos, J.C.; Bautista, M.J.; Sanchez-Cordon, P.J.; Carrasco, L. Pathology of African swine fever: The role of monocyte-macrophage. Virus Res. 2013, 173, 140–149. [Google Scholar] [CrossRef]
- Lacasta, A.; Monteagudo, P.L.; Jimenez-Marin, A.; Accensi, F.; Ballester, M.; Argilaguet, J.; Galindo-Cardiel, I.; Segales, J.; Salas, M.L.; Dominguez, J.; et al. Live attenuated African swine fever viruses as ideal tools to dissect the mechanisms involved in viral pathogenesis and immune protection. Vet. Res. 2015, 46, 135. [Google Scholar] [CrossRef] [Green Version]
- Carlson, J.; O’Donnell, V.; Alfano, M.; Velazquez Salinas, L.; Holinka, L.G.; Krug, P.W.; Gladue, D.P.; Higgs, S.; Borca, M.V. Association of the Host Immune Response with Protection Using a Live Attenuated African Swine Fever Virus Model. Viruses 2016, 8, 291. [Google Scholar] [CrossRef]
- Escribano, J.M.; Galindo, I.; Alonso, C. Antibody-mediated neutralization of African swine fever virus: Myths and facts. Virus Res. 2013, 173, 101–109. [Google Scholar] [CrossRef]
- Neilan, J.G.; Zsak, L.; Lu, Z.; Burrage, T.G.; Kutish, G.F.; Rock, D.L. Neutralizing antibodies to African swine fever virus proteins p30, p54, and p72 are not sufficient for antibody-mediated protection. Virology 2004, 319, 337–342. [Google Scholar] [CrossRef] [Green Version]
- Blome, S.; Gabriel, C.; Beer, M. Modern adjuvants do not enhance the efficacy of an inactivated African swine fever virus vaccine preparation. Vaccine 2014, 32, 3879–3882. [Google Scholar] [CrossRef] [PubMed]
- Cadenas-Fernández, E.; Sánchez-Vizcaíno, J.M.; van den Born, E.; Kosowska, A.; van Kilsdonk, E.; Fernández-Pacheco, P.; Gallardo, C.; Arias, M.; Barasona, J.A. High Doses of Inactivated African Swine Fever Virus Are Safe, but Do Not Confer Protection against a Virulent Challenge. Vaccines 2021, 9, 242. [Google Scholar] [CrossRef] [PubMed]
- Oura, C.A.; Denyer, M.S.; Takamatsu, H.; Parkhouse, R.M. In vivo depletion of CD8+ T lymphocytes abrogates protective immunity to African swine fever virus. J. Gen. Virol. 2005, 86 Pt 9, 2445–2450. [Google Scholar] [CrossRef] [PubMed]
- Banchereau, J.; Steinman, R.M. Dendritic cells and the control of immunity. Nature 1998, 392, 245–252. [Google Scholar] [CrossRef]
- Hume, D.A. The Many Alternative Faces of Macrophage Activation. Front. Immunol. 2015, 6, 370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janeway, C.A., Jr.; Medzhitov, R. Innate immune recognition. Annu. Rev. Immunol. 2002, 20, 197–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- den Haan, J.M.; Arens, R.; van Zelm, M.C. The activation of the adaptive immune system: Cross-talk between antigen-presenting cells, T cells and B cells. Immunol. Lett. 2014, 162, 103–112. [Google Scholar] [CrossRef]
- Bonneville, M.; O’Brien, R.L.; Born, W.K. γδ T cell effector functions: A blend of innate programming and acquired plasticity. Nat. Rev. Immunol. 2010, 10, 467–478. [Google Scholar] [CrossRef]
- Gerner, W.; Käser, T.; Saalmüller, A. Porcine T lymphocytes and NK cells--an update. Dev. Comp. Immunol. 2009, 33, 310–320. [Google Scholar] [CrossRef]
- Brandes, M.; Willimann, K.; Moser, B. Professional antigen-presentation function by human γδ T Cells. Science 2005, 309, 264–268. [Google Scholar] [CrossRef]
- Chambers, B.J. NK cell–T cell interactions. In Natural Killer Cells; Lotze, M.T., Thomson, A.W., Eds.; Academic Press: Cambridge, MA, USA, 2010; pp. 297–308. [Google Scholar]
- Mosser, D.M.; Edwards, J.P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008, 8, 958–969. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Cordón, P.J.; Romero-Trevejo, J.L.; Pedrera, M.; Sánchez-Vizcaíno, J.M.; Bautista, M.J.; Gomez-Villamandos, J.C. Role of hepatic macrophages during the viral haemorrhagic fever induced by African Swine Fever Virus. Histol. Histopathol. 2008, 23, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Castello, A.; Quintas, A.; Sanchez, E.G.; Sabina, P.; Nogal, M.; Carrasco, L.; Revilla, Y. Regulation of host translational machinery by African swine fever virus. PLoS Pathog. 2009, 5, e1000562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neefjes, J.; Jongsma, M.L.; Paul, P.; Bakke, O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 2011, 11, 823–836. [Google Scholar] [CrossRef]
- Franzoni, G.; Graham, S.P.; Giudici, S.D.; Bonelli, P.; Pilo, G.; Anfossi, A.G.; Pittau, M.; Nicolussi, P.S.; Laddomada, A.; Oggiano, A. Characterization of the interaction of African swine fever virus with monocytes and derived macrophage subsets. Vet. Microbiol. 2017, 198, 88–98. [Google Scholar] [CrossRef] [Green Version]
- Franzoni, G.; Razzuoli, E.; Dei Giudici, S.; Carta, T.; Galleri, G.; Zinellu, S.; Ledda, M.; Angioi, P.; Modesto, P.; Graham, S.P.; et al. Comparison of Macrophage Responses to African Swine Fever Viruses Reveals that the NH/P68 Strain is Associated with Enhanced Sensitivity to Type I IFN and Cytokine Responses from Classically Activated Macrophages. Pathogens 2020, 9, 209. [Google Scholar] [CrossRef] [Green Version]
