Potential Epha2 Receptor Blockers Involved in Cerebral Malaria from Taraxacum officinale, Tinospora cordifolia, Rosmarinus officinalis and Ocimum basilicum: A Computational Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Molecular Docking
2.2. Ligand Preparation
2.3. Target Preparation
3. Results and Discussion
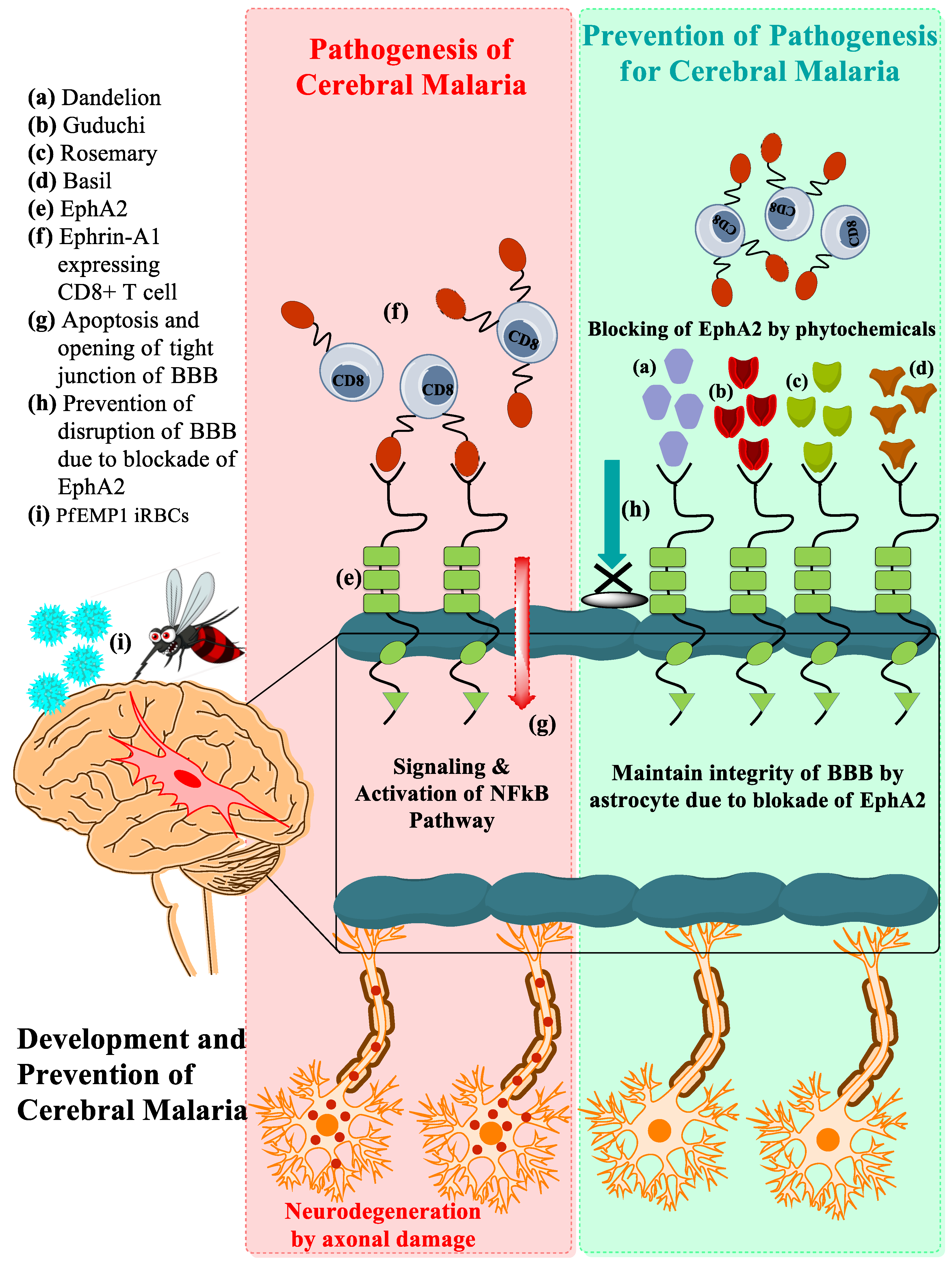
3.1. Development of CM Associated with EphA2
3.2. Prediction of ADME Parameters
3.3. In Silico Toxicity Prediction Study Employing ProTox-II Toxicity Explorer
3.4. Docking Studies Using PyRx
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schiess, N.; Villabona-Rueda, A.; Cottier, K.E.; Huether, K.; Chipeta, J.; Stins, M.F. Pathophysiology and neurologic sequelae of cerebral malaria. Malar. J. 2020, 19, 266. [Google Scholar] [CrossRef] [PubMed]
- Idro, R.; Marsh, K.; John, C.C.; Newton, C.R.J. Cerebral malaria: Mechanisms of brain injury and strategies for improved neurocognitive outcome. Pediatr. Res. 2010, 68, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Mandala, W.L.; Harawa, V.; Dzinjalamala, F.; Tembo, D. The role of different components of the immune system against Plasmodium falciparum malaria: Possible contribution towards malaria vaccine development. Mol. Biochem. Parasitol. 2021, 246, 111425. [Google Scholar] [CrossRef] [PubMed]
- Herbas, M.S.; Okazaki, M.; Terao, E.; Xuan, X.; Arai, H.; Suzuki, H. Alpha-Tocopherol transfer protein inhibition is effective in the prevention of cerebral malaria in mice. Am. J. Clin. Nutr. J. Clin. Nutr. 2010, 91, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Rénia, L.; Howland, S.W.; Claser, C.; Gruner, A.C.; Suwanarusk, R.; Teo, T.H.; Russell, B.; Lisa, N.P. Cerebral malaria Mysteries at the blood-brain barrier. Virulence 2012, 3, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Nishanth, G.; Schlüter, D. Blood–Brain Barrier in Cerebral Malaria: Pathogenesis and Therapeutic Intervention. Trends Parasitol. 2019, 35, 516–528. [Google Scholar] [CrossRef]
- Gallego-Delgado, J.; Rodriguez, A. Rupture and Release: A Role for Soluble Erythrocyte Content in the Pathology of Cerebral Malaria. Trends Parasitol. 2017, 33, 832–835. [Google Scholar] [CrossRef]
- Sorensen, E.W.; Lian, J.; Ozga, A.J.; Miyabe, Y.; Ji, S.W.; Bromley, S.K.; Mempel, T.R.; Luster, A.D. CXCL10 stabilizes T cell-brain endothelial cell adhesion leading to the induction of cerebral malaria. JCI Insight 2018, 3, e98911. [Google Scholar] [CrossRef]
- Vanka, R.; Nakka, V.P.; Kumar, S.P.; Baruah, U.K.; Babu, P.P. Molecular targets in cerebral malaria for developing novel therapeutic strategies. Brain Res. Bull. 2020, 157, 100–107. [Google Scholar] [CrossRef]
- Pino, P.; Taoufiq, Z.; Nitcheu, J.; Vouldoukis, I.; Mazier, D. Blood-brain barrier breakdown during cerebral malaria: Suicide or murder? Thromb. Haemost. 2005, 94, 336–340. [Google Scholar] [CrossRef]
