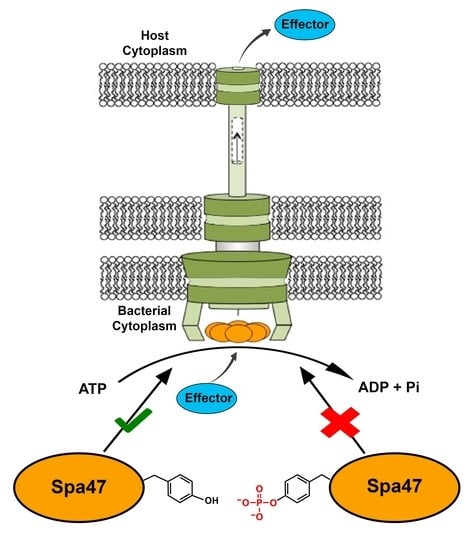
Phosphomimetic Tyrosine Mutations in Spa47 Inhibit Type Three Secretion ATPase Activity and Shigella Virulence Phenotype
Abstract
:1. Introduction
2. Results
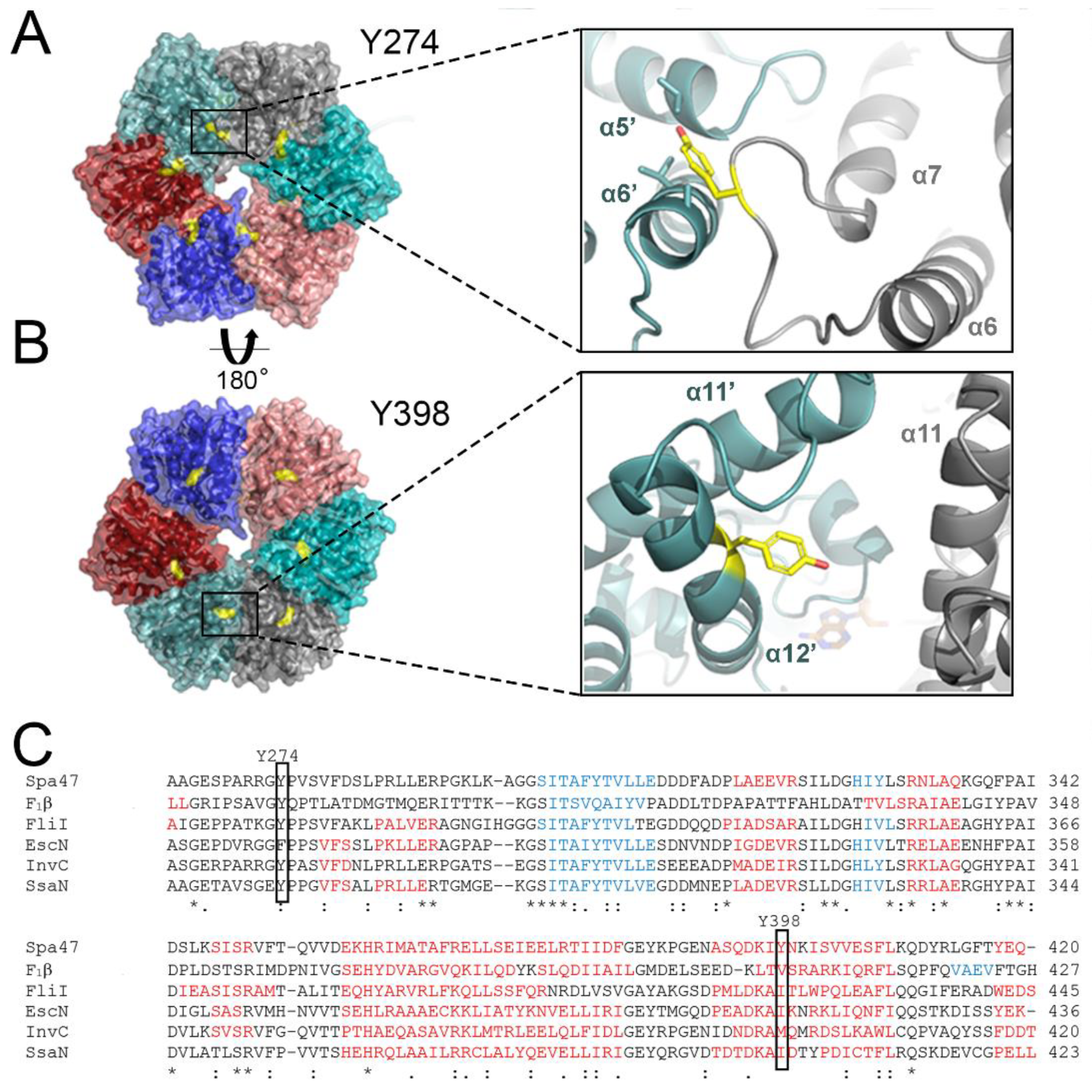
2.1. Structural Insights into the Roles of Post-Translationally Targeted Tyrosines in Spa47
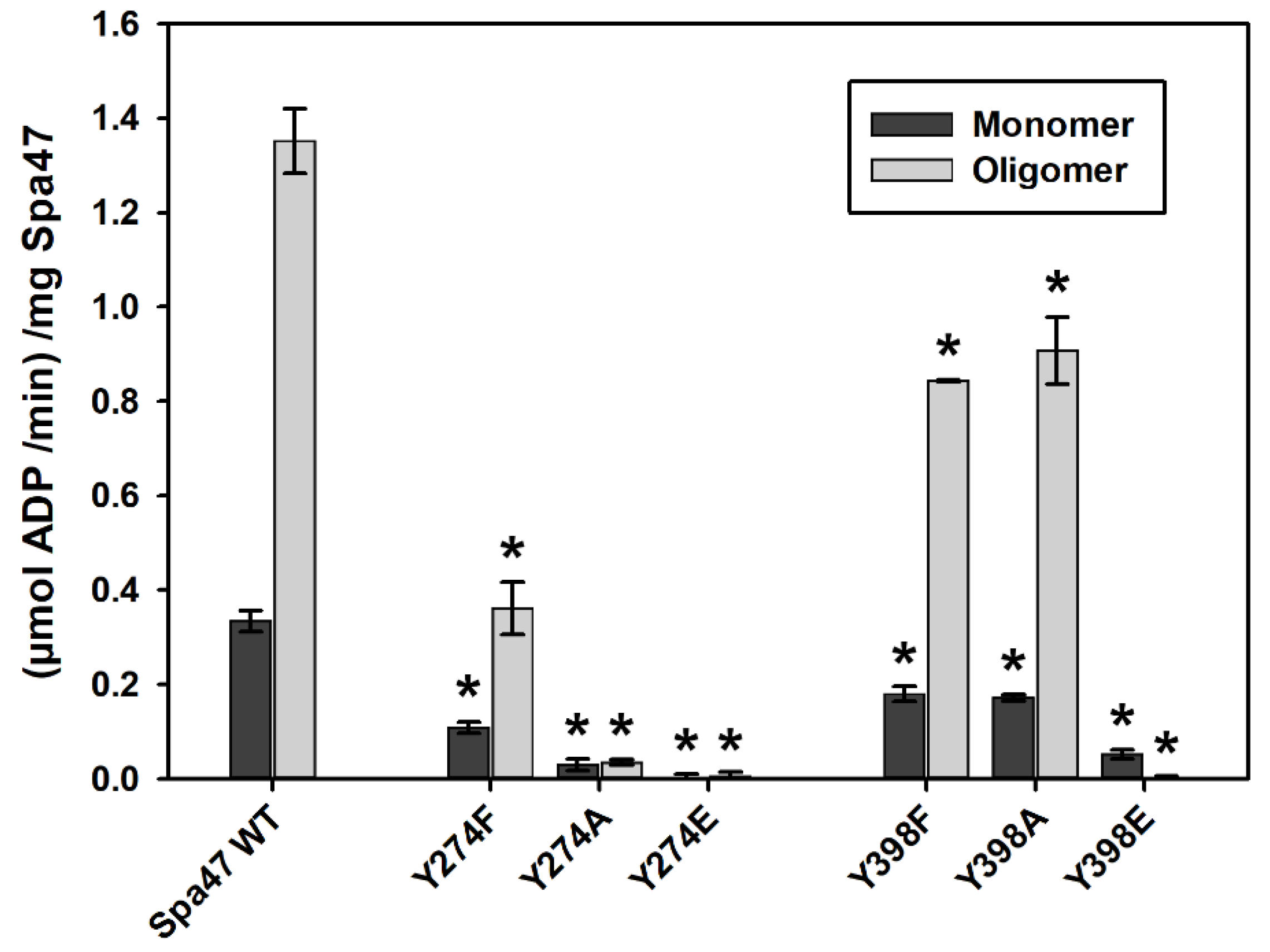
2.2. Phosphomimetic Tyrosine Mutations Impact Spa47 ATPase Activity
2.3. Phosphomimetic Tyrosine Mutations Impact Spa47 Thermal Stability
2.4. Phosphomimetic Substitutions to Spa47Y274, but Not Spa47Y398, Regulate Protein Secretion through the T3SS Injectisome
2.5. Spa47Y274, but Not Spa47Y398, Is Critical to Shigella Virulence Phenotype
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cloning
4.3. Protein Expression and Purification
4.4. ATP Hydrolysis Activity Assays
4.5. Far-UV Circular Dichroism (CD)
4.6. Shigella-Induced Erythrocyte Hemolysis
4.7. Bacterial Invasion of Epithelial Cells
4.8. Quantitation of S. flexneri T3SS Translocator Secretion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zaidi, M.B.; Estrada-Garcia, T. Shigella: A Highly Virulent and Elusive Pathogen. Curr. Trop. Med. Rep. 2014, 1, 81–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dupont, H.L.; Levine, M.M.; Hornick, R.B.; Formal, S.B. Inoculum Size in Shigellosis and Implications for Expected Mode of Transmission. J. Infect. Dis. 1989, 159, 1126–1128. [Google Scholar] [CrossRef] [PubMed]
- Khalil, I.A.; Troeger, C.; Blacker, B.F.; Rao, P.C.; Brown, A.; Atherly, D.E.; Brewer, T.G.; Engmann, C.M.; Houpt, E.R.; Kang, G.; et al. Morbidity and Mortality due to Shigella and Enterotoxigenic Escherichia Coli Diarrhoea: The Global Burden of Disease Study 1990–2016. Lancet Infect. Dis. 2018, 18, 1229–1240. [Google Scholar] [CrossRef] [Green Version]
- Puzari, M.; Sharma, M.; Chetia, P. Emergence of Antibiotic Resistant Shigella Species: A Matter of Concern. J. Infect. Public Health 2018, 11, 451–454. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Ghosh, A.; Ghosh, K.; Dutta, D.; Bhattacharya, S.K.; Nair, G.B.; Yoshida, S.-I. Newly Emerged Multiple-Antibiotic-Resistant Shigella Dysenteriae Type 1 Strains in and around Kolkata, India, Are Clonal. J. Clin. Microbiol. 2003, 41, 5833–5834. [Google Scholar] [CrossRef] [Green Version]
- Carayol, N.; Van Nhieu, G.T. The Inside Story of Shigella Invasion of Intestinal Epithelial Cells. Cold Spring Harb. Perspect. Med. 2013, 3, a016717. [Google Scholar] [CrossRef]
