The Epidemiology of Sudden Oak Death Disease Caused by Phytophthora ramorum in a Mixed Bay Laurel-Oak Woodland Provides Important Clues for Disease Management
Abstract
:1. Introduction
- (1)
- Which characteristics of the biotic and abiotic neighborhood are most important in predicting infection of the transmissive host bay laurel?
- (2)
- Are oak infection and oak mortality correlated with oak size, high levels of bay laurel infection, and/or close proximity to infected bay laurels?
- (3)
- Are there trees and sites that are epidemiologically more relevant?
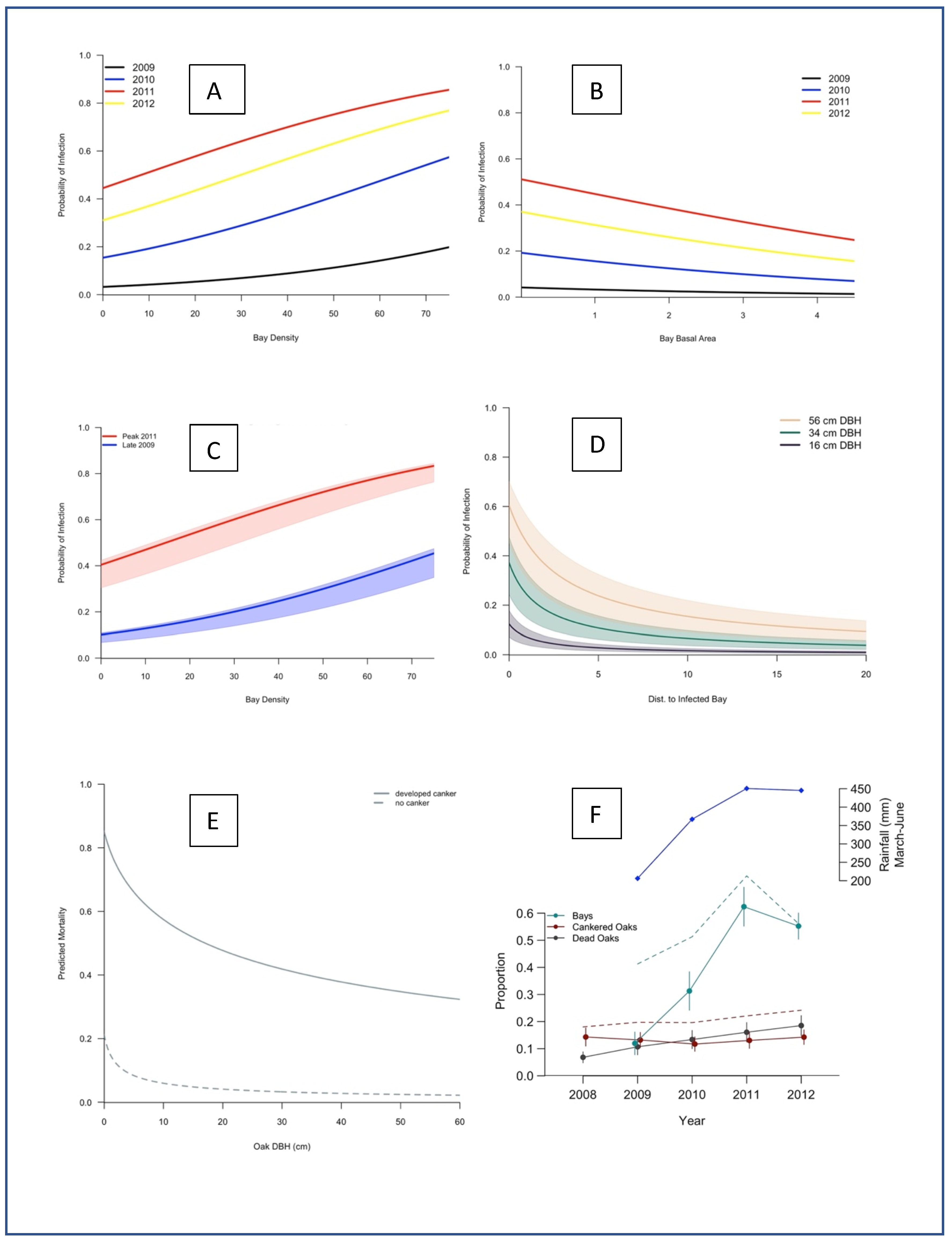
2. Results
3. Discussion
4. Materials and Methods
4.1. Site Selection and Experimental Design
4.2. Field Surveys
4.3. Pathogen Isolation and In Vitro Culturing
4.4. Molecular Diagnostics
4.5. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Madden, L.V.; Hughes, G. Plant disease incidence: Distributions, heterogeneity, and temporal analysis. Annu. Rev. Phytopathol. 1995, 33, 529–564. [Google Scholar] [CrossRef] [PubMed]
- Mundt, C.C.; Sackett, K.E.; Wallace, L.D. Landscape heterogeneity and disease spread: Experimental approaches with a plant pathogen. Ecol. Appl. 2011, 21, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Cobb, R.C.; Metz, M.R. Tree Diseases as a Cause and Consequence of Interacting Forest Disturbances. Forests 2017, 8, 147. [Google Scholar] [CrossRef]
- Garbelotto, M.; Gonthier, P. Variability and disturbances as key factors in forest pathology and plant health studies. Forests 2017, 8, 441. [Google Scholar] [CrossRef] [Green Version]
- Ostfeld, R.S.; Glass, G.E.; Keesing, F. Spatial epidemiology: An emerging (or re-emerging) discipline. Trends Ecol. Evol. 2005, 20, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Paull, S.H.; Song, S.; McClure, K.M.; Sackett, L.C.; Kilpatrick, A.M.; Johnson, P.T.J. From superspreaders to disease hotspots: Linking transmission across hosts and space. Front. Ecol. Environ. 2012, 10, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Stein, R.A. Super-spreaders in Infectious diseases. Int. J. Infect. Dis. 2011, 15, e510–e513. [Google Scholar] [CrossRef] [Green Version]
- Haydon, D.T.; Cleaveland, S.; Taylor, L.H.; Laurenson, M.K. Identifying reservoirs of infection: A conceptual and practical challenge. Emerg. Infect. Dis. 2002, 8, 1468–1473. [Google Scholar]
