First Isolation of Yarrowia lipolytica in a Granulomatous Pneumonia of a Spectacled Caiman, Caiman crocodilus Linnaeus, 1758
Abstract
:1. Introduction
2. Materials and Methods
2.1. Case
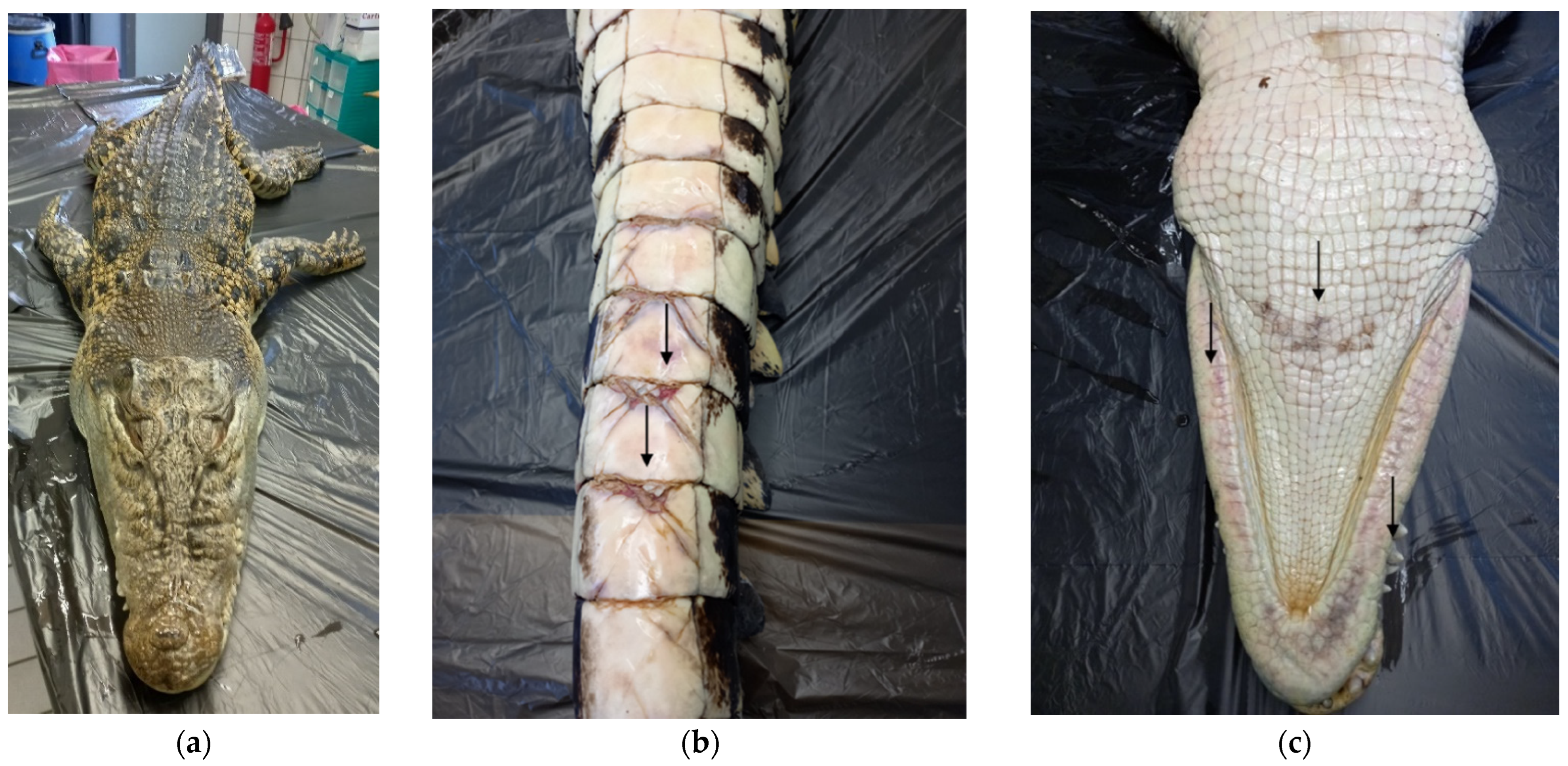
2.2. Gross Examination
2.3. Histological Examination
2.4. Bacteriological Examination
2.5. Parasitological Examination
2.6. Mycological Examination
2.7. Molecular Analysis
2.8. Library Preparation and Whole Genome Sequencing (WGS)
2.9. Bioinformatics Analysis
3. Results
3.1. Gross Examination
3.2. Histological Examination
3.3. Bacteriological and Parasitological Examination
3.4. Mycological Examination
3.5. Molecular Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meena, D.S.; Kumar, D. Candida Pneumonia: An Innocent Bystander or a Silent Killer? Med. Princ. Pract. 2022, 31, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Dermawan, J.K.T.; Ghosh, S.; Keating, M.K.; Gopalakrishna, K.V.; Mukhopadhyay, S. Candida Pneumonia with Severe Clinical Course, Recovery with Antifungal Therapy and Unusual Pathologic Findings. Medicine 2018, 97, e9650. [Google Scholar] [CrossRef] [PubMed]
- Misme-Aucouturier, B.; Albassier, M.; Alvarez-Rueda, N.; Le Pape, P. Specific Human and Candida Cellular Interactions Lead to Controlled or Persistent Infection Outcomes during Granuloma-like Formation. Infect. Immun. 2017, 85, e00807-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shweihat, Y.; Perry, J.; Shah, D. Isolated Candida Infection of the Lung. Respir. Med. Case Rep. 2015, 16, 18–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arobelidze, S.; Gopalakrishna, K.V. Necrotizing Granulomatous Pneumonia Caused by Candida Albicans: A Case Report. J. Hosp. Med. 2016, 11, 6–9. [Google Scholar]
- Seyedmousavi, S.; Bosco, S.; De Hoog, S.; Ebel, F.; Elad, D.; Gomes, R.R.; Jacobsen, I.D.; Martel, A.; Mignon, B.; Pasmans, F.; et al. Fungal Infections in Animals: A Patchwork of Different Situations. Med. Mycol. 2018, 56, S165–S187. [Google Scholar] [CrossRef] [Green Version]
- Lanteri, G.; Macrì, F.; Rapisarda, G.; Basile, F.; Reale, S.; Marino, F. Systemic Candidiasis in Farm-Reared Red-Legged Partridges (Alectoris rufa) Caused by Leucosporidium spp. BMC Vet. Res. 2012, 8, 1. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.S.; Kim, J.H. Systemic Candidiasis of a Wild Slaty-Backed Gull (Larus schistisagus) in Jeju, Korea. J. Vet. Clin. 2019, 36, 172–175. [Google Scholar] [CrossRef]