- Phalipon, A.; Sansonetti, P.J. Shigella’ s Ways of Manipulating the Host Intestinal Innate and Adaptive Immune System: A Tool Box for Survival? Immunol. Cell Biol. 2007, 85, 119–129. [Google Scholar] [CrossRef]
- Puhar, A.; Sansonetti, P.J. Type III Secretion System. Curr. Biol. 2014, 24, R784–R791. [Google Scholar] [CrossRef] [Green Version]
- Schroeder, G.; Hilbi, H. Molecular Pathogenesis of Shigella Spp.: Controlling Host Cell Signaling, Invasion, and Death by Type III Secretion. Clin. Microbiol. Rev. 2008, 21, 134–156. [Google Scholar] [CrossRef] [Green Version]
- Ashida, H.; Emimuro, H.; Esasakawa, C. Shigella Manipulates Host Immune Responses by Delivering Effector Proteins with Specific Roles. Front. Immunol. 2015, 6, 219. [Google Scholar] [CrossRef] [Green Version]
- Schroeder, G.N.; Jann, N.J.; Hilbi, H. Intracellular Type III Secretion by Cytoplasmic Shigella Flexneri Promotes Caspase-1-Dependent Macrophage Cell Death. Microbiology 2007, 153, 2862–2876. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Morado, D.R.; Margolin, W.; Rohde, J.R.; Arizmendi, O.; Picking, W.L.; Picking, W.D.; Liu, J. Visualization of the Type III Secretion Sorting Platform of Shigella Flexneri. Proc. Natl. Acad. Sci. USA 2015, 112, 1047–1052. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Worrall, L.J.; Hong, C.; Vuckovic, M.; Atkinson, C.E.; Caveney, N.; Yu, Z.; Strynadka, N.C.J. Cryo-EM Analysis of the T3S Injectisome Reveals the Structure of the Needle and Open Secretin. Nat. Commun. 2018, 9, 3840. [Google Scholar] [CrossRef] [PubMed]
- Guo, E.Z.; Galán, J.E. Cryo-EM Structure of the Needle Filament Tip Complex of the Salmonella Type III Secretion Injectisome. Proc. Natl. Acad. Sci. USA 2021, 118, e2114552118. [Google Scholar] [CrossRef]
- Chatterjee, S.; Chaudhury, S.; McShan, A.C.; Kaur, K.; De Guzman, R.N. Structure and Biophysics of Type III Secretion in Bacteria. Biochemistry 2013, 52, 2508–2517. [Google Scholar] [CrossRef] [Green Version]
- Case, H.B.; Dickenson, N.E. MxiN Differentially Regulates Monomeric and Oligomeric Species of the Shigella Type Three Secretion System ATPase Spa47. Biochemistry 2018, 57, 2266–2277. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.; Blocker, A. Characterization of Soluble Complexes of the Shigella Flexneritype III Secretion System ATPase. FEMS Microbiol. Lett. 2008, 286, 274–278. [Google Scholar] [CrossRef] [Green Version]