- Geils, B.W.; Hummer, K.E.; Hunt, R.S. White pines, Ribes, and blister rust: A review and synthesis. For. Pathol. 2010, 40, 147–185. [Google Scholar] [CrossRef]
- Gibbs, J.N. Intercontinental epidemiology of Dutch elm disease. Annu. Rev. Phytopathol. 1978, 16, 287–307. [Google Scholar] [CrossRef]
- Jung, T.; Vettraino, A.M.; Cech, T.L.; Vannini, A. The impact of invasive Phytophthora species on European forests. In Phytophthora: A Global Perspective; Lamour, K., Ed.; CABI: London, UK, 2013; Volume 2, pp. 146–158. [Google Scholar]
- Paillet, F. Chestnut: History and ecology of a transformed species. J. Biogeogr. 2002, 29, 1517–1530. [Google Scholar] [CrossRef] [Green Version]
- Rizzo, D.M.; Garbelotto, M.; Hansen, E.M. Phytophthora ramorum: Integrative research and management of an emerging pathogen in California and Oregon forests. Annu. Rev. Phytopathol. 2005, 43, 309–335. [Google Scholar] [CrossRef] [Green Version]
- Shearer, B.L.; Crane, C.E.; Barrett, S.; Cochrane, A. Phytophthora cinnamomi invasion, a major threatening process to conservation of flora diversity in the South-west Botanical Province of Western Australia. Aust. J. Bot. 2007, 55, 225–238. [Google Scholar] [CrossRef]
- Gilligan, C.A.; Truscott, J.E.; Stacey, A.J. Impact of scale on the effectiveness of disease control strategies for epidemics with cryptic infection in a dynamical landscape: An example for a crop disease. J. R. Soc. Interface 2007, 4, 925–934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filipe, J.A.N.; Cobb, R.C.; Meentemeyer, R.K.; Lee, C.A.; Valachovic, Y.S.; Cook, A.R.; Rizzo, D.M.; Gilligan, C.A. Landscape Epidemiology and Control of Pathogens with Cryptic and Long-Distance Dispersal: Sudden Oak Death in Northern Californian Forests. PLoS Comput. Biol. 2012, 8, e1002328. [Google Scholar] [CrossRef] [PubMed]
- Corredor-Moreno, P.; Saunders, D.G. Expecting the unexpected: Factors influencing the emergence of fungal and oomycete plant pathogens. New Phytol. 2020, 225, 118–125. [Google Scholar] [CrossRef]
- Rizzo, D.M.; Garbelotto, M. Sudden oak death: Endangering California and Oregon forest ecosystems. Front. Ecol. Environ. 2003, 1, 197–204. [Google Scholar] [CrossRef]
- USDA APHIS. List of Regulated Hosts and Plants Associated with Phytophthora ramorum. Available online: https://www.aphis.usda.gov/plant_health/plant_pest_info/pram/downloads/pdf_files/usdaprlist.pdf (accessed on 31 January 2022).