- Duchaussoy, A.C.; Rose, A.; Talbot, J.J.; Barrs, V.R. Gastrointestinal Granuloma Due to Candida Albicans in an Immunocompetent Cat. Med. Mycol. Case Rep. 2015, 10, 14–17. [Google Scholar] [CrossRef]
- Conti, H.R.; Huppler, A.R.; Whibley, N.; Gaffen, S.L. Animal Models for Candidiasis. Curr. Protoc. Immunol. 2014, 105, 19.6.1–19.6.17. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.L.; Sun, P.L.; Kao, C.F.; Li, W.T.; Cheng, I.J.; Yu, P.H. Disseminated Candidiasis and Candidemia Caused by Candida palmioleophila in a Green Sea Turtle (Chelonia mydas). Animals 2021, 11, 3480. [Google Scholar] [CrossRef] [PubMed]
- Paré, J.; Jacobson, E. Mycotic Diseases of Reptiles. Infect. Dis. Pathol. Reptil. 2007, 9, 527–570. [Google Scholar] [CrossRef]
- Hernandez-Divers, S.J. Pulmonary Candidiasis Caused by Candida Albicans in a Greek Tortoise (Testudo graeca) and Treatment with Intrapulmonary Amphotericin B. J. Zoo Wildl. Med. 2001, 32, 352–359. [Google Scholar] [CrossRef]
- Domiciano, I.G.; da Silva Gagliotti, G.F.P.; Domit, C.; Lorenzetti, E.; Bracarense, A.P.F.R.L. Bacterial and Fungal Pathogens in Granulomatous Lesions of Chelonia mydas in a Significant Foraging Ground off Southern Brazil. Vet. Res. Commun. 2022, 46, 859–870. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, Z.Y.; Ali, B.H.; Ali, R.K.; Jarad, A.S.; Farhan, W.H.; Hasan, M.S. Avian Candidiasis: A Review. Int. J. Pharm. Res. 2020, 12, 1088–1091. [Google Scholar] [CrossRef]
- Groenewald, M.; Boekhout, T.; Neuvéglise, C.; Gaillardin, C.; Van Dijck, P.W.M.; Wyss, M. Yarrowia lipolytica: Safety Assessment of an Oleaginous Yeast with a Great Industrial Potential. Crit. Rev. Microbiol. 2014, 40, 187–206. [Google Scholar] [CrossRef]
- Nagy, E.; Niss, M.; Dlauchy, D.; Arneborg, N.; Nielsen, D.S.; Péter, G. Yarrowia Divulgata f.a., Sp. Nov., a Yeast Species from Animal-Related and Marine Sources. Int. J. Syst. Evol. Microbiol. 2013, 63, 4818–4823. [Google Scholar] [CrossRef] [Green Version]