- Akeda, Y.; Galán, J.E. Chaperone Release and Unfolding of Substrates in Type III Secretion. Nature 2005, 437, 911–915. [Google Scholar] [CrossRef] [PubMed]
- Erhardt, M.; Mertens, M.E.; Fabiani, F.D.; Hughes, K.T. ATPase-Independent Type-III Protein Secretion in Salmonella Enterica. PLoS Genet. 2014, 10, e1004800. [Google Scholar] [CrossRef]
- Minamino, T.; Namba, K. Distinct Roles of the FliI ATPase and Proton Motive Force in Bacterial Flagellar Protein Export. Nature 2008, 451, 485–488. [Google Scholar] [CrossRef]
- Minamino, T. Protein Export through the Bacterial Flagellar Type III Export Pathway. Biochim. Biophys. Acta 2014, 1843, 1642–1648. [Google Scholar] [CrossRef] [Green Version]
- Muthuramalingam, M.; Whittier, S.; Picking, W.; Picking, W. The Shigella Type III Secretion System: An Overview from Top to Bottom. Microorganisms 2021, 9, 451. [Google Scholar] [CrossRef]
- Cordes, F.S.; Komoriya, K.; Larquet, E.; Yang, S.; Egelman, E.; Blocker, A.; Lea, S. Helical Structure of the Needle of the Type III Secretion System of Shigella Flexneri. J. Biol. Chem. 2003, 278, 17103–17107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blocker, A.; Gounon, P.; Larquet, E.; Niebuhr, K.; Cabiaux, V.; Parsot, C.; Sansonetti, P. The Tripartite Type III Secreton of Shigella Flexneri Inserts Ipab and Ipac into Host Membranes. J. Cell Biol. 1999, 147, 683–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Epler, C.R.; Dickenson, N.E.; Olive, A.J.; Picking, W.L.; Picking, W.D. Liposomes Recruit IpaC to the Shigella Flexneri Type III Secretion Apparatus Needle as a Final Step in Secretion Induction. Infect. Immun. 2009, 77, 2754–2761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stensrud, K.F.; Adam, P.R.; La Mar, C.D.; Olive, A.J.; Lushington, G.H.; Sudharsan, R.; Shelton, N.L.; Givens, R.S.; Picking, W.L.; Picking, W.D. Deoxycholate Interacts with IpaD of Shigella Flexneri in Inducing the Recruitment of IpaB to the Type III Secretion Apparatus Needle Tip. J. Biol. Chem. 2008, 283, 18646–18654. [Google Scholar] [CrossRef] [Green Version]
- Olive, A.J.; Kenjale, R.; Espina, M.; Moore, D.S.; Picking, W.L.; Picking, W.D. Bile Salts Stimulate Recruitment of IpaB to the Shigella flexneri Surface, Where It Colocalizes with IpaD at the Tip of the Type III Secretion Needle. Infect. Immun. 2007, 75, 2626–2629. [Google Scholar] [CrossRef] [Green Version]