- Garbelotto, M.; Hayden, K.J. Sudden Oak Death: Interactions of the Exotic Oomycete Phytophthora ramorum with Naïve North American Hosts. Eukaryot. Cell 2012, 11, 1313–1323. [Google Scholar] [CrossRef] [Green Version]
- Garbelotto, M.; Davidson, J.M.; Ivors, K.; Maloney, P.E.; Hüberli, D.; Koike, S.T.; Rizzo, D.M. Non-oak native plants are main hosts for sudden oak death pathogen in California. Calif. Agric. 2003, 57, 18–23. [Google Scholar] [CrossRef] [Green Version]
- Davidson, J.M.; Wickland, A.C.; Patterson, H.A.; Falk, K.R.; Rizzo, D.M. Transmission of Phytophthora ramorum in mixed evergreen forest in California. Phytopathology 2005, 95, 587–596. [Google Scholar] [CrossRef] [Green Version]
- Davidson, J.M.; Werres, S.; Garbelotto, M.; Hansen, E.M.; Rizzo, D.M. Sudden oak death and associated diseases caused by Phytophthora ramorum. Plant Health Prog. 2003, 4, 12. [Google Scholar] [CrossRef]
- Davidson, J.M.; Patterson, H.A.; Wickland, A.C.; Fichtner, E.J.; Rizzo, D.M. Forest type influences transmission of Phytophthora ramorum in California oak woodlands. Phytopathology 2011, 101, 492–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fichtner, E.J.; Lynch, S.C.; Rizzo, D.M. Survival, Dispersal, and Potential Soil-Mediated Suppression of Phytophthora ramorum in a California Redwood-Tanoak Forest. Phytopathology 2009, 99, 608–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garbelotto, M.; Schmidt, D.; Swain, S.; Hayden, K.; Lione, G. The ecology of infection between a transmissive and a dead-end host provides clues for the treatment of a plant disease. Ecosphere 2017, 8, e01815. [Google Scholar] [CrossRef] [Green Version]
- Rizzo, D.M.; Garbelotto, M.; Davidson, J.M.; Slaughter, G.W.; Koike, S.T. Phytophthora ramorum as the cause of extensive mortality of Quercus spp. and Lithocarpus densiflorus in California. Plant Dis. 2002, 86, 205–214. [Google Scholar] [CrossRef] [Green Version]
- Maloney, P.E.; Lynch, S.C.; Kane, S.F.; Jensen, C.E.; Rizzo, D.M. Establishment of an emerging generalist pathogen in redwood forest communities. J. Ecol. 2005, 93, 899–905. [Google Scholar] [CrossRef]
- Hüberli, D.; Hayden, K.J.; Calver, M.; Garbelotto, M. Intraspecific variation in host susceptibility and climatic factors mediate epidemics of sudden oak death in western US forests. Plant Pathol. 2011, 61, 579–592. [Google Scholar] [CrossRef] [Green Version]
- Anacker, B.L.; Rank, N.E.; Hüberli, D.; Garbelotto, M.; Gordon, S.; Harnik, T.; Whitkus, R.; Meentemeyer, R. Susceptibility to Phytophthora ramorum in a key infectious host: Landscape variation in host genotype, host phenotype, and environmental factors. New Phytol. 2008, 177, 756–766. [Google Scholar] [CrossRef]
- DiLeo, M.V.; Bostock, R.M.; Rizzo, D.M. Microclimate Impacts Survival and Prevalence of Phytophthora ramorum in Umbellularia californica, a Key Reservoir Host of Sudden Oak Death in Northern California Forests. PLoS ONE 2014, 9, e98195. [Google Scholar] [CrossRef] [Green Version]
- Lione, G.; Gonthier, P.; Garbelotto, M. Environmental Factors Driving the Recovery of Bay Laurels from Phytophthora ramorum Infections: An Application of Numerical Ecology to Citizen Science. Forests 2017, 8, 293. [Google Scholar] [CrossRef] [Green Version]
- Kozanitas, M.; Osmundson, T.O.; Linzer, R.; Garbelotto, M. Interspecific interactions between the Sudden Oak Death pathogen Phytophthora ramorum and two sympatric Phytophthora species in varying ecological conditions. Fungal Ecol. 2017, 28, 86–96. [Google Scholar] [CrossRef]
- Chimento, A.; Cacciola, S.O.; Garbelotto, M. Detection of mRNA by reverse-transcription PCR as an indicator of viability in Phytophthora ramorum. For. Pathol. 2012, 42, 14–21. [Google Scholar] [CrossRef]
- Garbelotto, M.; Rizzo, D. A California-based chronological review (1995–2004) of research on Phytophthora ramorum, the causal agent of sudden oak death. Phytopathol. Mediterr. 2005, 44, 127–143. [Google Scholar]
