Association of Gut Microbiota Composition in Pregnant Women Colonized with Group B Streptococcus with Maternal Blood Routine and Neonatal Blood-Gas Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Enrollment of Patients and Healthy Controls
2.2. Sample Collection of Feces
2.3. Measurement of CBC in Pregnant Women
2.4. Neonatal Umbilical Artery Blood-Gas Analysis
2.5. 16S rRNA Gene Sequence Analysis
2.6. Statistical Analysis
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hijona Elósegui, J.J.; Carballo García, A.L.; Fernández Rísquez, A.C.; Bermúdez Quintana, M.; Expósito Montes, J.F. Prevalence of maternal intrapartum colonization due to the Group-B Streptococus: Epidemiological analysis in the province of Jaén (Spain). Rev. Esp. Salud Publica 2022, 96, e202209064. [Google Scholar] [PubMed]
- Zhang, Y.; Liang, S.; Zhang, S.; Zhang, S.; Yu, Y.; Huochun, Y.; Liu, Y.; Zhang, W.; Liu, G. Development and evaluation of a multi-epitope subunit vaccine against group B Streptococcus infection. Emerg. Microbes Infect. 2022, 11, 2371–2382. [Google Scholar] [CrossRef] [PubMed]
- Rallu, F.; Barriga, P.; Scrivo, C.; Martel-Laferrière, V.; Laferrière, C. Sensitivities of antigen detection and PCR assays greatly increased compared to that of the standard culture method for screening for group B streptococcus carriage in pregnant women. J. Clin. Microbiol. 2006, 44, 725–728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tibussek, D.; Sinclair, A.; Yau, I.; Teatero, S.; Fittipaldi, N.; Richardson, S.E.; Mayatepek, E.; Jahn, P.; Askalan, R. Late-onset group B streptococcal meningitis has cerebrovascular complications. J. Pediatrics 2015, 166, 1187–1192. [Google Scholar] [CrossRef] [PubMed]