- Dickenson, N.E.; Choudhari, S.P.; Adam, P.R.; Kramer, R.M.; Joshi, S.B.; Middaugh, C.R.; Picking, W.L.; Picking, W.D. Oligomeric States of the Shigella Translocator Protein IpaB Provide Structural Insights into Formation of the Type III Secretion Translocon. Protein Sci. 2013, 22, 614–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dickenson, N.E.; Arizmendi, O.; Patil, M.K.; Toth, R.T.; Middaugh, C.R.; Picking, W.D.; Picking, W.L. N-Terminus of IpaB Provides a Potential Anchor to the Shigella Type III Secretion System Tip Complex Protein IpaD. Biochemistry 2013, 52, 8790–8799. [Google Scholar] [CrossRef] [Green Version]
- Sakai, T.; Sasakawa, C.; Yoshikawa, M. Expression of Four Virulence Antigens of Shigella Flexneri Is Positively Regulated at the Transcriptional Level by the 30 Kilo Dalton VirF Protein. Mol. Microbiol. 1988, 2, 589–597. [Google Scholar] [CrossRef]
- Tobe, T.; Nagai, S.; Okada, N.; Adter, B.; Yoshikawa, M.; Sasakawa, C. Temperature-Regulated Expression of Invasion Genes in Shigella Flexneriis Controlled through the Transcriptional Activation of the VirB Gene on the Large Plasmid. Mol. Microbiol. 1991, 5, 887–893. [Google Scholar] [CrossRef] [PubMed]
- Tobe, T.; Yoshikawa, M.; Mizuno, T.; Sasakawa, C. Transcriptional Control of the Invasion Regulatory Gene VirB of Shigella Flexneri: Activation by VirF and Repression by H-NS. J. Bacteriol. 1993, 175, 6142–6149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Goot, G.; van Nhieu, G.T.; Allaoui, A.; Sansonetti, P.; Lafont, F. Rafts Can Trigger Contact-mediated Secretion of Bacterial Effectors via a Lipid-Based Mechanism. J. Biol. Chem. 2004, 279, 47792–47798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Standish, A.J.; Teh, M.Y.; Tran, E.N.H.; Doyle, M.; Baker, P.J.; Morona, R. Unprecedented Abundance of Protein Tyrosine Phosphorylation Modulates Shigella Flexneri Virulence. J. Mol. Biol. 2016, 428, 4197–4208. [Google Scholar] [CrossRef]
- Burgess, J.L.; Burgess, R.A.; Morales, Y.; Bouvang, J.M.; Johnson, S.; Dickenson, N.E. Structural and Biochemical Characterization of Spa47 Provides Mechanistic Insight into Type III Secretion System ATPase Activation and Shigella Virulence Regulation. J. Biol. Chem. 2016, 291, 25837–25852. [Google Scholar] [CrossRef] [Green Version]