- Cobb, R.C.; Filipe, J.A.; Meentemeyer, R.K.; Gilligan, C.A.; Rizzo, D.M. Ecosystem transformation by emerging infectious disease: Loss of large tanoak from California forests. J. Ecol. 2012, 100, 712–722. [Google Scholar] [CrossRef]
- McPherson, B.A.; Mori, S.R.; Wood, D.L.; Kelly, M.; Storer, A.J.; Svihra, P.; Standiford, R.B. Responses of oaks and tanoaks to the sudden oak death pathogen after 8 y of monitoring in two coastal California forests. For. Ecol. Manag. 2010, 259, 2248–2255. [Google Scholar] [CrossRef]
- Garbelotto, M.; Svihra, P.; Rizzo, D. New pests and diseases: Sudden oak death syndrome fells 3 oak species. Calif. Agric. 2001, 55, 9–19. [Google Scholar] [CrossRef] [Green Version]
- Dodd, R.S.; Hüberli, D.; Mayer, W.; Harnik, T.Y.; Afzal-Rafii, Z.; Garbelotto, M. Evidence for the role of synchronicity between host phenology and pathogen activity in the distribution of sudden oak death canker disease. New Phytol. 2008, 179, 505–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cunniffe, N.J.; Cobb, R.C.; Meentemeyer, R.K.; Rizzo, D.M.; Gilligan, C.A. Modeling when, where, and how to manage a forest epidemic, motivated by sudden oak death in California. Proc. Natl. Acad. Sci. USA 2016, 113, 5640–5645. [Google Scholar] [CrossRef] [Green Version]
- Maloney, P.E.; Lynch, S.C.; Kane, S.F.; Rizzo, D.M. Disease progression of Phytophthora ramorum and Botryosphaeria dothidea on Pacific madrone. Plant Dis. 2004, 88, 852–857. [Google Scholar] [CrossRef]
- Croucher, P.J.P.; Mascheretti, S.; Garbelotto, M. Combining field epidemiological information and genetic data to comprehensively reconstruct the invasion history and the microevolution of the sudden oak death agent Phytophthora ramorum (Stramenopila: Oomycetes) in California. Biol. Invasions 2013, 15, 2281–2297. [Google Scholar] [CrossRef] [Green Version]
- De Martonne, E. L’indice d’aridité. Bull. Assoc. Geogr. Fr. 1926, 3, 3–5. [Google Scholar] [CrossRef]
- Mascheretti, S.; Croucher, P.J.P.; Vettraino, A.; Prospero, S.; Garbelotto, M. Reconstruction of the Sudden Oak Death epidemic in California through microsatellite analysis of the pathogen Phytophthora ramorum. Mol. Ecol. 2008, 17, 2755–2768. [Google Scholar] [CrossRef]
- Eyre, C.A.; Kozanitas, M.; Garbelotto, M. Population dynamics of aerial and terrestrial populations of Phytophthora ramorum in a California forest under different climatic conditions. Phytopathology 2013, 103, 1141–1152. [Google Scholar] [CrossRef] [Green Version]
- Hayden, K.; Ivors, K.; Wilkinson, C.; Garbelotto, M. TaqMan chemistry for Phytophthora ramorum detection and quantification, with a comparison of diagnostic methods. Phytopathology 2006, 96, 846–854. [Google Scholar] [CrossRef] [Green Version]
- Gallegly, M.E.; Hong, C.X. Phytophthora: Identifying Species by Morphology and DNA Fingerprints; APS Press: St. Paul, MN, USA, 2008; pp. 1–158. [Google Scholar]
- Lemmon, P.E. A new instrument for measuring forest overstory density. J. For. 1957, 55, 667–669. [Google Scholar]
- Hayden, K.J.; Rizzo, D.M.; Tse, J.; Garbelotto, M. Detection and quantification of Phytophthora ramorum from California forests using a real-time polymerase chain reaction assay. Phytopathology 2004, 94, 1075–1083. [Google Scholar] [CrossRef] [Green Version]
- Steiner, J.J.; Polemba, C.J.; Fjellstrom, R.G.; Elliott, L.F. A rapid one-tube genomic DNA extraction process for PCR and RAPD analyses. Nucleic Acids Res. 1995, 23, 2569–2570. [Google Scholar] [CrossRef] [Green Version]
- Osmundson, T.W.; Eyre, C.A.; Hayden, K.M.; Dhillon, J.; Garbelotto, M. Back to basics: An evaluation of NaOH and alternative rapid DNA extraction protocols for DNA barcoding, genotyping, and disease diagnostics from fungal and oomycete samples. Mol. Ecol. Resour. 2013, 13, 66–74. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2014. [Google Scholar]
- Bates, D.; Maechle, M. lme4: Linear Mixed-Effects Models using S4 Classes. R Package Version 0.999375-34. Available online: http://CRAN.R-project.org/package=lme4 (accessed on 31 January 2022).