- Makaranka, S.; Scutt, F.; Frixou, M.; Wensley, K.E.; Sharma, R.; Greenhowe, J. The gut microbiome and melanoma: A review. Exp. Dermatol. 2022, 31, 1292–1301. [Google Scholar] [CrossRef]
- Azad, M.B.; Konya, T.; Maughan, H.; Guttman, D.S.; Field, C.J.; Chari, R.S.; Sears, M.R.; Becker, A.B.; Scott, J.A.; Kozyrskyj, A.L. Gut microbiota of healthy Canadian infants: Profiles by mode of delivery and infant diet at 4 months. CMAJ: Can. Med. Assoc. J. J. L’association Med. Can. 2013, 185, 385–394. [Google Scholar] [CrossRef] [Green Version]
- Gschwendtner, S.; Kang, H.; Thiering, E.; Kublik, S.; Fösel, B.; Schulz, H.; Krauss-Etschmann, S.; Heinrich, J. Early life determinants induce sustainable changes in the gut microbiome of six-year-old children. Sci. Rep. 2019, 9, 12675. [Google Scholar] [CrossRef] [Green Version]
- Saavedra, J.M.; Dattilo, A.M. Early development of intestinal microbiota: Implications for future health. Gastroenterol. Clin. North Am. 2012, 41, 717–731. [Google Scholar] [CrossRef]
- Cassidy-Bushrow, A.E.; Sitarik, A.; Levin, A.M.; Lynch, S.V.; Havstad, S.; Ownby, D.R.; Johnson, C.C.; Wegienka, G. Maternal group B Streptococcus and the infant gut microbiota. J. Dev. Orig. Health Dis. 2016, 7, 45–53. [Google Scholar] [CrossRef] [Green Version]
- Kubota, T.; Nojima, M.; Itoh, S. Vaginal bacterial flora of pregnant women colonized with group B streptococcus. J. Infect. Chemother. Off. J. Jpn. Soc. Chemother. 2002, 8, 326–330. [Google Scholar] [CrossRef]