- Lithgow, P.; Takamatsu, H.; Werling, D.; Dixon, L.; Chapman, D. Correlation of cell surface marker expression with African swine fever virus infection. Vet. Microbiol. 2014, 168, 413–419. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Torres, C.; Gómez-Puertas, P.; Gómez-del-Moral, M.; Alonso, F.; Escribano, J.M.; Ezquerra, A.; Domínguez, J. Expression of porcine CD163 on monocytes/macrophages correlates with permissiveness to African swine fever infection. Arch. Virol. 2003, 148, 2307–2323. [Google Scholar] [CrossRef]
- Netherton, C.L.; McCrossan, M.C.; Denyer, M.; Ponnambalam, S.; Armstrong, J.; Takamatsu, H.H.; Wileman, T.E. African swine fever virus causes microtubule-dependent dispersal of the trans-golgi network and slows delivery of membrane protein to the plasma membrane. J. Virol. 2006, 80, 11385–11392. [Google Scholar] [CrossRef] [Green Version]
- Lanier, L.L. NK cell recognition. Annu. Rev. Immunol. 2005, 23, 225–274. [Google Scholar] [CrossRef]
- Leitão, A.; Cartaxeiro, C.; Coelho, R.; Cruz, B.; Parkhouse, R.M.E.; Portugal, F.C.; Vigario, J.D.; Martins, C.L.V. The non-haemadsorbing African swine fever virus isolate ASFV/NH/P68 provides a model for defining the protective anti-virus immune response. J. Gen. Virol. 2001, 82, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Fishbourne, E.; Abrams, C.C.; Takamatsu, H.H.; Dixon, L.K. Modulation of chemokine and chemokine receptor expression following infection of porcine macrophages with African swine fever virus. Vet. Microbiol. 2013, 162, 937–943. [Google Scholar] [CrossRef] [Green Version]
- García-Belmonte, R.; Pérez-Núñez, D.; Pittau, M.; Richt, J.A.; Revilla, Y. African Swine Fever Virus Armenia/07 Virulent Strain Controls Interferon Beta Production through the cGAS-STING Pathway. J. Virol. 2019, 93, e02298-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gil, S.; Sepúlveda, N.; Albina, E.; Leitao, A.; Martins, C. The low-virulent African swine fever virus (ASFV/NH/P68) induces enhanced expression and production of relevant regulatory cytokines (IFNα, TNFα and IL12p40) on porcine macrophages in comparison to the highly virulent ASFV/L60. Arch. Virol. 2008, 153, 1845–1854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Razzuoli, E.; Franzoni, G.; Carta, T.; Zinellu, S.; Amadori, M.; Modesto, P.; Oggiano, A. Modulation of Type I Interferon System by African Swine Fever Virus. Pathogens 2020, 9, 361. [Google Scholar] [CrossRef]
- Reis, A.L.; Abrams, C.C.; Goatley, L.C.; Netherton, C.; Chapman, D.G.; Sánchez-Cordón, P.J.; Dixon, L.K. Deletion of African swine fever virus interferon inhibitors from the genome of a virulent isolate reduces virulence in domestic pigs and induces a protective response. Vaccine 2016, 34, 4698–4705. [Google Scholar] [CrossRef] [Green Version]
- Afonso, C.L.; Piccone, M.E.; Zaffuto, K.M.; Neilan, J.; Kutish, G.F.; Lu, Z.; Balinsky, C.A.; Gibb, T.R.; Bean, T.J.; Zsak, L.; et al. African swine fever virus multigene family 360 and 530 genes affect host interferon response. J. Virol. 2004, 78, 1858–1864. [Google Scholar] [CrossRef] [Green Version]
- Golding, J.P.; Goatley, L.; Goodbourn, S.; Dixon, L.K.; Taylor, G.; Netherton, C.L. Sensitivity of African swine fever virus to type I interferon is linked to genes within multigene families 360 and 505. Virology 2016, 493, 154–161. [Google Scholar] [CrossRef] [Green Version]
- Summerfield, A. Viewpoint: Factors involved in type I interferon responses during porcine virus infections. Vet. Immunol. Immunopathol. 2012, 148, 168–171. [Google Scholar] [CrossRef]
- Theofilopoulos, A.N.; Baccala, R.; Beutler, B.; Kono, D.H. Type I interferons (α/β) in immunity and autoimmunity. Annu. Rev. Immunol. 2005, 23, 307–336. [Google Scholar] [CrossRef]
- Franzoni, G.; Edwards, J.C.; Kurkure, N.V.; Edgar, D.S.; Sanchez-Cordon, P.J.; Haines, F.J.; Salguero, F.J.; Everett, H.E.; Bodman-Smith, K.B.; Crooke, H.R.; et al. Partial Activation of natural killer and γδ T cells by classical swine fever viruses is associated with type I interferon elicited from plasmacytoid dendritic cells. Clin. Vaccine Immunol. 2014, 21, 1410–1420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zanotti, C.; Razzuoli, E.; Crooke, H.; Soule, O.; Pezzoni, G.; Ferraris, M.; Ferrari, A.; Amadori, M. Differential Biological Activities of Swine Interferon-α Subtypes. J. Interferon Cytokine Res. 2015, 35, 990–1002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Correia, S.; Ventura, S.; Parkhouse, R.M. Identification and utility of innate immune system evasion mechanisms of ASFV. Virus Res. 2013, 173, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Duque, G.A.; Descoteaux, A. Macrophage cytokines: Involvement in immunity and infectious diseases. Front. Immunol. 2014, 5, 491. [Google Scholar] [CrossRef] [Green Version]
- Ben-Sasson, S.Z.; Hu-Li, J.; Quiel, J.; Cauchetaux, S.; Ratner, M.; Shapira, I.; Dinarello, C.A.; Paul, W.E. IL-1 acts directly on CD4 T cells to enhance their antigen-driven expansion and differentiation. Proc. Natl. Acad. Sci. USA 2009, 106, 7119–7124. [Google Scholar] [CrossRef] [Green Version]