- Burgess, J.L.; Jones, H.B.; Kumar, P.; Iv, R.T.T.; Middaugh, C.R.; Antony, E.; Dickenson, N.E. Spa47 Is an Oligomerization-Activated Type Three Secretion System (T3SS) ATPase from Shigella Flexneri. Protein Sci. 2016, 25, 1037–1048. [Google Scholar] [CrossRef]
- Demler, H.J.; Case, H.B.; Morales, Y.; Bernard, A.R.; Johnson, S.J.; Dickenson, N.E. Interfacial Amino Acids Support Spa47 Oligomerization and Shigella Type Three Secretion System Activation. Proteins Struct. Funct. Bioinform. 2019, 87, 931–942. [Google Scholar] [CrossRef]
- Dissmeyer, N.; Schnittger, A. Use of Phospho-Site Substitutions to Analyze the Biological Relevance of Phosphorylation Events in Regulatory Networks. Methods Mol. Biol. 2011, 779, 93–138. [Google Scholar] [CrossRef]
- Stateva, S.R.; Salas, V.; Benaim, G.; Menéndez, M.; Solís, D.; Villalobo, A. Characterization of Phospho-(Tyrosine)-Mimetic Calmodulin Mutants. PLoS ONE 2015, 10, e0120798. [Google Scholar] [CrossRef] [Green Version]
- Burgess, J.L.; Case, H.B.; Burgess, R.A.; Dickenson, N.E. Dominant Negative Effects by Inactive Spa47 Mutants Inhibit T3SS Function and Shigella Virulence. PLoS ONE 2020, 15, e0228227. [Google Scholar] [CrossRef]
- Mounier, J.; Popoff, M.R.; Enninga, J.; Frame, M.C.; Sansonetti, P.J.; Van Nhieu, G.T. The IpaC Carboxyterminal Effector Domain Mediates Src-Dependent Actin Polymerization during Shigella Invasion of Epithelial Cells. PLoS Pathog. 2009, 5, e1000271. [Google Scholar] [CrossRef] [Green Version]
- Harrington, A.T.; Hearn, P.D.; Picking, W.L.; Barker, J.R.; Wessel, A.; Picking, W.D. Structural Characterization of the N Terminus of IpaC from Shigella Flexneri. Infect. Immun. 2003, 71, 1255–1264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Gijsegem, F.; Genin, S.; Boucher, C. Conservation of Secretion Pathways for Pathogenicity Determinants of Plant and Animal Bacteria. Trends Microbiol. 1993, 1, 175–180. [Google Scholar] [CrossRef]
- Salmond, G.P.; Reeves, P.J. Membrance Traffic Wardens and Protein Secretion in Gram-Negative Bacteria. Trends Biochem. Sci. 1993, 18, 7–12. [Google Scholar] [CrossRef]
- Parsot, C.; Ménard, R.; Gounon, P.; Sansonetti, P.J. Enhanced Secretion through the Shigella Flexneri Mxi-Spa Rranslocon Leads to Assembly of Extracellular Proteins into Macromolecular Structures. Mol. Microbiol. 1995, 16, 291–300. [Google Scholar] [CrossRef]
- Epler, C.R.; Dickenson, N.E.; Bullitt, E.; Picking, W.L. Ultrastructural Analysis of IpaD at the Tip of the Nascent MxiH Type III Secretion Apparatus of Shigella Flexneri. J. Mol. Biol. 2012, 420, 29–39. [Google Scholar] [CrossRef] [Green Version]