- Bayó, M.; Berlanga, M.; Agut, M. Vaginal microbiota in healthy pregnant women and prenatal screening of group B streptococci (GBS). Int. Microbiol. Off. J. Span. Soc. Microbiol. 2002, 5, 87–90. [Google Scholar] [CrossRef]
- Stearns, J.C.; Simioni, J.; Gunn, E.; McDonald, H.; Holloway, A.C.; Thabane, L.; Mousseau, A.; Schertzer, J.D.; Ratcliffe, E.M.; Rossi, L.; et al. Intrapartum antibiotics for GBS prophylaxis alter colonization patterns in the early infant gut microbiome of low risk infants. Sci. Rep. 2017, 7, 16527. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010, 7, 335–336. [Google Scholar]
- Schuchat, A.; Whitney, C.G.; Zangwill, K. Prevention of perinatal group B streptococcal disease: A public health perspective. Centers for Disease Control and Prevention. MMWR Recomm. Rep. 1996, 45, 1–24. [Google Scholar]
- Verani, J.R.; McGee, L.; Schrag, S.J. Prevention of perinatal group B streptococcal diseas—Revised guidelines from CDC. MMWR Recomm. Rep. 2010, 59, 1–36. [Google Scholar]
- Mu, X.; Zhao, C.; Yang, J.; Wei, X.; Zhang, J.; Liang, C.; Gai, Z.; Zhang, C.; Zhu, D.; Wang, Y.; et al. Group B Streptococcus colonization induces Prevotella and Megasphaera abundance-featured vaginal microbiome compositional change in non-pregnant women. PeerJ 2019, 7, e7474. [Google Scholar] [CrossRef] [Green Version]
- Cruciani, F.; Biagi, E.; Severgnini, M.; Consolandi, C.; Calanni, F.; Donders, G.; Brigidi, P.; Vitali, B. Development of a microarray-based tool to characterize vaginal bacterial fluctuations and application to a novel antibiotic treatment for bacterial vaginosis. Antimicrob. Agents Chemother. 