- Borish, L.C.; Steinke, J.W. 2. Cytokines and chemokines. J. Allergy Clin. Immunol. 2003, 111 (Suppl. S2), S460–S475. [Google Scholar] [CrossRef]
- Zhu, J.J.; Ramanathan, P.; Bishop, E.A.; O’Donnell, V.; Gladue, D.P.; Borca, M.V. Mechanisms of African swine fever virus pathogenesis and immune evasion inferred from gene expression changes in infected swine macrophages. PLoS ONE 2019, 14, e0223955. [Google Scholar] [CrossRef]
- Mosser, D.M. The many faces of macrophage activation. J. Leukoc. Biol. 2003, 73, 209–212. [Google Scholar] [CrossRef]
- Fan, W.; Jiao, P.; Zhang, H.; Chen, T.; Zhou, X.; Qi, Y.; Sun, L.; Shang, Y.; Zhu, H.; Hu, R.; et al. Inhibition of African Swine Fever Virus Replication by Porcine Type I and Type II Interferons. Front. Microbiol. 2020, 11, 1203. [Google Scholar] [CrossRef]
- Ai, Q.; Lin, X.; Xie, H.; Li, B.; Liao, M.; Fan, H. Proteome Analysis in PAM Cells Reveals That African Swine Fever Virus Can Regulate the Level of Intracellular Polyamines to Facilitate Its Own Replication through ARG1. Viruses 2021, 13, 1236. [Google Scholar] [CrossRef]
- Sautter, C.A.; Auray, G.; Python, S.; Liniger, M.; Summerfield, A. Phenotypic and functional modulations of porcine macrophages by interferons and interleukin-4. Dev. Comp. Immunol. 2018, 84, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Carta, T.; Razzuoli, E.; Fruscione, F.; Zinellu, S.; Meloni, D.; Anfossi, A.; Chessa, B.; Dei Giudici, S.; Graham, S.P.; Oggiano, A.; et al. Comparative Phenotypic and Functional Analyses of the Effects of IL-10 or TGF-beta on Porcine Macrophages. Animal 2021, 11, 1098. [Google Scholar] [CrossRef] [PubMed]
- Franzoni, G.; Graham, S.P.; Dei Giudici, S.; Oggiano, A. Porcine Dendritic Cells and Viruses: An Update. Viruses 2019, 11, 445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Summerfield, A.; McCullough, K.C. The porcine dendritic cell family. Dev. Comp. Immunol. 2009, 33, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Carrasco, C.P.; Rigden, R.C.; Schaffner, R.; Gerber, H.; Neuhaus, V.; Inumaru, S.; Takamatsu, H.; Bertoni, G.; McCullough, K.C.; Summerfield, A. Porcine dendritic cells generated in vitro: Morphological, phenotypic and functional properties. Immunology 2001, 104, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Franzoni, G.; Graham, S.P.; Sanna, G.; Angioi, P.; Fiori, M.S.; Anfossi, A.; Amadori, M.; Dei Giudici, S.; Oggiano, A. Interaction of porcine monocyte-derived dendritic cells with African swine fever viruses of diverse virulence. Vet. Microbiol. 2018, 216, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Drummond, R.; Johnson, L.; Netherton, C.; Reis, A.; Morgan, S.; Graham, S.; Dixon, L. Understanding the role of porcine dendritic cells in African swine fever infection. In Proceedings of the 6th European Veterinary Immunology Workshop, Utrecht, The Netherlands, 5–7 September 2018. [Google Scholar]
- Gregg, D.A.; Mebus, C.A.; Schlafer, D.H. Early infection of interdigitating dendritic cells in the pig lymph node with African swine fever viruses of high and low virulence: Immunohistochemical and ultrastructural studies. J. Vet. Diagn. Investig. 1995, 7, 23–30. [Google Scholar] [CrossRef] [Green Version]
- Sehl, J.; Pikalo, J.; Schäfer, A.; Franzke, K.; Pannhorst, K.; Elnagar, A.; Blohm, U.; Blome, S.; Breithaupt, A. Comparative Pathology of Domestic Pigs and Wild Boar Infected with the Moderately Virulent African Swine Fever Virus Strain “Estonia 2014”. Pathogens 2020, 9, 662. [Google Scholar] [CrossRef]
- Heath, W.R.; Carbone, F.R. Cross-presentation in viral immunity and self-tolerance. Nat. Rev. Immunol. 2001, 1, 126–134. [Google Scholar] [CrossRef]
- De Pelsmaeker, S.; Devriendt, B.; Leclercq, G.; Favoreel, H.W. Porcine NK cells display features associated with antigen-presenting cells. J. Leukoc. Biol. 2018, 103, 129–140. [Google Scholar] [CrossRef] [Green Version]
- Schäfer, A.; Hühr, J.; Schwaiger, T.; Dorhoi, A.; Mettenleiter, T.C.; Blome, S.; Schroder, C.; Blohm, U. Porcine Invariant Natural Killer T Cells: Functional Profiling and Dynamics in Steady State and Viral Infections. Front. Immunol. 2019, 10, 1380. [Google Scholar] [CrossRef] [PubMed]
- Takamatsu, H.H.; Denyer, M.S.; Stirling, C.; Cox, S.; Aggarwal, N.; Dash, P.; Wileman, T.E.; Barnett, P.V. Porcine γδ T cells: Possible roles on the innate and adaptive immune responses following virus infection. Vet. Immunol. Immunopathol. 2006, 112, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Argilaguet, J.M.; Perez-Martin, E.; Nofrarias, M.; Gallardo, C.; Accensi, F.; Lacasta, A.; Mora, M.; Ballester, M.; Galindo-Cardiel, I.; Lopez-Soria, S.; et al. DNA vaccination partially protects against African swine fever virus lethal challenge in the absence of antibodies. PLoS ONE 2012, 7, e40942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onisk, D.V.; Borca, M.V.; Kutish, G.; Kramer, E.; Irusta, P.; Rock, D.L. Passively transferred African swine fever virus antibodies protect swine against lethal infection. Virology 1994, 198, 350–354. [Google Scholar] [CrossRef]