- Combes, V.; Coltel, N.; Faille, D.; Wassmer, S.C.; Grau, G.E. Cerebral malaria: Role of microparticles and platelets in alterations of the blood-brain barrier. Int. J. Parasitol. 2006, 36, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Hunt, N.H.; Golenser, J.; Chan-Ling, T.; Parekh, S.; Rae, C.; Potter, S.; Medana, I.M.; Miu, J.; Ball, H.J. Immunopathogenesis of cerebral malaria. Int. J. Parasitol. 2006, 36, 569–582. [Google Scholar] [CrossRef] [PubMed]
- Darling, T.K.; Mimche, P.N.; Bray, C.; Umaru, B.; Brady, L.M.; Stone, C.; Moukoko, C.E.E.; Lane, T.E.; Ayong, L.S.; Lamb, T.J. EphA2 contributes to disruption of the blood-brain barrier in cerebral malaria. PLoS Pathog. 2020, 16, e1008261. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, W.; Song, J.; Zhang, M.; Huang, T.; Wei, X. High expression of EphA2 led to secondary injury by destruction of BBB integrity though the ROCK pathway after diffuse axonal injury. Neurosci. Lett. 2020, 736, 135234. [Google Scholar] [CrossRef]
- Emran, T.B.; Iyori, M.; Ono, Y.; Amelia, F.; Yusuf, Y.; Islam, A.; Alam, A.; Tamura, M.; Ogawa, R.; Matsuoka, H.; et al. Baculovirus-induced fast-acting innate immunity kills liver-stage Plasmodium. J. Immun. 2018, 201, 2441–2451. [Google Scholar] [CrossRef]
- Shaikh, M.S.; Kale, M.A.; Sharuk, K. Discovery of Heterocyclic Analogs of Diaminopimelic Acid as Promising Antibacterial Agents Through Enzyme Targeted Inhibition of Lysine Bio-synthesis. Res. Artic. 2018, 14, 120–130. [Google Scholar] [CrossRef]
- Khan, S.L.; Siddiqui, F.A.; Shaikh, M.S.; Nema, N.V.; Shaikh, A.A. Discovery of potential inhibitors of the receptor-binding domain (RBD) of pandemic disease-causing SARS-CoV-2 Spike Glycoprotein from Triphala through molecular docking. Curr. Chin. Chem. 2021, 2, e220321192390. [Google Scholar] [CrossRef]
- Khan, A.; Unnisa, A.; Sohel, M.; Date, M.; Panpaliya, N.; Saboo, S.G.; Siddiqui, F.; Khan, S. Investigation of phytoconstituents of Enicostemma littorale as potential glucokinase activators through molecular docking for the treatment of type 2 diabetes mellitus. Silico Pharmacol. 2022, 10, 1. [Google Scholar] [CrossRef]
- Aita, S.; Badavath, V.N.; Gundluru, M.; Sudileti, M.; Nemallapudi, B.R.; Gundala, S.; Zyryanov, G.V.; Chamarti, N.R.; Cirandur, S.R. Novel α-Aminophosphonates of imatinib Intermediate: Synthesis, anticancer Activity, human Abl tyrosine kinase Inhibition, ADME and toxicity prediction. Bioorg. Chem. 2021, 109, 104718. [Google Scholar] [CrossRef]
- Darling, T.K.; Lamb, T.J. Emerging roles for Eph receptors and ephrin ligands in immunity. Front. Immunol. 2019, 10, 1473. [Google Scholar] [CrossRef]
- Lis, B.; Olas, B. Pro-health activity of dandelion (Taraxacum officinale L.) and its food products—History and present. J. Funct. Foods 2019, 59, 40–48. [Google Scholar] [CrossRef]