2015, 59, 2825–2834. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.F.; Gong, X.L.; Chen, S.X.; Wang, K.; Jiang, Y.H. Deviations in the gut microbiota of neonates affected by maternal group B Streptococcus colonization. BMC Microbiol. 2021, 21, 140. [Google Scholar] [CrossRef]
- Bonifácio Andrade, E.; Lorga, I.; Roque, S.; Geraldo, R.; Mesquita, P.; Castro, R.; Simões-Costa, L.; Costa, M.; Faustino, A.; Ribeiro, A.; et al. Maternal vaccination against group B Streptococcus glyceraldehyde-3-phosphate dehydrogenase leads to gut dysbiosis in the offspring. Brain Behav. Immun. 2022, 103, 186–201. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Zhou, Y.; Liu, B.; Jin, Z.; Zhuang, X.; Dai, W.; Yang, Z.; Feng, X.; Zhou, Q.; Liu, Y.; et al. Perinatal Antibiotic Exposure Affects the Transmission between Maternal and Neonatal Microbiota and Is Associated with Early-Onset Sepsis. mSphere 2020, 5, e00984-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, N.; Zhou, J.; Mo, H.; Mu, Q.; Su, H.; Li, M.; Yu, Y.; Liu, A.; Zhang, Q.; Xu, J.; et al. The Gut Microbial Signature of Gestational Diabetes Mellitus and the Association With Diet Intervention. Front. Cell. Infect. Microbiol. 2021, 11, 800865. [Google Scholar] [CrossRef] [PubMed]
- Primec, M.; Klemenak, M.; Di Gioia, D.; Aloisio, I.; Bozzi Cionci, N.; Quagliariello, A.; Gorenjak, M.; Mičetić-Turk, D.; Langerholc, T. Clinical intervention using Bifidobacterium strains in celiac disease children reveals novel microbial modulators of TNF-α and short-chain fatty acids. Clin. Nutr. (Edinb. Scotl.) 2019, 38, 1373–1381. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Xu, F.; Wan, W.; Yu, B.; Tang, L.; Yang, Y.; Du, Y.; Chen, Z. Gut microbial characteristics of adult patients with allergy rhinitis. Microb. Cell Fact. 2020, 19, 171. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Li, B.; Zhang, Y.; Ma, S.; Liu, C.; Liang, F.; Wei, Z.; Huang, T.; Wang, R. Influence of Pelvic Intensity-Modulated Radiation Therapy With Concurrent Cisplatin-Based Chemotherapy of Cervical Cancer on the Vaginal Microbiome. Front. Oncol. 2021, 11, 615439. [Google Scholar] [CrossRef]
- Nardelli, C.; Granata, I.; D’Argenio, V. Characterization of the Duodenal Mucosal Microbiome in Obese Adult Subjects by 16S rRNA Sequencing. Microorganisms 2020, 8, 485. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Xu, X.; Wu, X.; Cao, H.; Li, X.; Hou, Z.; Wang, B.; Liu, J.; Ji, X.; Zhang, P.; et al. A comparison of the composition and functions of the oral and gut microbiotas in Alzheimer ’s patients. Front. Cell. Infect. Microbiol. 2022, 12, 942460. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, H.; Tang, X.; Jiang, X.; Yan, X.; Liu, X.; Gong, J.; Mew, K.; Sun, H.; Chen, X.; et al. Distinct Metagenomic Signatures in the SARS-CoV-2 Infection. Front. Cell. Infect. Microbiol. 2021, 11, 706970. [Google Scholar] [CrossRef]
- Johnson, K.; Stephen, M. Early Onset Sepsis. S D Med. 2016, 69, 29–33. [Google Scholar] [PubMed]
- Van Doorn Marle, B.; van der Voorn, J.P.; Tanger, H.L.; van Weissenbruch, M.M.; Visser, D.H. Exposure to intrauterine inflammation and late-onset sepsis in very preterm infants. Pediatric Res. 2021, 91, 230–234. [Google Scholar] [CrossRef]