- Schlafer, D.H.; Mebus, C.A.; McVicar, J.W. African swine fever in neonatal pigs: Passively acquired protection from colostrum or serum of recovered pigs. Am. J. Vet. Res. 1984, 45, 1367–1372. [Google Scholar]
- Saalmüller, A.; Gerner, W. The immune system of swine. In Encyclopedia of immunology; Ratcliffe, M.J.H., Ed.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 538–548. [Google Scholar]
- Norley, S.G.; Wardley, R.C. Cytotoxic lymphocytes induced by African swine fever infection. Res. Vet. Sci. 1984, 37, 255–257. [Google Scholar] [CrossRef]
- Wardley, R.C.; Wilkinson, P.J. Lymphocyte responses to African swine fever virus infection. Res. Vet. Sci. 1980, 28, 185–189. [Google Scholar] [CrossRef]
- Martins, C.L.; Leitao, A.C. Porcine immune responses to African swine fever virus (ASFV) infection. Vet. Immunol. Immunopathol. 1994, 43, 99–106. [Google Scholar] [CrossRef]
- Martins, C.L.; Lawman, M.J.; Scholl, T.; Mebus, C.A.; Lunney, J.K. African swine fever virus specific porcine cytotoxic T cell activity. Arch. Virol. 1993, 129, 211–225. [Google Scholar] [CrossRef]
- Vigário, J.D.; Terrinha, A.M.; Moura Nunes, J.F. Antigenic relationships among strains of African swine fecre virus. Arch. Gesamte Virusforsch. 1974, 45, 272–277. [Google Scholar] [CrossRef]
- Gerner, W.; Talker, S.C.; Koinig, H.C.; Sedlak, C.; Mair, K.H.; Saalmüller, A. Phenotypic and functional differentiation of porcine αβ T cells: Current knowledge and available tools. Mol. Immunol. 2015, 66, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, A.; Zani, L.; Pikalo, J.; Hühr, J.; Sehl, J.; Mettenleiter, T.C.; Breithaupt, A.; Blome, S.; Blohm, U. T-cell responses in domestic pigs and wild boar upon infection with the moderately virulent African swine fever virus strain ’Estonia2014’. Transbound. Emerg. Dis. 2021, 68, 2733–2749. [Google Scholar] [CrossRef] [PubMed]
- Hühr, J.; Schäfer, A.; Schwaiger, T.; Zani, L.; Sehl, J.; Mettenleiter, T.C.; Blome, S.; Blohm, U. Impaired T-cell responses in domestic pigs and wild boar upon infection with a highly virulent African swine fever virus strain. Transbound. Emerg. Dis. 2020, 67, 3016–3032. [Google Scholar] [CrossRef] [PubMed]
- Prager, I.; Watzl, C. Mechanisms of natural killer cell-mediated cellular cytotoxicity. J. Leukoc. Biol. 2019, 105, 1319–1329. [Google Scholar] [CrossRef]
- Meiraz, A.; Garber, O.G.; Harari, S.; Hassin, D.; Berke, G. Switch from perforin-expressing to perforin-deficient CD8(+) T cells accounts for two distinct types of effector cytotoxic T lymphocytes in vivo. Immunology 2009, 128, 69–82. [Google Scholar] [CrossRef]
- Afonso, C.L.; Neilan, J.G.; Kutish, G.F.; Rock, D.L. An African swine fever virus Bc1-2 homolog, 5-HL, suppresses apoptotic cell death. J. Virol. 1996, 70, 4858–4863. [Google Scholar] [CrossRef] [Green Version]
- Galindo, I.; Hernaez, B.; Diaz-Gil, G.; Escribano, J.M.; Alonso, C. A179L, a viral Bcl-2 homologue, targets the core Bcl-2 apoptotic machinery and its upstream BH3 activators with selective binding restrictions for Bid and Noxa. Virology 2008, 375, 561–572. [Google Scholar] [CrossRef] [Green Version]
- Sutton, V.R.; Vaux, D.L.; Trapani, J.A. Bcl-2 prevents apoptosis induced by perforin and granzyme B, but not that mediated by whole cytotoxic lymphocytes. J. Immunol. 1997, 158, 5783–5790. [Google Scholar]
- Welz, M.; Eickhoff, S.; Abdullah, Z.; Trebicka, J.; Gartlan, K.H.; Spicer, J.A.; Demetris, A.J.; Akhlaghi, H.; Anton, M.; Manske, K.; et al. Perforin inhibition protects from lethal endothelial damage during fulminant viral hepatitis. Nat. Commun. 2018, 9, 4805. [Google Scholar] [CrossRef]
- Hersperger, A.R.; Makedonas, G.; Betts, M.R. Flow cytometric detection of perforin upregulation in human CD8 T cells. Cytom. A 2008, 73, 1050–1057. [Google Scholar] [CrossRef]
- Erickson, J.J.; Lu, P.; Wen, S.; Hastings, A.K.; Gilchuk, P.; Joyce, S.; Shyr, Y.; Williams, J.V. Acute Viral Respiratory Infection Rapidly Induces a CD8+ T Cell Exhaustion-like Phenotype. J. Immunol. 2015, 195, 4319–4330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bismarck, D.; Schütze, N.; Moore, P.; Buttner, M.; Alber, G.; Buttlar, H. Canine CD4+CD8+ double positive T cells in peripheral blood have features of activated T cells. Vet. Immunol. Immunopathol. 2012, 149, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Overgaard, N.H.; Jung, J.W.; Steptoe, R.J.; Wells, J.W. CD4+/CD8+ double-positive T cells: More than just a developmental stage? J. Leukoc. Biol. 2015, 97, 31–38. [Google Scholar] [CrossRef]
- Canals, A.; Alonso, F.; Tomillo, J.; Dominguez, J. Analysis of T lymphocyte subsets proliferating in response to infective and UV-inactivated African swine fever viruses. Vet. Microbiol. 1992, 33, 117–127. [Google Scholar] [CrossRef]
- Casal, I.; Vinuela, E.; Enjuanes, L. Synthesis of African swine fever (ASF) virus-specific antibodies in vitro in a porcine leucocyte system. Immunology 1987, 62, 207–213. [Google Scholar]