- You, Y.; Yoo, S.; Yoon, H.G.; Park, J.; Lee, Y.H.; Kim, S.; Oh, K.T.; Lee, J.; Cho, H.Y.; Jun, W. In vitro and in vivo hepatoprotective effects of the aqueous extract from Taraxacum officinale (dandelion) root against alcohol-induced oxidative stress. Food Chem. Toxicol. 2010, 48, 1632–1637. [Google Scholar] [CrossRef] [PubMed]
- Saeed, M.; Naveed, M.; Leskovec, J.; Ali kamboh, A.; Kakar, I.; Ullah, K.; Ahmad, F.; Sharif, M.; Javaid, A.; Rauf, M.; et al. Using Guduchi (Tinospora cordifolia) as an eco-friendly feed supplement in human and poultry nutrition. Poult. Sci. 2020, 99, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, P.; Nayak, P.; Prusty, S.K.; Sahu, P.K. Phytochemistry and pharmacology of tinospora cordifolia: A review. Syst. Rev. Pharm. 2018, 9, 70–78. [Google Scholar] [CrossRef]
- Singh, H.; Sharma, A.K.; Gupta, M.; Singh, A.P.; Kaur, G. Tinospora cordifolia attenuates high fat diet-induced obesity and associated hepatic and renal dysfunctions in rats. PharmaNutrition 2020, 13, 100189. [Google Scholar] [CrossRef]
- De Oliveira, J.R.; Esteves, S.; Camargo, A. Rosmarinus officinalis L. ( rosemary ) as therapeutic and prophylactic agent. J. Biomed. Sci. 2019, 8, 5. [Google Scholar] [CrossRef]
- Andrade, J.M.; Faustino, C.; García, C.; Ladeiras, D.; Reis, C.P.; Rijo, P. Rosmarinus officinalis L.: An update review of its phytochemistry and biological activity. Futur. Sci. 2018, 4, FSO283. [Google Scholar] [CrossRef]
- Akshay, K.; Swathi, K.; Bakshi, V.; Boggula, N. Journal of Drug Delivery and Therapeutics Rosmarinus officinalis L.: An update review of its phytochemistry and biological activity. J. Drug Deliv. Ther. 2019, 9, 323–330. [Google Scholar]
- Jafir, M.; Ahmad, J.N.; Arif, M.J.; Ali, S.; Ahmad, S.J.N. Characterization of Ocimum basilicum synthesized silver nanoparticles and its relative toxicity to some insecticides against tobacco cutworm, Spodoptera litura Feb. (Lepidoptera; Noctuidae). Ecotoxicol. Environ. Saf. 2021, 218, 112278. [Google Scholar] [CrossRef]
- Mohammed, A.B.A.; Yagi, S.; Tzanova, T.; Schohn, H.; Abdelgadir, H.; Stefanucci, A.; Mollica, A.; Mahomoodally, M.F.; Adlan, T.A.; Zengin, G. Chemical profile, antiproliferative, antioxidant and enzyme inhibition activities of Ocimum basilicum L. and Pulicaria undulata (L.) C.A. Mey. grown in Sudan. S. Afr. J. Bot. 2020, 132, 403–409. [Google Scholar] [CrossRef]


| Sl. No. | Parameter and Compound Name | GI * Absorption | BBB * Permeability | P. gp* Substrate | CYP1A2 Inhibitor | CYP219 Inhibitor | CYP2C9 Inhibitor | CYP2D6 Inhibitor | CYP3A4 Inhibitor | Log Kp(Skin Permeation | Ghose | Egan | Muegge | Bioavailability |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Linalool | High | Yes | No | No | No | No | No | No | −5.13 cm/gm | 01 MW < 160 | Yes | 2 MW < 2000, heteroatoms < 2 | 0.55 |
| 2 | Methyl Eugenol | High | Yes | No | Yes | No | No | No | No | −5.60 cm/s | Yes | Yes | No | 0.55 |
| 3 | Methyl Chavicool (Basil) | High | Yes | No | Yes | Yes | No | No | No | −5.34 cm/s | Yes | Yes | Yes | 0.55 |
| 4 | Palmatine | High | Yes | Yes | Yes | No | No | Yes | Yes | −5.79 cm/s | Yes | Yes | yes | 0.55 |