- Neonatal Resuscitation Group, Perinatal Medicine Branch, Chinese Medical Association. Expert consensus on the clinical application of umbilical artery in newborn blood gas analysis. Chin. J. Perinat. Med. 2021, 24, 401–405. [Google Scholar] [CrossRef]
- Yu, R.; Ye, H.; Zhu, J.; Zhu, X. Expert Consensus on the diagnosis of neonatal asphyxia. Chin. J. Perinat. Med. 2016, 19, 3–6. [Google Scholar]
- Krivec, J.L.; Manfreda, K.L.; Paro-Panjan, D. Clinical Factors Influencing Endogenous Carbon Monoxide Production and Carboxyhemoglobin Levels in Neonates. J. Pediatric Hematol. Oncol. 2022, 44, e84–e90. [Google Scholar] [CrossRef]
- Schaer, D.J.; Buehler, P.W.; Alayash, A.I.; Belcher, J.D.; Vercellotti, G.M. Hemolysis and free hemoglobin revisited: Exploring hemoglobin and hemin scavengers as a novel class of therapeutic proteins. Blood 2013, 121, 1276–1284. [Google Scholar] [CrossRef]
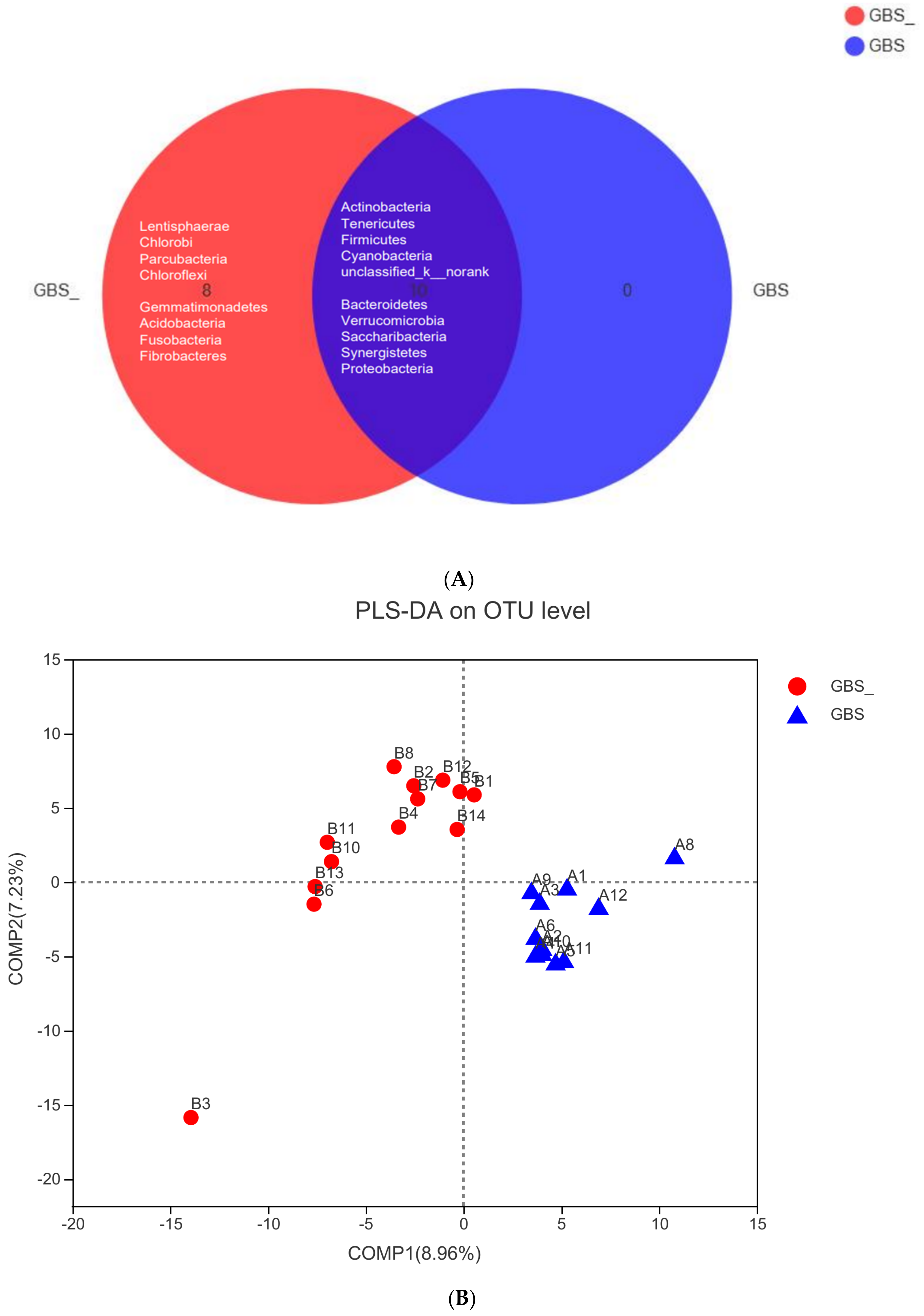

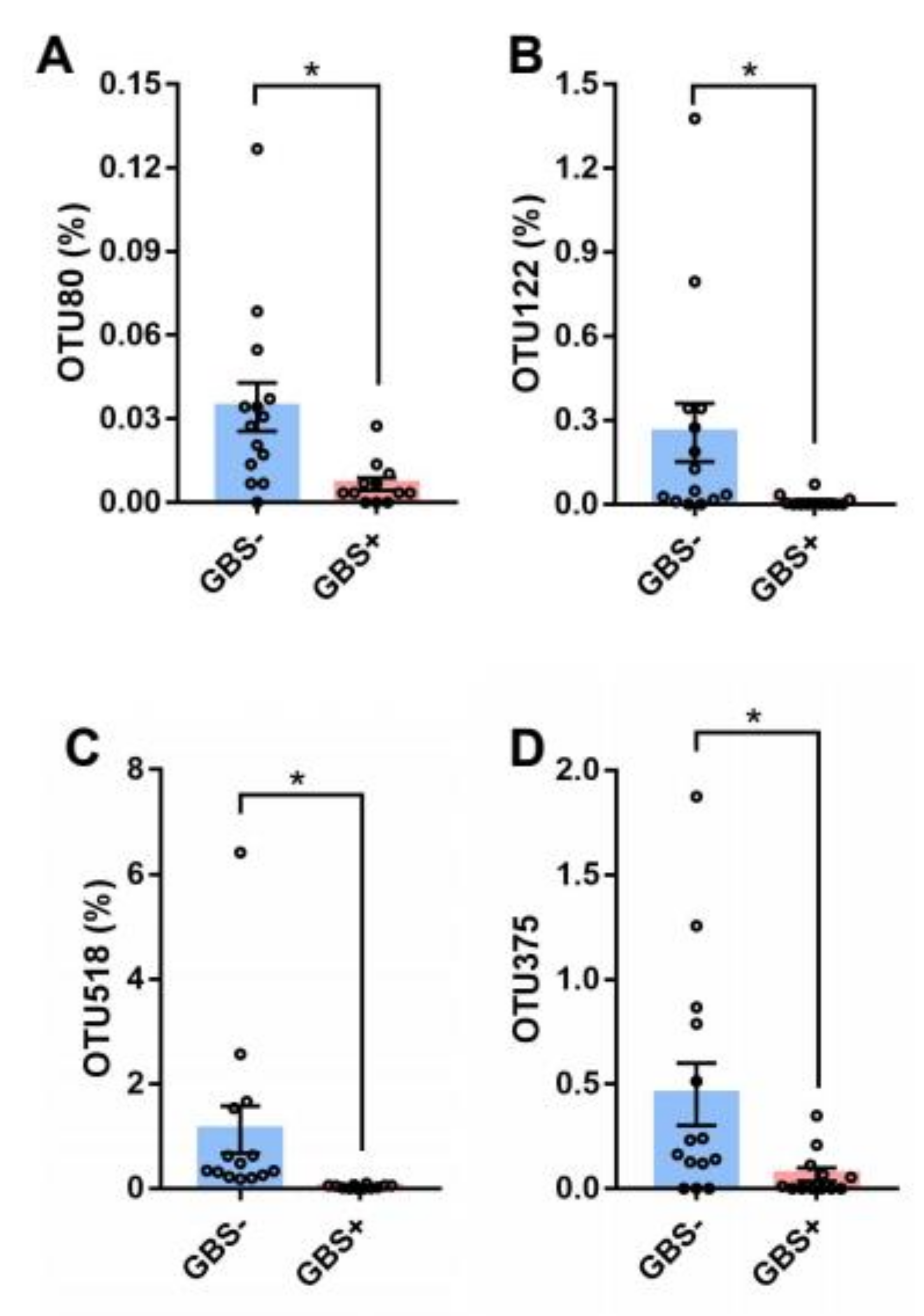
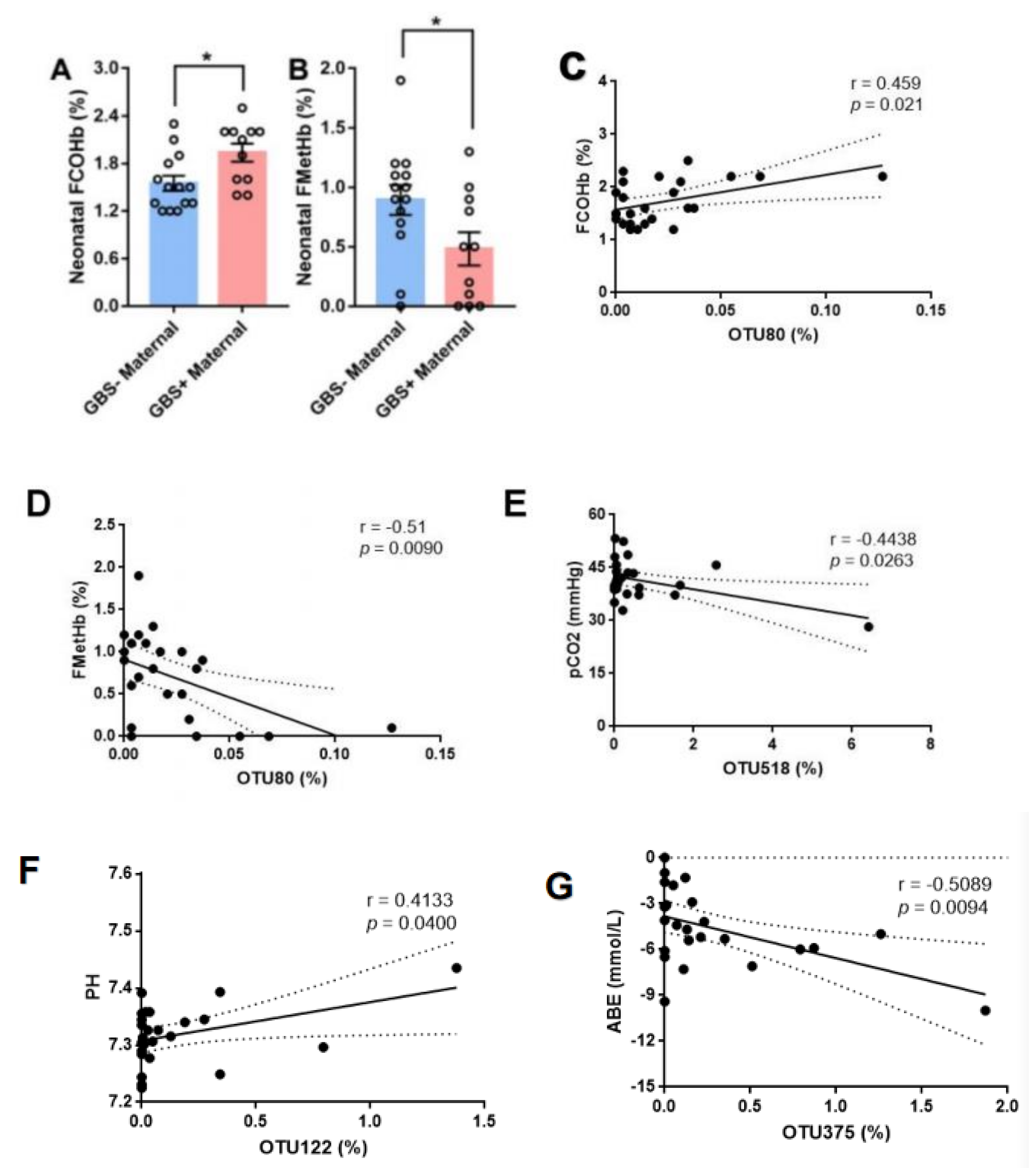
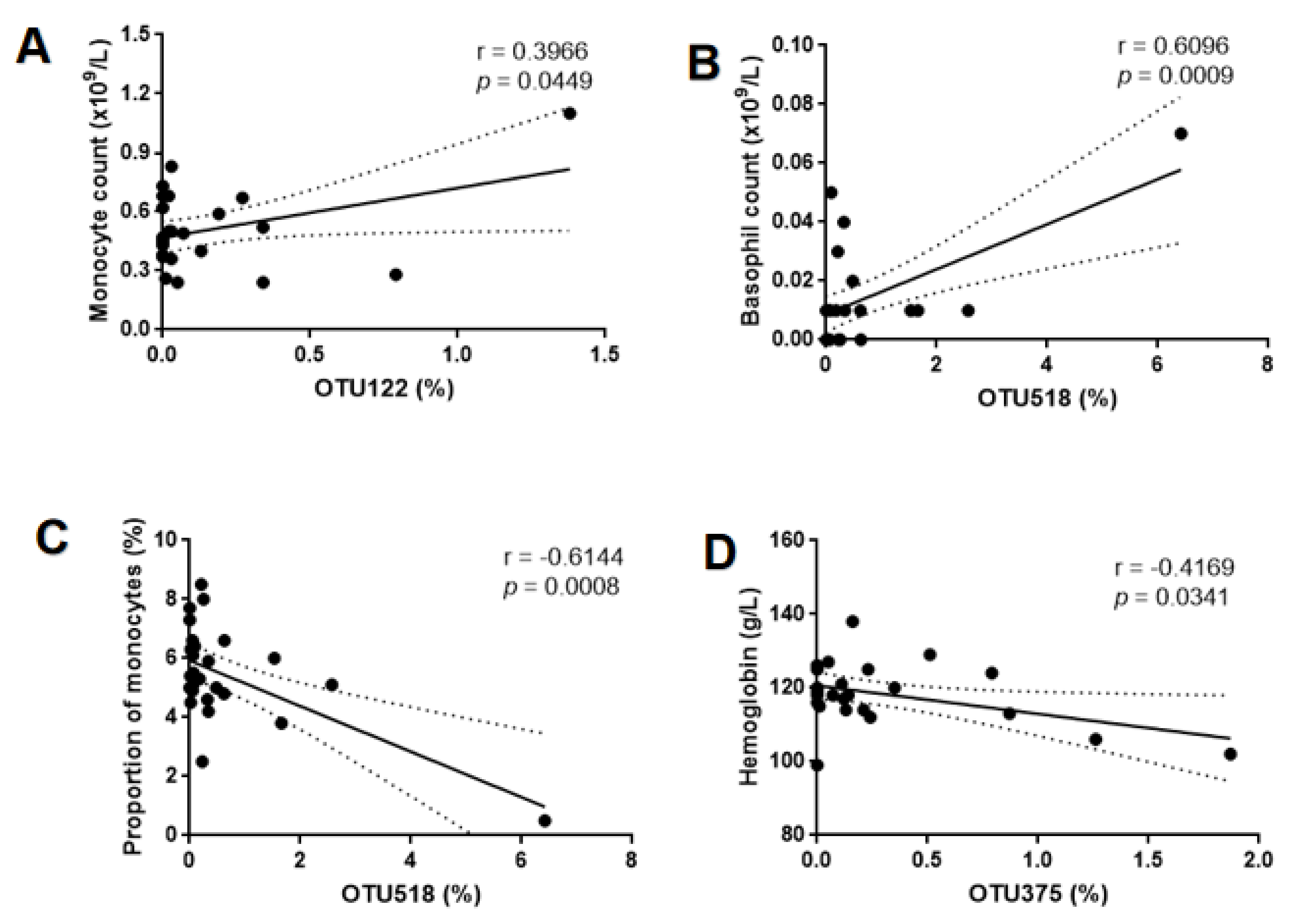
| Characteristic | GBS− (n = 14) | GBS+ (n = 12) | p-Value |
|---|---|---|---|
| Age (years) | 29.79 ± 2.97 | 30.58 ± 3.96 | 0.563 |
| Gender | Female | Female | - |
| WBC (×109/L) | 8.53 ± 1.20 | 9.37 ± 2.76 | 0.309 |
| HGB (g/L) | 118.43 ± 8.21 | 118.67 ± 8.90 | 0.944 |
| Neutrophil (%) | 72.21 ± 5.20 | 71.64 ± 9.48 | 0.849 |
| Basophil (%) | 0.20 ± 0.30 | 0.17 ± 0.16 | 0.819 |
| Lymphocyte (%) | 20.75 ± 4.87 | 21.46 ± 8.41 | 0. 792 |
| Monocyte (%) | 5.75 ±0.10 | 5.09 ± 0.22 | 0.327 |
| Eosinophil (%) | 0.97 ± 0.46 | 1.18 ± 1.11 | 0.536 |