- Takamatsu, H.H.; Denyer, M.S.; Lacasta, A.; Stirling, C.M.; Argilaguet, J.M.; Netherton, C.L.; Oura, C.A.; Martins, C.; Rodriguez, F. Cellular immunity in ASFV responses. Virus Res. 2013, 173, 110–121. [Google Scholar] [CrossRef]
- Mosser, D.M.; Zhang, X. Interleukin-10: New perspectives on an old cytokine. Immunol. Rev. 2008, 226, 205–218. [Google Scholar] [CrossRef]
- Silva-Campa, E.; Mata-Haro, V.; Mateu, E.; Hernandez, J. Porcine reproductive and respiratory syndrome virus induces CD4+CD8+CD25+Foxp3+ regulatory T cells (Tregs). Virology 2012, 430, 73–80. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Cordón, P.J.; Chapman, D.; Jabbar, T.; Reis, A.L.; Goatley, L.; Netherton, C.L.; Taylor, G.; Montoya, M.; Dixon, L. Different routes and doses influence protection in pigs immunised with the naturally attenuated African swine fever virus isolate OURT88/3. Antivir. Res 2017, 138, 1–8. [Google Scholar] [CrossRef]
- Sanchez-Cordon, P.J.; Jabbar, T.; Berrezaie, M.; Chapman, D.; Reis, A.; Sastre, P.; Rueda, P.; Goatley, L.; Dixon, L.K. Evaluation of protection induced by immunisation of domestic pigs with deletion mutant African swine fever virus BeninΔMGF by different doses and routes. Vaccine 2018, 36, 707–715. [Google Scholar] [CrossRef]
- Sánchez-Cordón, P.J.; Jabbar, T.; Chapman, D.; Dixon, L.K.; Montoya, M. Absence of Long-Term Protection in Domestic Pigs Immunized with Attenuated African Swine Fever Virus Isolate OURT88/3 or BeninΔMGF Correlates with Increased Levels of Regulatory T Cells and Interleukin-10. J. Virol. 2020, 94, e00350-20. [Google Scholar] [CrossRef] [PubMed]
- Cabezon, O.; Munoz-Gonzalez, S.; Colom-Cadena, A.; Perez-Simo, M.; Rosell, R.; Lavin, S.; Marco, I.; Fraile, L.; de la Riva, P.M.; Rodriguez, F.; et al. African swine fever virus infection in Classical swine fever subclinically infected wild boars. BMC Vet. Res. 2017, 13, 227. [Google Scholar] [CrossRef] [PubMed]
- Barroso-Arévalo, S.; Barasona, J.A.; Cadenas-Fernández, E.; Sánchez-Vizcaíno, J.M. The Role of Interleukine-10 and Interferon-gamma as Potential Markers of the Evolution of African Swine Fever Virus Infection in Wild Boar. Pathogens 2021, 10, 757. [Google Scholar] [CrossRef] [PubMed]
- Reis, A.L.; Goatley, L.C.; Jabbar, T.; Lopez, E.; Rathakrishnan, A.; Dixon, L.K. Deletion of the Gene for the Type I Interferon Inhibitor I329L from the Attenuated African Swine Fever Virus OURT88/3 Strain Reduces Protection Induced in Pigs. Vaccines 2020, 8, 262. [Google Scholar] [CrossRef]
- Reis, A.L.; Goatley, L.C.; Jabbar, T.; Sanchez-Cordon, P.J.; Netherton, C.L.; Chapman, D.A.G.; Dixon, L.K. Deletion of the African Swine Fever Virus Gene DP148R Does Not Reduce Virus Replication in Culture but Reduces Virus Virulence in Pigs and Induces High Levels of Protection against Challenge. J. Virol. 2017, 91, e01428-17. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Zhang, J.; Zhang, Y.; Yang, J.; Wang, L.; Qi, Y.; Han, X.; Zhou, X.; Miao, F.; Chen, T.; et al. Cytokine Storm in Domestic Pigs Induced by Infection of Virulent African Swine Fever Virus. Front. Vet. Sci. 2020, 7, 601641. [Google Scholar] [CrossRef]
- Post, J.; Weesendorp, E.; Montoya, M.; Loeffen, W.L. Influence of Age and Dose of African Swine Fever Virus Infections on Clinical Outcome and Blood Parameters in Pigs. Viral Immunol. 2017, 30, 58–69. [Google Scholar] [CrossRef]
- Chien, Y.H.; Meyer, C.; Bonneville, M. γδ T cells: First line of defense and beyond. Annu. Rev. Immunol. 2014, 32, 121–155. [Google Scholar] [CrossRef]
- Morita, C.T.; Mariuzza, R.A.; Brenner, M.B. Antigen recognition by human γδ T cells: Pattern recognition by the adaptive immune system. Springer Semin. Immunopathol. 2000, 22, 191–217. [Google Scholar] [CrossRef]
- Halary, F.; Pitard, V.; Dlubek, D.; Krzysiek, R.; de la Salle, H.; Merville, P.; Dromer, C.; Emilie, D.; Moreau, J.F.; Dechanet-Merville, J. Shared reactivity of Vδ2neg γδ T cells against cytomegalovirus-infected cells and tumor intestinal epithelial cells. J. Exp. Med. 2005, 201, 1567–1578. [Google Scholar] [CrossRef] [Green Version]
- Sciammas, R.; Johnson, R.M.; Sperling, A.I.; Brady, W.; Linsley, P.S.; Spear, P.G.; Fitch, F.W.; Bluestone, J.A. Unique antigen recognition by a herpesvirus-specific TCR-γδ cell. J. Immunol. 1994, 152, 5392–5397. [Google Scholar] [PubMed]
- Holderness, J.; Hedges, J.F.; Ramstead, A.; Jutila, M.A. Comparative biology of γδ T cell function in humans, mice, and domestic animals. Annu. Rev. Anim. Biosci. 2013, 1, 99–124. [Google Scholar] [CrossRef] [PubMed]
- Sedlak, C.; Patzl, M.; Saalmüller, A.; Gerner, W. CD2 and CD8α define porcine γδ T cells with distinct cytokine production profiles. Dev. Comp. Immunol. 2014, 45, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, K.; Sinkora, M. The expression of CD25, CD11b, SWC1, SWC7, MHC-II, and family of CD45 molecules can be used to characterize different stages of γδ T lymphocytes in pigs. Dev. Comp. Immunol. 2012, 36, 728–740. [Google Scholar] [CrossRef]