| 5 | Magnoflorine | High | Yes | Yes | No | No | No | Yes | Yes | −6.44 cm/s | Yes | Yes | yes | 0.55 |
| 6 | Isocolumbin | High | No | Yes | No | No | No | No | No | −6.95 cm/s | Yes | Yes | Yes | 0.55 |
| 7 | Carnosol | High | Yes | Yes | No | No | Yes | No | Yes | −5.01 cm/s | Yes | Yes | Yes | 0.55 |
| 8 | Rosmarinic Acid | Low | No | No | No | No | No | No | No | −6.82 cm/s | Yes | 1 TPSA > 131.6 | Yes | 0.58 |
| 9 | Carnosic Acid | High | No | No | No | No | Yes | No | No | −4.86 cm/s | Yes | Yes | Yes | 0.56 |
| 10 | Taraxasterol | Low | No | NO | NO | No | No | No | No | −2.42 cm/s | 3 WLOGP > 5.6, MR > 130, atoms > 70 | 1 WLOGP > 5.88 | 2 XLOGP3 > 5, heteroatoms < 2 | 0.55 |
| 11 | ||||||||||||||
| Luteolin (Dandelion) | High | No | No | Yes | No | No | Yes | Yes | −6.25 cm/s | Yes | Yes | Yes | 0.55 | |
| 12 | Taraxinic Acid (Dandelion) | High | yes | NO | NO | No | NO | NO | No | −6.89 cm/s | Yes | Yes | Yes | 0.85 |
| Sr. No. | Compounds | Molecular Formula | Lipinski Rule of 5 | Veber’s Rule | |||||
|---|---|---|---|---|---|---|---|---|---|
| Molecular Weight | HBA * | HBD * | Log P | Violation | Total Polar Surface Area (TPSA) (A2) | Number of Rotatable Bonds | |||
| 1 | Linalool (Basil) | C10H18O | 154.25 | 1 | 1 | 2.66 | 00 | 20.23 | 4 |
| 2 | Methyl Eugenol (Basil) | C11H14O2 | 178.23 | 2 | 0 | 2.58 | 00 | 18.48 | 4 |
| 3 | Methyl Chavicool (Basil) | C11H12O3 | 192.21 | 3 | 0 | 2.30 | 00 | 35.53 | 4 |
| 4 | Palmatine (Guduchi) | C21H22NO4+ | 352.40 | 4 | 0 | 2.53 | 00 | 41.85 | 4 |
| 5 | Magnoflorine (Guduchi) | C20H24NO4+ | 342.41 | 4 | 2 | 1.88 | 00 | 62.16 | 2 |
| 6 | Isocolumbin (Guduchi) | C20H22O6 | 358.4 | 6 | 1 | 2.13 | 00 | 85.97 | 1 |
| 7 | Carnosol (Rosemary) | C21H28O4 | 344.44 | 4 | 2 | 4.03 | 00 | 66.76 | 1 |
| 8 | Rosmarinic Acid (Rosemary) | C18H16O8 | 360.31 | 8 | 5 | 1.52 | 00 | 144.52 | 7 |
| 9 | Carnosic Acid (Rosemary) | C20H28O4 | 332.43 | 4 | 3 | 3.82 | 00 | 77.76 | 2 |
| 10 | Taraxasterol (dandelion) | C30H50O | 426.72 | 1 | 1 | 7.11 | 01, MLOGP > 4.15 | 20.23 | 0 |
| 11 | Luteolin (Dandelion) | C15H10O6 | 286.24 | 6 | 4 | 1.73 | 00 | 111.13 | 1 |
| 12 | Taraxinic Acid (Dandelion) | C15H18O4 | 262.30 | 4 | 1 | 2.12 | 00 | 63.60 | 1 |
| Sl. No | Compound | Predicted LD50 Value (mg/Kg)/Toxicity Class | Hepatotoxicity | Carcinogenicity | Immunotoxicity | Cytotoxicity | Nuclear Receptor Signaling Pathways Active | ||
|---|---|---|---|---|---|---|---|---|---|
| Androgen Receptor (AR) | Aromatase Active | Estrogen Receptor Alpha (ER) | |||||||
| 1 | Linalool (Basil) | 2200/5 | None | None | None | None | None | None | None |
| 2 | Methyl Eugenol (Basil) | 810/4 | None | Yes | None | None | None | None | None |
| 3 | Methyl Chavicool (Basil) | 7900/6 | None | None | Yes | None | None | None | None |
| 4 | Palmatine (Guduchi) | 200/3 | None | None | Yes | None | None | None | None |