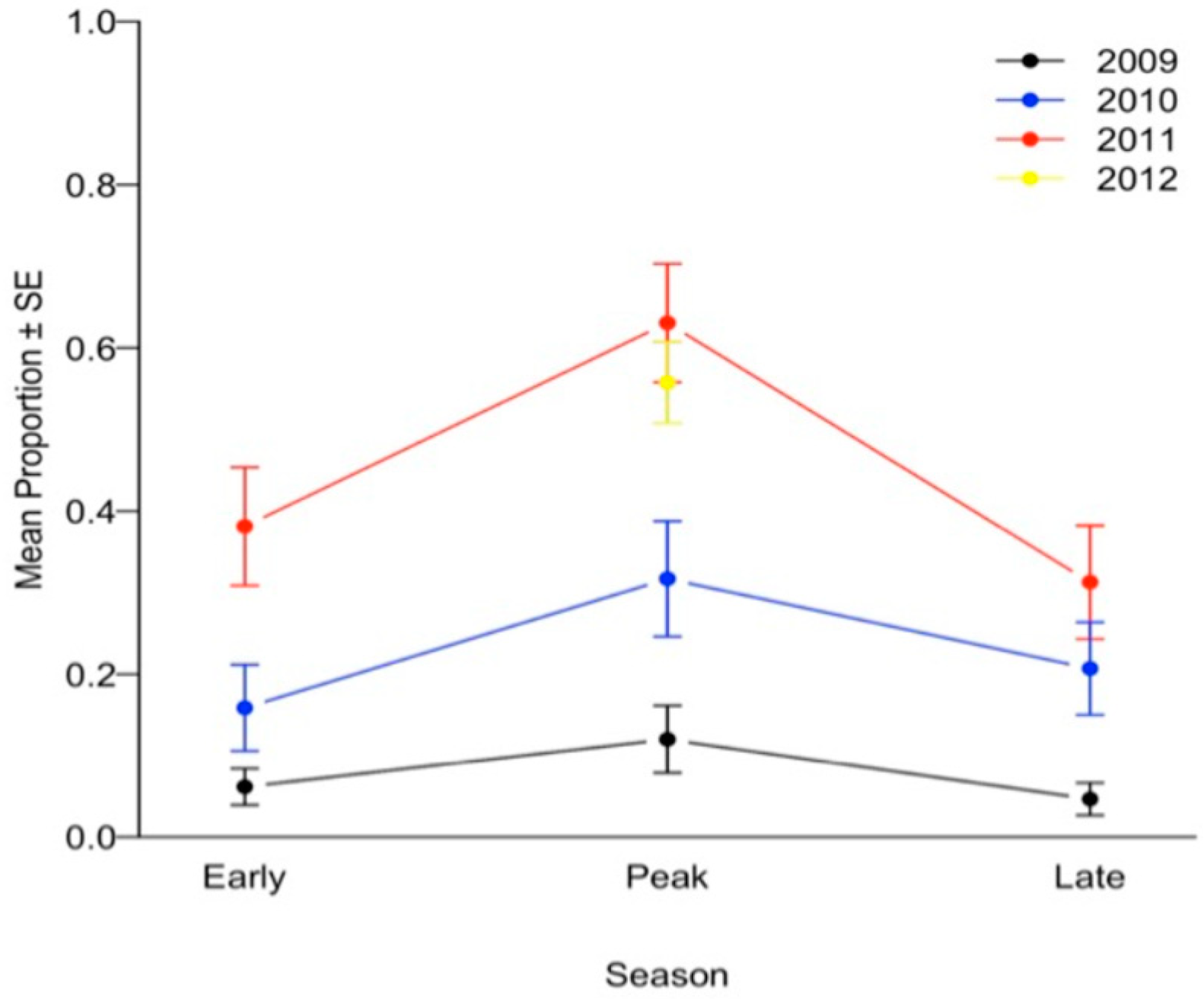
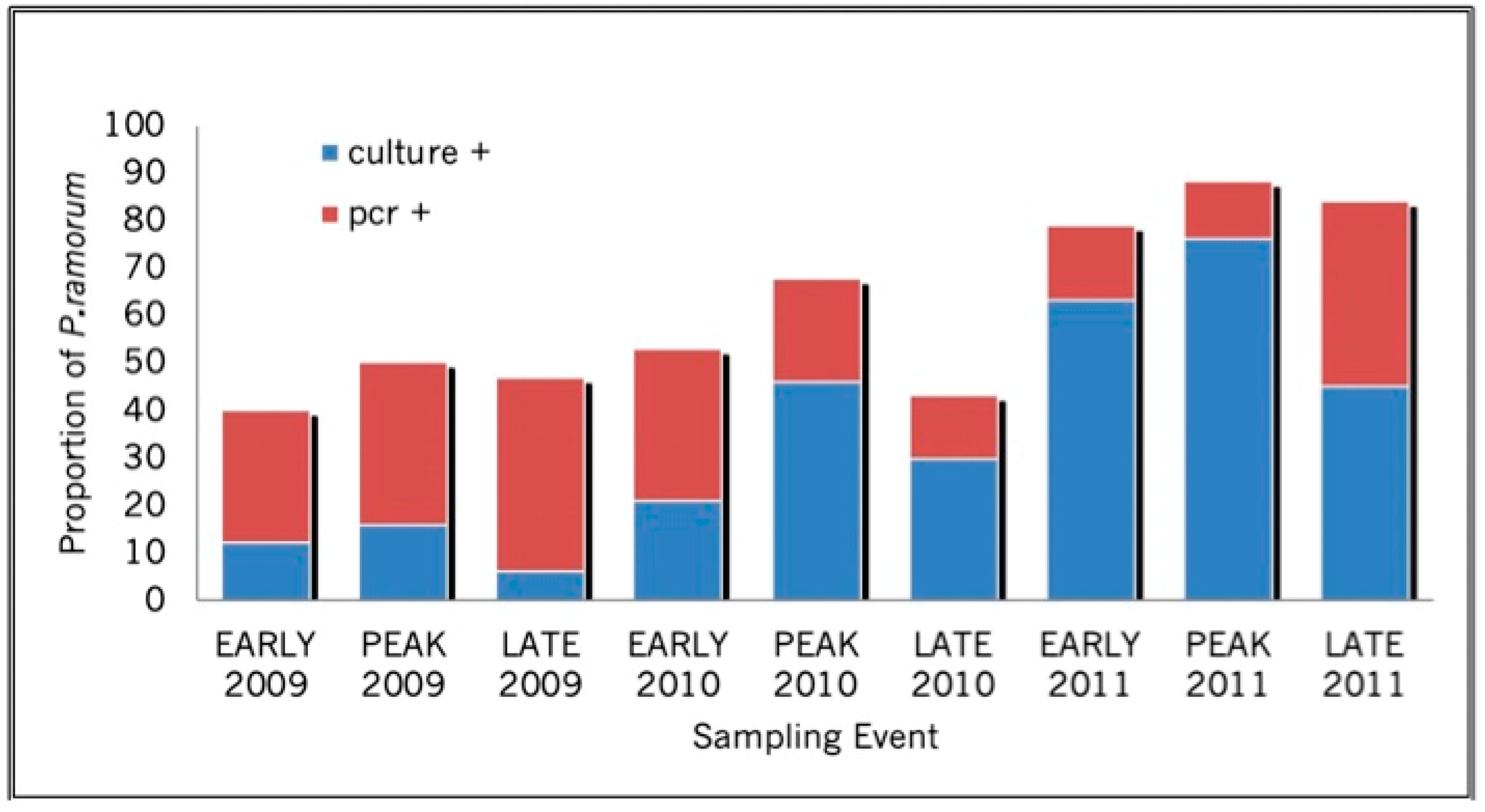

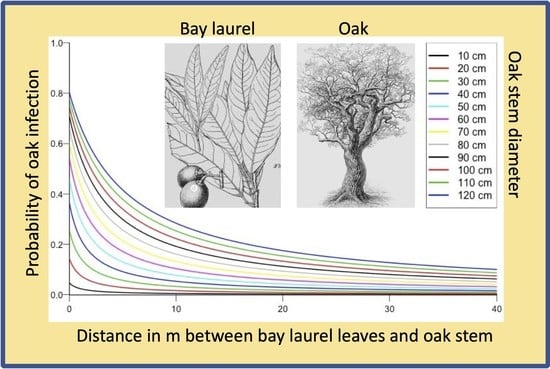
| INFECTION PREDICTION | SURVIVAL PREDICTION | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group | U. californica | Q. agrifolia | Q. agrifolia | |||||||||||||||
| RANDOM EFFECTS | Variance | (Std. Dev.) | Variance | (Std. Dev.) | Variance | (Std. Dev.) | ||||||||||||
| Tree ID | 0.4499 | 0.6707 | ||||||||||||||||
| Transect (Plot) | 0.3325 | 0.5766 | 0.8221 | 0.9067 | 0.0000 | 0.0000 | ||||||||||||
| Plot | 0.8729 | 0.9343 | 0.2211 | 0.4702 | 1.1172 | 0.3424 | ||||||||||||
| FIXED EFFECTS | Estimate | (SE) | p Value | Estimate | (SE) | p Value | Estimate | (SE) | p Value | |||||||||
| (Intercept) | 1.05 | (0.40) | 0.009 | |||||||||||||||
| Bay Laurel DBH | −0.08 | (0.07) | 0.235 | |||||||||||||||
| Bay Basal Area | −0.31 | (0.14) | 0.031 | −0.03 | (0.33) | 0.923 | 0.05 | (0.22) | 0.83 | |||||||||
| Bay Density | 0.44 | (0.17) | 0.010 | 0.83 | (0.35) | 0.017 | 0.09 | (0.23) | 0.71 | |||||||||