- Takamatsu, H.H.; Denyer, M.S.; Wileman, T.E. A sub-population of circulating porcine γδ T cells can act as professional antigen presenting cells. Vet. Immunol. Immunopathol. 2002, 87, 223–224. [Google Scholar] [CrossRef]
- Zani, L.; Forth, J.H.; Forth, L.; Nurmoja, I.; Leidenberger, S.; Henke, J.; Carlson, J.; Breidenstein, C.; Viltrop, A.; Höper, D.; et al. Deletion at the 5’-end of Estonian ASFV strains associated with an attenuated phenotype. Sci. Rep. 2018, 8, 6510. [Google Scholar] [CrossRef]
- Kohlgruber, A.C.; Donado, C.A.; LaMarche, N.M.; Brenner, M.B.; Brennan, P.J. Activation strategies for invariant natural killer T cells. Immunogenetics 2016, 68, 649–663. [Google Scholar] [CrossRef]
- Renukaradhya, G.J.; Manickam, C.; Khatri, M.; Rauf, A.; Li, X.; Tsuji, M.; Rajashekara, G.; Dwivedi, V. Functional invariant NKT cells in pig lungs regulate the airway hyperreactivity: A potential animal model. J. Clin. Immunol. 2011, 31, 228–239. [Google Scholar] [CrossRef] [Green Version]
- Thierry, A.; Robin, A.; Giraud, S.; Minouflet, S.; Barra, A.; Bridoux, F.; Hauet, T.; Touchard, G.; Herbelin, A.; Gombert, J.M. Identification of invariant natural killer T cells in porcine peripheral blood. Vet. Immunol. Immunopathol. 2012, 149, 272–279. [Google Scholar] [CrossRef]
- Denyer, M.S.; Wileman, T.E.; Stirling, C.M.; Zuber, B.; Takamatsu, H.H. Perforin expression can define CD8 positive lymphocyte subsets in pigs allowing phenotypic and functional analysis of natural killer, cytotoxic T, natural killer T and MHC un-restricted cytotoxic T-cells. Vet. Immunol. Immunopathol. 2006, 110, 279–292. [Google Scholar] [CrossRef]
- Akbari, O.; Stock, P.; Meyer, E.H.; Freeman, G.J.; Sharpe, A.H.; Umetsu, D.T.; DeKruyff, R.H. ICOS/ICOSL interaction is required for CD4+ invariant NKT cell function and homeostatic survival. J. Immunol. 2008, 180, 5448–5456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaneda, H.; Takeda, K.; Ota, T.; Kaduka, Y.; Akiba, H.; Ikarashi, Y.; Wakasugi, H.; Kronenberg, M.; Kinoshita, K.; Yagita, H.; et al. ICOS costimulates invariant NKT cell activation. Biochem. Biophys. Res. Commun. 2005, 327, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Garner, L.C.; Klenerman, P.; Provine, N.M. Insights Into Mucosal-Associated Invariant T Cell Biology From Studies of Invariant Natural Killer T Cells. Front. Immunol. 2018, 9, 1478. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Li, K.; Ma, X.; Liu, B.; He, X.; Yang, S.; Wang, W.; Jiang, B.; Cai, J. Mucosal-Associated Invariant T Cells Expressing the TRAV1-TRAJ33 Chain Are Present in Pigs. Front. Immunol. 2019, 10, 2070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Netherton, C.L.; Goatley, L.C.; Reis, A.L.; Portugal, R.; Nash, R.H.; Morgan, S.B.; Gault, L.; Nieto, R.; Norlin, V.; Gallardo, C.; et al. Identification and Immunogenicity of African Swine Fever Virus Antigens. Front. Immunol. 2019, 10, 1318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Donnell, V.; Risatti, G.R.; Holinka, L.G.; Krug, P.W.; Carlson, J.; Velazquez-Salinas, L.; Azzinaro, P.A.; Gladue, D.P.; Borca, M.V. Simultaneous Deletion of the 9GL and UK Genes from the African Swine Fever Virus Georgia 2007 Isolate Offers Increased Safety and Protection against Homologous Challenge. J. Virol. 2017, 91, e01760-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muñoz-Pérez, C.; Jurado, C.; Sánchez-Vizcaíno, J.M. African swine fever vaccine: Turning a dream into reality. Transbound. Emerg. Dis. 2021, 68, 2657–2668. [Google Scholar] [CrossRef]
- O’Donnell, V.; Holinka, L.G.; Gladue, D.P.; Sanford, B.; Krug, P.W.; Lu, X.; Arzt, J.; Reese, B.; Carrillo, C.; Risatti, G.R.; et al. African Swine Fever Virus Georgia Isolate Harboring Deletions of MGF360 and MGF505 Genes Is Attenuated in Swine and Confers Protection against Challenge with Virulent Parental Virus. J. Virol. 2015, 89, 6048–6056. [Google Scholar] [CrossRef] [Green Version]
- Goatley, L.C.; Reis, A.L.; Portugal, R.; Goldswain, H.; Shimmon, G.L.; Hargreaves, Z.; Ho, C.-S.; Montoya, M.; Sánchez-Cordón, P.J.; Taylor, G.; et al. A Pool of Eight Virally Vectored African Swine Fever Antigens Protect Pigs Against Fatal Disease. Vaccines 2020, 8, 234. [Google Scholar] [CrossRef]
- Cackett, G.; Matelska, D.; Sykora, M.; Portugal, R.; Malecki, M.; Bahler, J.; Dixon, L.; Werner, F. The African Swine Fever Virus Transcriptome. J. Virol. 2020, 94, e00119-20. [Google Scholar] [CrossRef] [Green Version]
- Alonso, F.; Dominguez, J.; Vinuela, E.; Revilla, Y. African swine fever virus-specific cytotoxic T lymphocytes recognize the 32 kDa immediate early protein (vp32). Virus Res. 1997, 49, 123–130. [Google Scholar] [CrossRef]
- Leitao, A.; Malur, A.; Cornelis, P.; Martins, C.L. Identification of a 25-aminoacid sequence from the major African swine fever virus structural protein VP72 recognised by porcine cytotoxic T lymphocytes using a lipoprotein based expression system. J. Virol. Methods 1998, 75, 113–119. [Google Scholar] [CrossRef]