| 5 | Magnoflorine (Guduchi) | 401/4 | None | None | Yes | None | None | None | None |
| 6 | Isocolumbin (Guduchi) | 280/3 | None | None | None | None | None | None | None |
| 7 | Carnosol (Rosemary) | 287/3 | None | None | Yes | None | None | None | None |
| 8 | Rosmarinic Acid (Rosemary) | 5000/5 | None | None | Yes | None | None | None | None |
| 9 | Carnosic Acid (Rosemary) | 287/3 | None | None | None | None | None | None | None |
| 10 | Taraxasterol (Dandelion) | 5000/5 | None | None | Yes | None | None | None | None |
| 11 | Luteolin (Dandelion) | 3919/5 | None | None | None | None | None | None | Yes |
| 12 | Taraxinic Acid (Dandelion) | 900/4 | None | None | Yes | None | None | None | None |
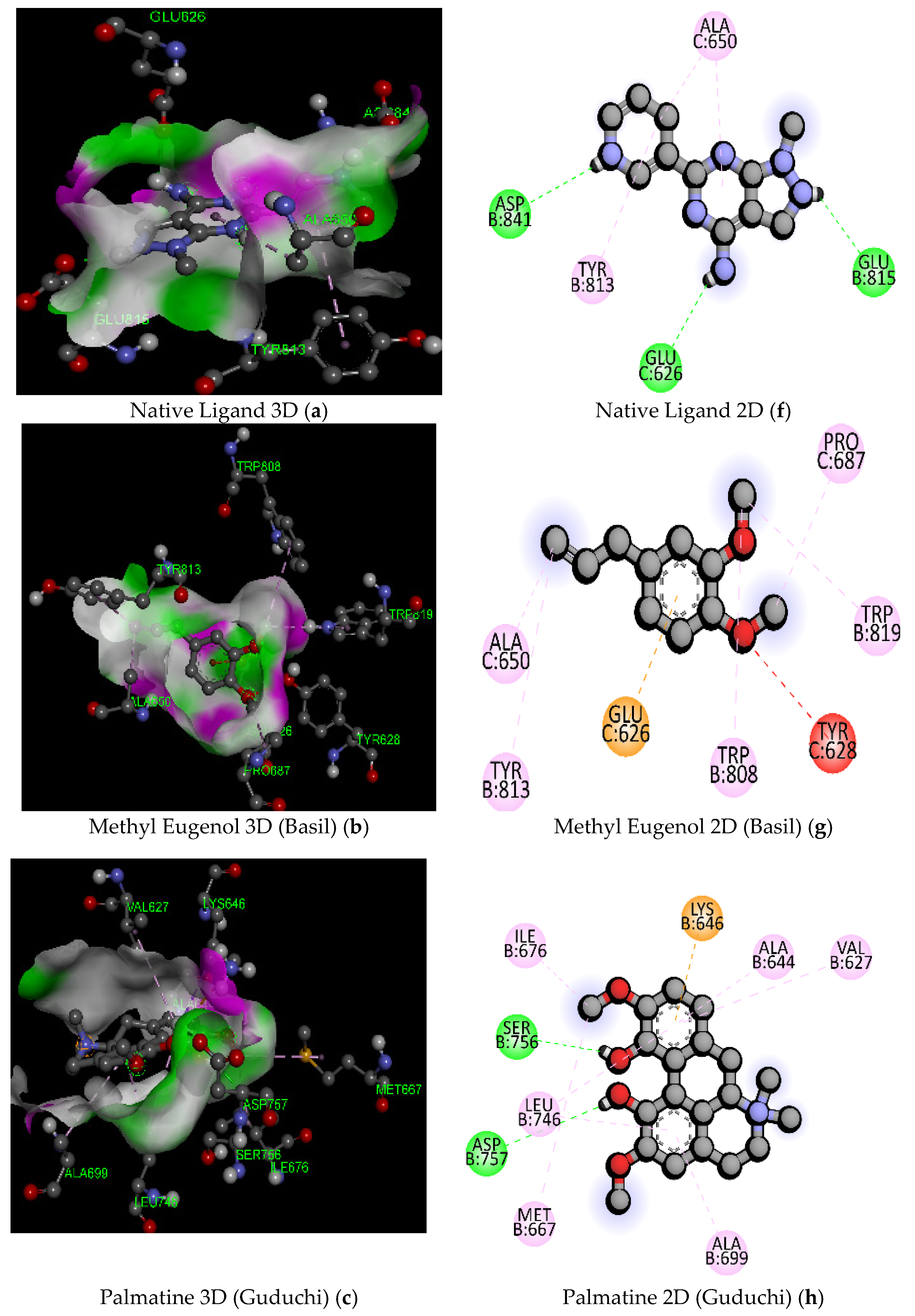
| Sl. No. | Compound Name | Molecular Formula/Molecular Weight (gm/mol) | Docking Score/Binding Affinity (kcal/mol) | Active Amino Acid Residue | Bond Length (A°) | Bond Category | Bond Types |
|---|---|---|---|---|---|---|---|
| 1 | Native Ligand (DXK) (PubChem ID: 134693866) | C11 H10 N6/226.24 | −7.3 | GLU815 | 2.17 | Hydrogen Bond | Conventional Hydrogen Bond |
| ASP841 | 2.65 | ||||||
| GLU626 | 2.28 | ||||||
| ALA650 | 3.94 | Hydrophobic | Alkyl | ||||
| ALA650 | 4.69 | Pi-Alkyl | |||||
| TYR813 | 5.27 | ||||||
| 2 | Linalool (Basil) | C10H11O/154.25 | −4.9 | VAL627 | 5.03 | Hydrophobic | Alkyl |
| LEU746 | 4.81 | ||||||
| MET695 | 5.25 | ||||||
| LEU746 | 4.14 | ||||||
| ALA644 | 3.99 | ||||||
| 3 | Methyl Eugenol (Basil) | C11H14O2/178.23 | −5.6 | GLU626 | 3.40 | Electrostatic | Pi-Anion |
| ALA650 | 3.86 | Hydrophobic | Alkyl | ||||
| PRO687 | 4.01 | Hydrophobic | Alkyl | ||||
| TRP808 | 4.62 | Hydrophobic | Pi-Alkyl | ||||
| TYR813 | 4.95 | Hydrophobic | Pi-Alkyl | ||||
| TRP819 | 4.77 | Hydrophobic | Pi-Alkyl | ||||
| 4 | Methyl Chavicool (Basil) | C10H12O/148.20 | −5.4 | GLU815 | 3.77 | Hydrogen Bond | Carbon Hydrogen Bond |
| GLU626 | 3.53 | Electrostatic | Pi-Anion | ||||
| ALA650 | 4.08 | Hydrophobic | Alkyl | ||||
| PRO687 | 4.10 | Hydrophobic | Alkyl | ||||
| MET840 | 5.46 | Hydrophobic | Pi-Alkyl | ||||
| TYR813 | 5.04 | Hydrophobic | Pi-Alkyl | ||||
| TYR628 | 5.41 | Hydrophobic | Pi-Alkyl | ||||
| 5 | Palmatine (Guduchi) | C21H22NO4+/352.4 | −8.4 | GLN855 | 2.43 | Hydrogen Bond | Conventional Hydrogen Bond |
| MET733 | 5.02 | Hydrophobic | Alkyl | ||||
| MET851 | 4.26 | Hydrophobic | Alkyl | ||||
| MET851 | 5.21 | Hydrophobic | Pi-Alkyl | ||||
| PHE604 | 4.57 | Hydrophobic | Pi-Alkyl | ||||
| 6 | Magnoflorine (Guduchi) | C20H24NO4+/342.4 | −7.9 | ASP757 | 2.75 | Hydrogen Bond | Conventional Hydrogen Bond |
| SER756 | 2.00 | Hydrogen Bond | Conventional Hydrogen Bond | ||||
| LYS646 | 4.63 | Electrostatic | Pi-Cation | ||||
| LYS646 | 3.90 | Hydrophobic | Pi-Sigma | ||||
| MET667 | 4.74 | Hydrophobic | Alkyl | ||||
| ILE676 | 3.64 | Hydrophobic | Alkyl | ||||
| ALA699 | 5.16 | Hydrophobic | Pi-Alkyl | ||||
| LEU746 | 4.89 | Hydrophobic | Pi-Alkyl | ||||