| Northness | 0.09 | (0.08) | 0.303 | −0.19 | (0.17) | 0.250 | −0.22 | (0.17) | 0.19 | |||||||||
| Eastness | 0.20 | (0.09) | 0.022 | −0.17 | (0.16) | 0.311 | 0.05 | (0.17) | 0.78 | |||||||||
| Canopy Cover | −0.003 | (0.003) | 0.347 | |||||||||||||||
| Oak DBH | 0.94 | (0.15) | 4.41 × 10−10 | −0.29 | (0.14) | 0.04 | ||||||||||||
| Proximity to Infected Bay | −1.2 | (0.15) | 1.81 × 10−15 | −0.10 | (0.16) | 0.511 | ||||||||||||
| Cankered | 3.027 | (0.30) | <2 × 10−16 | |||||||||||||||
| Season Peak | ||||||||||||||||||
| Season Early | −1.21 | (0.12) | <2 × 10−16 | |||||||||||||||
| Season Late | −1.38 | (0.12) | <2 × 10−16 | |||||||||||||||
| Year 2009 | −3.18 | (0.14) | <2 × 10−16 | |||||||||||||||
| Year 2010 | −1.48 | (0.11) | <2 × 10−16 | |||||||||||||||
| Year 2011 | −0.57 | (0.15) | 0.0002 | |||||||||||||||
| Model AIC | 3434 | 617.5 | 518.4 | |||||||||||||||
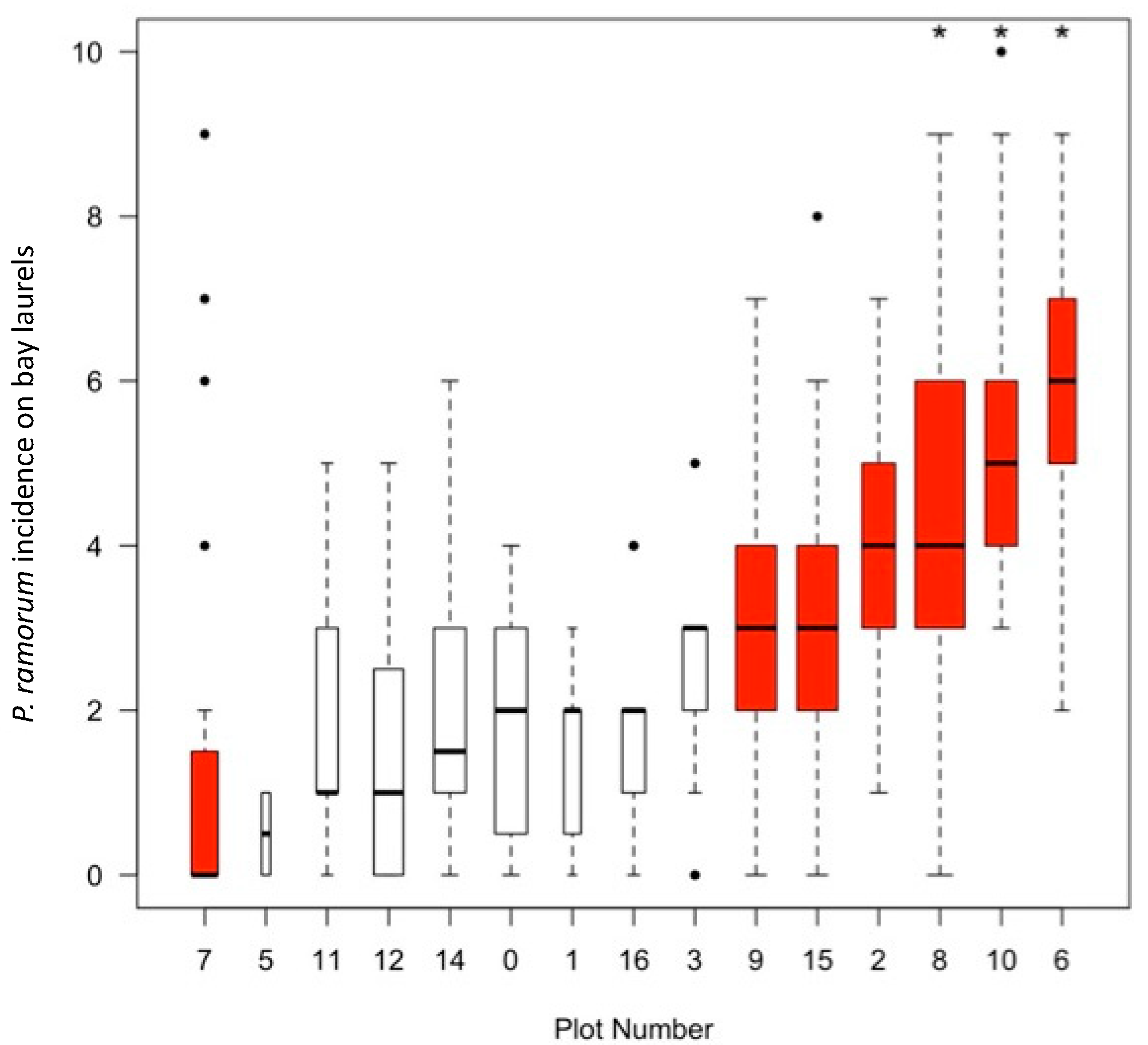
| Metric | Plot Type | p Value | |
|---|---|---|---|
| Hot Spots (7 Plots) | Cold Spots (8 Plots) | ||