- Leitao, A.; Malur, A.; Cartaxeiro, C.; Vasco, G.; Cruz, B.; Cornelis, P.; Martins, C.L. Bacterial lipoprotein based expression vectors as tools for the characterisation of African swine fever virus (ASFV) antigens. Arch. Virol. 2000, 145, 1639–1657. [Google Scholar] [CrossRef] [PubMed]
- Lokhandwala, S.; Waghela, S.D.; Bray, J.; Martin, C.L.; Sangewar, N.; Charendoff, C.; Shetti, R.; Ashley, C.; Chen, C.H.; Berghman, L.R.; et al. Induction of Robust Immune Responses in Swine by Using a Cocktail of Adenovirus-Vectored African Swine Fever Virus Antigens. Clin. Vaccine Immunol. 2016, 23, 888–900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Revilla, Y.; Pena, L.; Vinuela, E. Interferon-gamma production by African swine fever virus-specific lymphocytes. Scand. J. Immunol. 1992, 35, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Bosch-Camós, L.; López, E.; Navas, M.J.; Pina-Pedrero, S.; Accensi, F.; Correa-Fiz, F.; Park, C.; Carrascal, M.; Domínguez, J.; Salas, M.L.; et al. Identification of Promiscuous African Swine Fever Virus T-Cell Determinants Using a Multiple Technical Approach. Vaccines 2021, 9, 29. [Google Scholar] [CrossRef]
- Jenson, J.S.; Childerstone, A.; Takamatsu, H.; Dixon, L.K.; Parkhouse, R.M. The cellular immune recognition of proteins expressed by an African swine fever virus random genomic library. J. Immunol. Methods 2000, 242, 33–42. [Google Scholar] [CrossRef]
- Rock, K.L.; Farfan-Arribas, D.J.; Colbert, J.D.; Goldberg, A.L. Re-examining class-I presentation and the DRiP hypothesis. Trends Immunol. 2014, 35, 144–152. [Google Scholar] [CrossRef] [Green Version]
- Boinas, F.S.; Hutchings, G.H.; Dixon, L.K.; Wilkinson, P.J. Characterization of pathogenic and non-pathogenic African swine fever virus isolates from Ornithodoros erraticus inhabiting pig premises in Portugal. J. Gen. Virol. 2004, 85, 2177–2187. [Google Scholar] [CrossRef]
- Bosch-Camos, L.; Lopez, E.; Collado, J.; Navas, M.J.; Blanco-Fuertes, M.; Pina-Pedrero, S.; Accensi, F.; Salas, M.L.; Mundt, E.; Nikolin, V.; et al. M448R and MGF505-7R: Two African Swine Fever Virus Antigens Commonly Recognized by ASFV-Specific T-Cells and with Protective Potential. Vaccines 2021, 9, 508. [Google Scholar] [CrossRef]
- Burmakina, G.; Malogolovkin, A.; Tulman, E.R.; Xu, W.; Delhon, G.; Kolbasov, D.; Rock, D.L. Identification of T-cell epitopes in African swine fever virus CD2v and C-type lectin proteins. J. Gen. Virol. 2019, 100, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Malogolovkin, A.; Burmakina, G.; Titov, I.; Sereda, A.; Gogin, A.; Baryshnikova, E.; Kolbasov, D. Comparative analysis of African swine fever virus genotypes and serogroups. Emerg. Infect Dis. 2015, 21, 312–315. [Google Scholar] [CrossRef] [PubMed]
- Malogolovkin, A.; Burmakina, G.; Tulman, E.R.; Delhon, G.; Diel, D.G.; Salnikov, N.; Kutish, G.F.; Kolbasov, D.; Rock, D.L. African swine fever virus CD2v and C-type lectin gene loci mediate serological specificity. J. Gen. Virol. 2015, 96 Pt 4, 866–873. [Google Scholar] [CrossRef] [PubMed]
- Gaudreault, N.N.; Madden, D.W.; Wilson, W.C.; Trujillo, J.D.; Richt, J.A. African Swine Fever Virus: An Emerging DNA Arbovirus. Front. Vet. Sci. 2020, 7, 215. [Google Scholar] [CrossRef]
- Oura, C.A.L.; Powell, P.P.; Anderson, E.; Parkhouse, R.M.E. The pathogenesis of African swine fever in the resistant bushpig. J. Gen. Virol. 1998, 79, 1439–1443. [Google Scholar] [CrossRef] [Green Version]
- Frant, M.; Wozniakowski, G.; Pejsak, Z. African Swine Fever (ASF) and Ticks. No Risk of Tick-mediated ASF Spread in Poland and Baltic States. J. Vet. Res. 2017, 61, 375–380. [Google Scholar] [CrossRef] [Green Version]
- Guinat, C.; Gogin, A.; Blome, S.; Keil, G.; Pollin, R.; Pfeiffer, D.U.; Dixon, L. Transmission routes of African swine fever virus to domestic pigs: Current knowledge and future research directions. Vet. Rec. 2016, 178, 262–267. [Google Scholar] [CrossRef] [Green Version]
- Jori, F.; Bastos, A.D. Role of wild suids in the epidemiology of African swine fever. Ecohealth 2009, 6, 296–310. [Google Scholar] [CrossRef] [Green Version]
- Pietschmann, J.; Mur, L.; Blome, S.; Beer, M.; Perez-Sanchez, R.; Oleaga, A.; Sanchez-Vizcaino, J.M. African swine fever virus transmission cycles in Central Europe: Evaluation of wild boar-soft tick contacts through detection of antibodies against Ornithodoros erraticus saliva antigen. BMC Vet. Res. 2016, 12, 1. [Google Scholar] [CrossRef] [Green Version]
- European Food Safety, A.; Alvarez, J.; Bicout, D.; Boklund, A.; Botner, A.; Depner, K.; More, S.J.; Roberts, H.; Stahl, K.; Thulke, H.H.; et al. Research gap analysis on African swine fever. EFSA J. 2019, 17, e05811. [Google Scholar] [CrossRef] [Green Version]