| VAL627 | 5.47 | Hydrophobic | Pi-Alkyl | ||||
| ALA644 | 5.08 | Hydrophobic | Pi-Alkyl | ||||
| LEU746 | 5.18 | Hydrophobic | Pi-Alkyl | ||||
| 7 | Isocolumbin (Guduchi) | C20H22O6/358.4 | −9.3 | GLN848 | 2.55 | Hydrogen Bond | Conventional Hydrogen Bond |
| PHE604 | 3.73 | Hydrogen Bond | Carbon Hydrogen Bond | ||||
| ARG860 | 5.19 | Hydrophobic | Alkyl | ||||
| 8 | Carnosol (Rosemary) | C20H26O4/330.40 | −9.0 | GLN669 | 2.43 | Hydrogen Bond | Conventional Hydrogen Bond |
| GLN855 | 2.43 | Hydrogen Bond | Conventional Hydrogen Bond | ||||
| GLN852 | 3.03 | Hydrogen Bond | Conventional Hydrogen Bond | ||||
| GLN855 | 2.26 | Hydrogen Bond | Conventional Hydrogen Bond | ||||
| SER671 | 3.57 | Hydrogen Bond | Carbon Hydrogen Bond | ||||
| 9 | Rosmarinic Acid (Rosemary) | C18H16O8/360.3 | −7.6 | SER756 | 2.54 | Hydrogen Bond | Conventional Hydrogen Bond |
| TYR694 | 3.25 | Hydrogen Bond | Pi-Donor Hydrogen Bond | ||||
| LYS646 | 3.80 | Hydrophobic | Pi-Sigma | ||||
| N:UNK1 | 5.27 | Hydrophobic | Pi-Pi Stacked | ||||
| ALA644 | 5.30 | Hydrophobic | Pi-Alkyl | ||||
| LEU746 | 5.27 | Hydrophobic | Pi-Alkyl | ||||
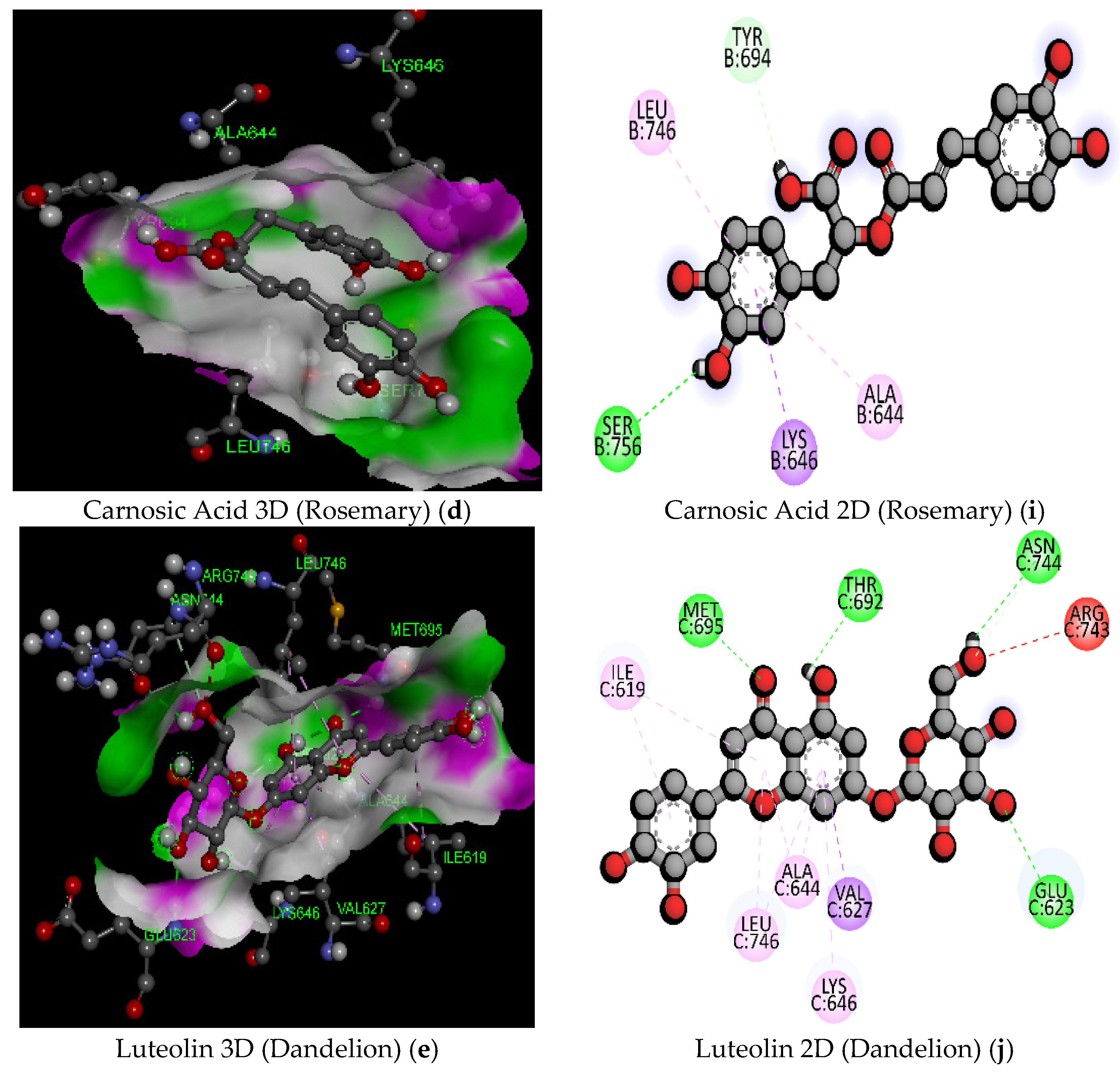
| 10 | Carnosic Acid (Rosemary) | C20H28O4/332.4 | −7.3 | THR605 | 2.20 | Hydrogen Bond | Conventional Hydrogen Bond |
| LYS603 | 2.35 | Hydrogen Bond | Conventional Hydrogen Bond | ||||
| GLN848 | 2.58 | Hydrogen Bond | Conventional Hydrogen Bond | ||||
| PHE604 | 3.06 | Hydrogen Bond | Carbon Hydrogen Bond | ||||
| ILE870 | 5.41 | Hydrophobic | Alkyl | ||||
| 11 | Taraxasterol (Dandelion) | C30H50O/426.7 | −8.9 | ARG743 | 3.66 | Hydrogen Bond | Carbon Hydrogen Bond |
| VAL627 | 4.89 | Hydrophobic | Alkyl | ||||
| ALA644 | 4.40 | Hydrophobic | Alkyl | ||||
| LEU746 | 4.68 | Hydrophobic | Alkyl | ||||
| 12 | Luteolin (Dandelion) | C21H20O11/448.4 | −9.5 | ASN744 | 2.67 | Hydrogen Bond | Conventional Hydrogen Bond |
| H-O | 2.26 | Hydrogen Bond | Conventional Hydrogen Bond | ||||
| THR692 | 2.54 | Hydrogen Bond | Conventional Hydrogen Bond | ||||
| GLU623 | 2.22 | Hydrogen Bond | Conventional Hydrogen Bond | ||||
| MET695 | 2.41 | Hydrogen Bond | Conventional Hydrogen Bond | ||||
| ASN744 | 3.54 | Hydrogen Bond | Carbon Hydrogen Bond | ||||
| VAL627 | 3.92 | Hydrophobic | Pi-Sigma | ||||
| ILE619 | 5.35 | Hydrophobic | Pi-Alkyl | ||||
| ALA644 | 4.43 | Hydrophobic | Pi-Alkyl | ||||
| LEU746 | 4.74 | Hydrophobic | Pi-Alkyl | ||||
| ALA644 | 4.46 | Hydrophobic | Pi-Alkyl | ||||
| LYS646 | 5.49 | Hydrophobic | Pi-Alkyl | ||||
| LEU746 | 5.14 | Hydrophobic | Pi-Alkyl | ||||
| ILE619 | 4.51 | Hydrophobic | Pi-Alkyl | ||||
| 13 | Taraxinic Acid (Dandelion) | C21H28O9/424.4 | −6.8 | LEU746 | 4.32 | Hydrophobic | Alkyl |
| Sl. No. | Compound Name | Molecular Formula/Molecular Weight (gm/mol) | Docking Score/Binding Affinity (kcal/mol) | Active Amino Acid Residue | Bond Length (A°) | Bond Category | Bond Types |
|---|---|---|---|---|---|---|---|