| Average bay laurel density (number of trees/ha), SE | 126.47, 47.8 | 70.83, 14.5 | 0.04 |
| Average bay laurel basal area (square m/ha), SE | 11.4, 2 | 5.54, 1.7 | 0.05 |
| CDI * by PCR and culturing, % positive (sample size) | 61.5% (2540) | 41.57% (1340) | 0.0001 |
| CDI by culturing only, % positive (sample size) | 40% (2540) | 18.43% (1340) | 0.0001 |
| Fall 2009 DI by PCR and culturing (% positive of sample size) | 47.64% (254) | 44.03% (134) | 0.52 |
| Fall 2009 DI by culturing only (% positive of sample size) | 8.27% (254) | 1.49% (134) | 0.0059 |
| Spring 2011 DI by PCR and culturing (% positive of sample size) | 88.19% (254) | 70.15% (134) | 0.0001 |
| Spring 2011 DI by culturing only (% positive of sample size) | 77.17% (254) | 54.48% (134) | 0.0001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kozanitas, M.; Metz, M.R.; Osmundson, T.W.; Serrano, M.S.; Garbelotto, M. The Epidemiology of Sudden Oak Death Disease Caused by Phytophthora ramorum in a Mixed Bay Laurel-Oak Woodland Provides Important Clues for Disease Management. Pathogens 2022, 11, 250. https://doi.org/10.3390/pathogens11020250
Kozanitas M, Metz MR, Osmundson TW, Serrano MS, Garbelotto M. The Epidemiology of Sudden Oak Death Disease Caused by Phytophthora ramorum in a Mixed Bay Laurel-Oak Woodland Provides Important Clues for Disease Management. Pathogens. 2022; 11(2):250. https://doi.org/10.3390/pathogens11020250
Chicago/Turabian StyleKozanitas, Melina, Margaret R. Metz, Todd W. Osmundson, Maria Socorro Serrano, and Matteo Garbelotto. 2022. "The Epidemiology of Sudden Oak Death Disease Caused by Phytophthora ramorum in a Mixed Bay Laurel-Oak Woodland Provides Important Clues for Disease Management" Pathogens 11, no. 2: 250. https://doi.org/10.3390/pathogens11020250
APA StyleKozanitas, M., Metz, M. R., Osmundson, T. W., Serrano, M. S., & Garbelotto, M. (2022). The Epidemiology of Sudden Oak Death Disease Caused by Phytophthora ramorum in a Mixed Bay Laurel-Oak Woodland Provides Important Clues for Disease Management. Pathogens, 11(2), 250. https://doi.org/10.3390/pathogens11020250