- Nurmoja, I.; Petrov, A.; Breidenstein, C.; Zani, L.; Forth, J.H.; Beer, M.; Kristian, M.; Viltrop, A.; Blome, S. Biological characterization of African swine fever virus genotype II strains from north-eastern Estonia in European wild boar. Transbound. Emerg. Dis. 2017, 64, 2034–2041. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety, A.; Nielsen, S.S.; Alvarez, J.; Bicout, D.J.; Calistri, P.; Depner, K.; Drewe, J.A.; Garin-Bastuji, B.; Rojas, J.L.G.; Schmidt, C.G.; et al. ASF Exit Strategy: Providing cumulative evidence of the absence of African swine fever virus circulation in wild boar populations using standard surveillance measures. EFSA J. 2021, 19, e06419. [Google Scholar] [CrossRef] [PubMed]
- Pautienius, A.; Schulz, K.; Staubach, C.; Grigas, J.; Zagrabskaite, R.; Buitkuviene, J.; Stankevicius, R.; Streimikyte, Z.; Oberauskas, V.; Zienius, D.; et al. African swine fever in the Lithuanian wild boar population in 2018: A snapshot. Virol. J. 2020, 17, 148. [Google Scholar] [CrossRef] [PubMed]
- Schulz, K.; Staubach, C.; Blome, S.; Nurmoja, I.; Viltrop, A.; Conraths, F.J.; Kristian, M.; Sauter-Louis, C. How to Demonstrate Freedom from African Swine Fever in Wild Boar-Estonia as an Example. Vaccines 2020, 8, 336. [Google Scholar] [CrossRef]
- Groenen, M.A.; Archibald, A.L.; Uenishi, H.; Tuggle, C.K.; Takeuchi, Y.; Rothschild, M.F.; Rogel-Gaillard, C.; Park, C.; Milan, D.; Megens, H.J.; et al. Analyses of pig genomes provide insight into porcine demography and evolution. Nature 2012, 491, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Ballesteros, C.; Garrido, J.M.; Vicente, J.; Romero, B.; Galindo, R.C.; Minguijon, E.; Villar, M.; Martin-Hernando, M.P.; Sevilla, I.; Juste, R.; et al. First data on Eurasian wild boar response to oral immunization with BCG and challenge with a Mycobacterium bovis field strain. Vaccine 2009, 27, 6662–6668. [Google Scholar] [CrossRef]
- Beltran-Beck, B.; de la Fuente, J.; Garrido, J.M.; Aranaz, A.; Sevilla, I.; Villar, M.; Boadella, M.; Galindo, R.C.; de la Lastra, J.M.P.; Moreno-Cid, J.A.; et al. Oral vaccination with heat inactivated Mycobacterium bovis activates the complement system to protect against tuberculosis. PLoS ONE 2014, 9, e98048. [Google Scholar] [CrossRef] [Green Version]
- Lastra, J.M.; Galindo, R.C.; Gortazar, C.; Ruiz-Fons, F.; Aranaz, A.; de la Fuente, J. Expression of immunoregulatory genes in peripheral blood mononuclear cells of European wild boar immunized with BCG. Vet. Microbiol. 2009, 134, 334–339. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Gomes, R.; Costa, V.; Santos, P.; Charneca, R.; Zhang, Y.P.; Liu, X.H.; Wang, S.Q.; Bento, P.; Nunes, J.L.; et al. How immunogenetically different are domestic pigs from wild boars: A perspective from single-nucleotide polymorphisms of 19 immunity-related candidate genes. Immunogenetics 2013, 65, 737–748. [Google Scholar] [CrossRef] [Green Version]
- O’Donnell, V.; Holinka, L.G.; Krug, P.W.; Gladue, D.P.; Carlson, J.; Sanford, B.; Alfano, M.; Kramer, E.; Lu, Z.; Arzt, J.; et al. African Swine Fever Virus Georgia 2007 with a Deletion of Virulence-Associated Gene 9GL (B119L), when Administered at Low Doses, Leads to Virus Attenuation in Swine and Induces an Effective Protection against Homologous Challenge. J. Virol. 2015, 89, 8556–8566. [Google Scholar] [CrossRef] [Green Version]
- Sunwoo, S.-Y.; Pérez-Núñez, D.; Morozov, I.; Sánchez, E.G.; Gaudreault, N.N.; Trujillo, J.D.; Mur, L.; Nogal, M.; Madden, D.; Urbaniak, K.; et al. DNA-Protein Vaccination Strategy Does Not Protect from Challenge with African Swine Fever Virus Armenia 2007 Strain. Vaccines 2019, 7, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, K.; Chapman, D.; Argilaguet, J.M.; Fishbourne, E.; Hutet, E.; Cariolet, R.; Hutchings, G.; Oura, C.A.; Netherton, C.L.; Moffat, K.; et al. Protection of European domestic pigs from virulent African isolates of African swine fever virus by experimental immunisation. Vaccine 2011, 29, 4593–4600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez, E.; van Heerden, J.; Bosch-Camós, L.; Accensi, F.; Navas, M.J.; López-Monteagudo, P.; Argilaguet, J.; Gallardo, C.; Pina-Pedrero, S.; Salas, M.L.; et al. Live Attenuated African Swine Fever Viruses as Ideal Tools to Dissect the Mechanisms Involved in Cross-Protection. Viruses 2020, 12, 1474. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schäfer, A.; Franzoni, G.; Netherton, C.L.; Hartmann, L.; Blome, S.; Blohm, U. Adaptive Cellular Immunity against African Swine Fever Virus Infections. Pathogens 2022, 11, 274. https://doi.org/10.3390/pathogens11020274
Schäfer A, Franzoni G, Netherton CL, Hartmann L, Blome S, Blohm U. Adaptive Cellular Immunity against African Swine Fever Virus Infections. Pathogens. 2022; 11(2):274. https://doi.org/10.3390/pathogens11020274
Chicago/Turabian StyleSchäfer, Alexander, Giulia Franzoni, Christopher L. Netherton, Luise Hartmann, Sandra Blome, and Ulrike Blohm. 2022. "Adaptive Cellular Immunity against African Swine Fever Virus Infections" Pathogens 11, no. 2: 274. https://doi.org/10.3390/pathogens11020274
APA StyleSchäfer, A., Franzoni, G., Netherton, C. L., Hartmann, L., Blome, S., & Blohm, U. (2022). Adaptive Cellular Immunity against African Swine Fever Virus Infections. Pathogens, 11(2), 274. https://doi.org/10.3390/pathogens11020274