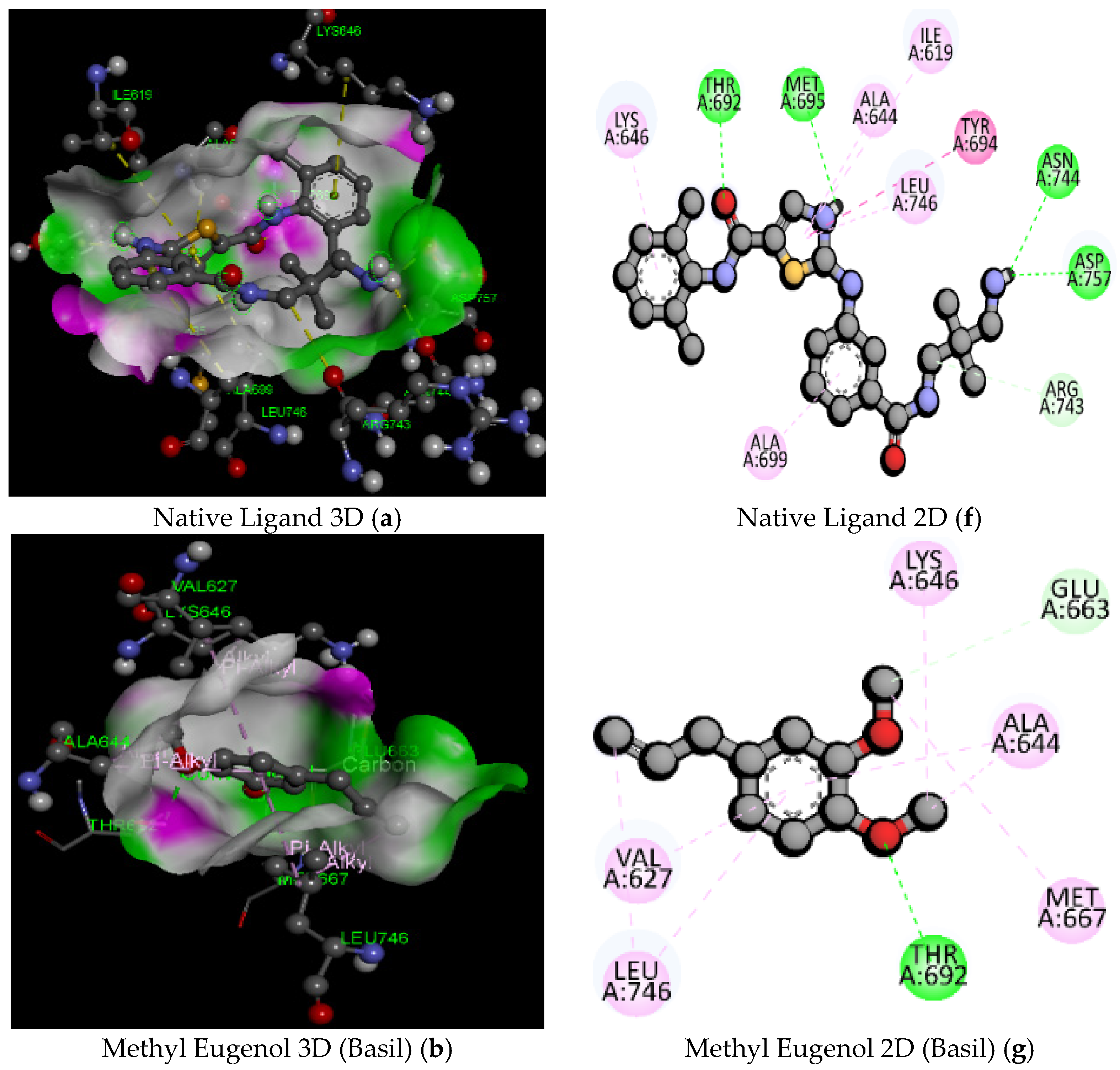
| 1 | Native Ligand (91E) (PubChem ID: 127053578) | C23H26ClN5O2S/472.00 | −7.6 | ASN744 | 2.83 | Hydrogen Bond | Conventional Hydrogen Bond |
| ASP757 | 2.15 | ||||||
| MET695 | 2.15 | ||||||
| THR692 | 2.00 | ||||||
| ARG743 | 3.38 | Carbon Hydrogen Bond | |||||
| TYR694 | 5.54 | Hydrophobic | Pi-Pi Stacked | ||||
| ALA699 | 5.03 | Pi-Alkyl | |||||
| ILE619 | 5.40 | ||||||
| ALA644 | 4.15 | ||||||
| LEU746 | 4.56 | ||||||
| LYS646 | 4.76 | ||||||
| 2 | Linalool (Basil) | C10H11O/154.25 | −5.1 | LEU746 | 4.78 | Hydrophobic | Alkyl |
| ILE619 | 4.53 | ||||||
| LEU746 | 5.25 | ||||||
| ALA644 | 4.34 | ||||||
| MET695 | 5.26 | ||||||
| LEU746 | 4.38 | ||||||
| ALA644 | 4.01 | ||||||
| LYS627 | 4.44 | ||||||
| VAL627 | 4.03 | ||||||
| ALA644 | 4.88 | ||||||
| TYR694 | 4.97 | Hydrophobic | Pi-Alkyl | ||||
| 3 | MethylEugenol (Basil) | C11H14O2/178.23 | −5.2 | THR692 | 2.44 | Hydrogen Bond | Conventional Hydrogen Bond |
| GLU663 | 3.73 | Carbon Hydrogen Bond | |||||
| LEU746 | 4.02 | Hydrophobic | Alkyl | ||||
| MET667 | 5.06 | ||||||
| ALA644 | 3.76 | ||||||
| LYS646 | 4.45 | ||||||
| VAL627 | 5.41 | Pi-Alkyl | |||||
| ALA644 | 5.07 | ||||||
| LEU746 | 5.22 | ||||||
| 4 | Methyl Chavicool (Basil) | C10H12O/148.20 | −5.1 | LYS646 | 4.49 | Hydrophobic | Alkyl |
| MET667 | 5.15 | ||||||
| ILE690 | 3.73 | ||||||
| VAL627 | 5.23 | ||||||
| LEU746 | 4.45 | ||||||
| VAL627 | 4.76 | Pi-Alkyl | |||||
| ALA644 | 4.61 | ||||||
| LYS646 | 5.06 | ||||||
| 5 | Palmatine (Guduchi) | C21H22NO4+/352.4 | −7.9 | THR692 | 2.61 | Hydrogen Bond | Conventional Hydrogen Bond |
| ALA699 | 2.27 | ||||||
| ILE690 | 3.70 | Carbon Hydrogen Bond | |||||
| LEU746 | 3.57 | Hydrophobic | Pi-Sigma | ||||
| ALA699 | 3.77 | Alkyl | |||||
| ALA644 | 4.02 | ||||||
| LYS646 | 4.30 | ||||||
| LYS646 | 4.57 | ||||||
| MET667 | 5.02 | ||||||
| ILE690 | 3.77 | ||||||
| ALA699 | 4.66 | Pi-Alkyl | |||||
| 6 | Magnoflorine (Guduchi) | C20H24NO4+/342.4 | −7.8 | H-O | 1.90 | Hydrogen Bond | Conventional Hydrogen Bond |
| ARG743 | 3.76 | Carbon Hydrogen Bond | |||||
| ASP757 | 3.68 | ||||||
| TYR694 | 3.71 | ||||||
| GLY698 | 3.36 | ||||||
| ILE619 | 3.97 | Hydrophobic | Pi-Sigma | ||||
| ILE619 | 3.98 | Alkyl | |||||
| VAL627 | 5.17 | Pi-Alkyl | |||||
| LEU746 | 5.28 | ||||||
| ALA644 | 4.63 | ||||||
| LEU746 | 4.58 | ||||||
| TYR694 | 4.89 | ||||||
| 7 | Isocolumbin (Guduchi) | C20H22O6/358.4 | −8.2 | GLU663 | 2.52 | Hydrogen Bond | Conventional Hydrogen Bond |
| LYS646 | 2.55 | ||||||
| ILE619 | 3.61 | Hydrophobic | Pi-Sigma | ||||
| 8 | Carnosol (Rosemary) | C20H26O4/330.40 | −7.9 | MET667 | 2.82 | Hydrogen Bond | Conventional Hydrogen Bond |
| LYS646 | 5.21 | Hydrophobic | Pi- Alkyl | ||||
| 9 | Rosmarinic Acid (Rosemary) | C18H16O8/360.3 | −7.6 | ILE690 | 2.12 | Hydrogen Bond | Conventional Hydrogen Bond |
| ASP757 | 3.81 | Electrostatic | Pi-Anion | ||||
| LEU746 | 3.66 | Hydrophobic | Pi-Sigma | ||||