- Dickenson, N.E.; Zhang, L.; Epler, C.R.; Adam, P.R.; Picking, W.L.; Picking, W.D. Conformational Changes in IpaD from Shigella Flexneri upon Binding Bile Salts Provide Insight into the Second Step of Type III Secretion. Biochemistry 2010, 50, 172–180. [Google Scholar] [CrossRef] [Green Version]
- Adam, P.R.; Dickenson, N.E.; Greenwood, J.C.; Picking, W.L.; Picking, W.D. Influence of Oligomerization State on the Structural Properties of Invasion Plasmid Antigen B from Shigella Flexneri in the Presence and Absence of Phospholipid Membranes. Proteins Struct. Funct. Bioinform. 2014, 82, 3013–3022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adam, P.R.; Patil, M.K.; Dickenson, N.E.; Choudhari, S.; Barta, M.; Geisbrecht, B.V.; Picking, W.L.; Picking, W.D. Binding Affects the Tertiary and Quaternary Structures of the Shigella Translocator Protein IpaB and Its Chaperone IpgC. Biochemistry 2012, 51, 4062–4071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Case, H.B.; Mattock, D.S.; Dickenson, N.E. Shutting Down Shigella Secretion: Characterizing Small Molecule Type Three Secretion System ATPase Inhibitors. Biochemistry 2018, 57, 6906–6916. [Google Scholar] [CrossRef] [PubMed]
- Case, H.B.; Mattock, D.; Miller, B.R.; Dickenson, N.E. Novel Noncompetitive Type Three Secretion System ATPase Inhibitors Shut Down Shigella Effector Secretion. Biochemistry 2020, 59, 2667–2678. [Google Scholar] [CrossRef] [PubMed]
- Jouihri, N.; Sory, M.-P.; Page, A.-L.; Gounon, P.; Parsot, C.; Allaoui, A. MxiK and MxiN Interact with the Spa47 ATPase and Are Required for Transit of the Needle Components MxiH and MxiI, but not of Ipa Proteins, through the Type III Secretion Apparatus of Shigella Flexneri. Mol. Microbiol. 2004, 49, 755–767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Case, H.B.; Dickenson, N.E. Kinetic Characterization of the Shigella Type Three Secretion System ATPase Spa47 Using α-32P ATP. Bio-Protoc. 2018, 8, e3074. [Google Scholar] [CrossRef] [PubMed]
- Sansonetti, P.J.; Ryter, A.; Clerc, P.; Maurelli, A.T.; Mounier, J. Multiplication of Shigella Flexneri within HeLa Cells: Lysis of the Phagocytic Vacuole and Plasmid-Mediated Contact Hemolysis. Infect. Immun. 1986, 51, 461–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niesel, D.W.; Chambers, C.E.; Stockman, S.L. Quantitation of HeLa Cell Monolayer Invasion by Shigella and Salmonella Species. J. Clin. Microbiol. 1985, 22, 897–902. [Google Scholar] [CrossRef] [Green Version]