| Neutrophil count (×109/L) | 6.19 ± 1.17 | 6.80 ± 2.29 | 0.393 |
| Basophil count (×109/L) | 0.01 ± 0.01 | 0.02 ± 0.02 | 0.704 |
| Lymphocyte count (×109/L) | 1.74 ± 0.32 | 1.92 ± 0.67 | 0.370 |
| Monocyte count (×109/L) | 0.49 ± 0.11 | 0.52 ± 0.27 | 0.668 |
| Eosinophil count (×109/L) | 0.10 ± 0.04 | 0.10 ± 0.07 | 0.944 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Pu, W.; Liu, Q.; Zhu, M.; Chen, Q.; Xu, Y.; Zhou, C. Association of Gut Microbiota Composition in Pregnant Women Colonized with Group B Streptococcus with Maternal Blood Routine and Neonatal Blood-Gas Analysis. Pathogens 2022, 11, 1297. https://doi.org/10.3390/pathogens11111297
Wang Z, Pu W, Liu Q, Zhu M, Chen Q, Xu Y, Zhou C. Association of Gut Microbiota Composition in Pregnant Women Colonized with Group B Streptococcus with Maternal Blood Routine and Neonatal Blood-Gas Analysis. Pathogens. 2022; 11(11):1297. https://doi.org/10.3390/pathogens11111297
Chicago/Turabian StyleWang, Zhixia, Wenyuan Pu, Qi Liu, Meifeng Zhu, Qinlei Chen, Yingchun Xu, and Chunxiang Zhou. 2022. "Association of Gut Microbiota Composition in Pregnant Women Colonized with Group B Streptococcus with Maternal Blood Routine and Neonatal Blood-Gas Analysis" Pathogens 11, no. 11: 1297. https://doi.org/10.3390/pathogens11111297
APA StyleWang, Z., Pu, W., Liu, Q., Zhu, M., Chen, Q., Xu, Y., & Zhou, C. (2022). Association of Gut Microbiota Composition in Pregnant Women Colonized with Group B Streptococcus with Maternal Blood Routine and Neonatal Blood-Gas Analysis. Pathogens, 11(11), 1297. https://doi.org/10.3390/pathogens11111297