| ASP757 | 3.93 | ||||||
| LEU746 | 5.15 | Pi-Alkyl | |||||
| VAL627 | 5.25 | ||||||
| ALA644 | 3.83 | ||||||
| 10 | Carnosic Acid (Rosemary) | C20H28O4/332.4 | −7.7 | LEU746 | 3.70 | Hydrophobic | Pi-Sigma |
| LEU746 | 4.73 | Hydrophobic | Alkyl | ||||
| 11 | Taraxasterol (Dandelion) | C30H50O/426.7 | −9.2 | THR692 | 2.01 | Hydrogen Bond | Conventional Hydrogen Bond |
| VAL627 | 4.80 | Hydrophobic | Alkyl | ||||
| ALA644 | 5.37 | ||||||
| LYS646 | 4.22 | ||||||
| 12 | Luteolin (Dandelion) | C21H20O11/448.4 | −9 | H-O | 1.90 | Hydrogen Bond | Conventional Hydrogen Bond |
| SER756 | 2.18 | ||||||
| LYS646 | 2.82 | ||||||
| THR692 | 2.48 | ||||||
| TYR694 | 3.24 | Pi-Donor Hydrogen Bond | |||||
| ILE619 | 3.75 | Hydrophobic | Pi-Sigma | ||||
| LEU746 | 3.63 | ||||||
| VAL627 | 5.04 | Pi-Alkyl | |||||
| LEU746 | 5.41 | ||||||
| ALA644 | 4.00 | ||||||
| VAL627 | 5.43 | ||||||
| 13 | Taraxinic Acid (Dandelion) | C21H28O9/424.4 | −8.7 | H-O | 2.45 | Hydrogen Bond | Conventional Hydrogen Bond |
| GLU696 | 2.60 | ||||||
| LYS646 | 2.98 | ||||||
| LYS646 | 2.87 | ||||||
| VAL627 | 4.34 | Hydrophobic | Alkyl | ||||
| ALA644 | 3.66 | ||||||
| LYS646 | 4.24 | ||||||
| VAL627 | 4.55 | ||||||
| ALA644 | 5.24 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shaikh, M.S.; Islam, F.; Gargote, P.P.; Gaikwad, R.R.; Dhupe, K.C.; Khan, S.L.; Siddiqui, F.A.; Tapadiya, G.G.; Ali, S.S.; Dey, A.; et al. Potential Epha2 Receptor Blockers Involved in Cerebral Malaria from Taraxacum officinale, Tinospora cordifolia, Rosmarinus officinalis and Ocimum basilicum: A Computational Approach. Pathogens 2022, 11, 1296. https://doi.org/10.3390/pathogens11111296
Shaikh MS, Islam F, Gargote PP, Gaikwad RR, Dhupe KC, Khan SL, Siddiqui FA, Tapadiya GG, Ali SS, Dey A, et al. Potential Epha2 Receptor Blockers Involved in Cerebral Malaria from Taraxacum officinale, Tinospora cordifolia, Rosmarinus officinalis and Ocimum basilicum: A Computational Approach. Pathogens. 2022; 11(11):1296. https://doi.org/10.3390/pathogens11111296
Chicago/Turabian StyleShaikh, Mohd Sayeed, Fahadul Islam, Parag P. Gargote, Rutuja R. Gaikwad, Kalpana C. Dhupe, Sharuk L. Khan, Falak A. Siddiqui, Ganesh G. Tapadiya, Syed Sarfaraz Ali, Abhijit Dey, and et al. 2022. "Potential Epha2 Receptor Blockers Involved in Cerebral Malaria from Taraxacum officinale, Tinospora cordifolia, Rosmarinus officinalis and Ocimum basilicum: A Computational Approach" Pathogens 11, no. 11: 1296. https://doi.org/10.3390/pathogens11111296
APA StyleShaikh, M. S., Islam, F., Gargote, P. P., Gaikwad, R. R., Dhupe, K. C., Khan, S. L., Siddiqui, F. A., Tapadiya, G. G., Ali, S. S., Dey, A., & Emran, T. B. (2022). Potential Epha2 Receptor Blockers Involved in Cerebral Malaria from Taraxacum officinale, Tinospora cordifolia, Rosmarinus officinalis and Ocimum basilicum: A Computational Approach. Pathogens, 11(11), 1296. https://doi.org/10.3390/pathogens11111296