- Bahrani, F.K.; Sansonetti, P.J.; Parsot, C. Secretion of Ipa Proteins by Shigella Flexneri: Inducer Molecules and Kinetics of Activation. Infect. Immun. 1997, 65, 4005–4010. [Google Scholar] [CrossRef] [Green Version]
- Kenjale, R.; Wilson, J.; Zenk, S.F.; Saurya, S.; Picking, W.L.; Picking, W.D.; Blocker, A. The Needle Component of the Type III Secreton of Shigella Regulates the Activity of the Secretion Apparatus. J. Biol. Chem. 2005, 280, 42929–42937. [Google Scholar] [CrossRef] [Green Version]

| Spa47 | Melt Temperature (Tm) a (°C ± S.D.) | |
|---|---|---|
| Monomer | Oligomer | |
| Wild-Type Spa47 | 44.4 ± 0.2 | 43.5 ± 1.5 |
| Spa47Y274F | 42.1 ± 0.5 * | 41.6 ± 0.1 |
| Spa47Y274A | 39.5 ± 0.4 * | 41.8 ± 1.4 |
| Spa47Y274E | 40.1 ± 0.2 * | 45.1 ± 0.5 |
| Spa47Y398F | 43.9 ± 0.5 * | 44.1 ± 1.7 |
| Spa47Y398A | 41.3 ± 0.2 * | 40.5 ± 1.9 * |
| Spa47Y398E | 36.0 ± 0.6 * | N/A |
| Spa47 | Relative Phenotype (% ± S.D.) | |
|---|---|---|
| Hemolysis a | Invasion b | |
| Spa47 null | 3 ± 0 * | 1 ± 1 * |
| Vector Control | 3 ± 0 * | 0 ± 0 * |
| Wild-Type Spa47 | 100 ± 0 | 100 ± 0 |
| Spa47Y274F | 96 ± 7 | 99 ± 13 |
| Spa47Y274A | 78 ± 8 * | 51 ± 9 * |
| Spa47Y274E | 46 ± 3 * | 32 ± 11 * |
| Spa47Y398F | 119 ± 3 * | 90 ± 10 |
| Spa47Y398A | 132 ± 6 * | 110 ± 20 |
| Spa47Y398E | 117 ± 8 * | 119 ± 16 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hardy, K.D.; Dickenson, N.E. Phosphomimetic Tyrosine Mutations in Spa47 Inhibit Type Three Secretion ATPase Activity and Shigella Virulence Phenotype. Pathogens 2022, 11, 202. https://doi.org/10.3390/pathogens11020202
Hardy KD, Dickenson NE. Phosphomimetic Tyrosine Mutations in Spa47 Inhibit Type Three Secretion ATPase Activity and Shigella Virulence Phenotype. Pathogens. 2022; 11(2):202. https://doi.org/10.3390/pathogens11020202
Chicago/Turabian StyleHardy, Koleton D., and Nicholas E. Dickenson. 2022. "Phosphomimetic Tyrosine Mutations in Spa47 Inhibit Type Three Secretion ATPase Activity and Shigella Virulence Phenotype" Pathogens 11, no. 2: 202. https://doi.org/10.3390/pathogens11020202
APA StyleHardy, K. D., & Dickenson, N. E. (2022). Phosphomimetic Tyrosine Mutations in Spa47 Inhibit Type Three Secretion ATPase Activity and Shigella Virulence Phenotype. Pathogens, 11(2), 202. https://doi.org/10.3390/pathogens11020202