- Zieniuk, B.; Fabiszewska, A. Yarrowia lipolytica: A Beneficious Yeast in Biotechnology as a Rare Opportunistic Fungal Pathogen: A Minireview. World J. Microbiol. Biotechnol. 2019, 35, 10. [Google Scholar] [CrossRef] [Green Version]
- Patsios, S.I.; Dedousi, A.; Sossidou, E.N.; Zdragas, A. Sustainable Animal Feed Protein through the Cultivation of Yarrowia lipolytica on Agro-Industrial Wastes and by-Products. Sustainability 2020, 12, 1398. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Kumar, A.; Roudbary, M.; Mohammadi, R.; Černáková, L.; Rodrigues, C.F. Overview on the Infections Related to Rare Candida Species. Pathogens 2022, 11, 963. [Google Scholar] [CrossRef]
- Mamaev, D.; Zvyagilskaya, R. Yarrowia lipolytica: A Multitalented Yeast Species of Ecological Significance. FEMS Yeast Res. 2021, 21, foab008. [Google Scholar] [CrossRef] [PubMed]
- Bergey, D.H.; Holt, J.G. Bergey’s Manual of Determinative Bacteriology, 9th ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2000. [Google Scholar]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR—Protocols and Applications—A Laboratory Manual; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press, Inc.: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Romanelli, A.M.; Sutton, D.A.; Thompson, E.H.; Rinaldi, M.G.; Wickes, B.L. Sequence-Based Identification of Filamentous Basidiomycetous Fungi from Clinical Specimens: A Cautionary Note. J. Clin. Microbiol. 2010, 48, 741–752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alba, P.; Taddei, R.; Cordaro, G.; Fontana, M.C.; Toschi, E.; Gaibani, P.; Marani, I.; Giacomi, A.; Diaconu, E.L.; Iurescia, M.; et al. Carbapenemase IncF-Borne BlaNDM-5 Gene in the E. coli ST167 High-Risk Clone from Canine Clinical Infection, Italy. Vet. Microbiol. 2021, 256, 109045. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA Hybridization Values and Their Relationship to Whole-Genome Sequence Similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [Green Version]
- Roach, D.J.; Burton, J.N.; Lee, C.; Stackhouse, B.; Butler-Wu, S.M.; Cookson, B.T.; Shendure, J.; Salipante, S.J. A Year of Infection in the Intensive Care Unit: Prospective Whole Genome Sequencing of Bacterial Clinical Isolates Reveals Cryptic Transmissions and Novel Microbiota. PLoS Genet. 2015, 11, e1005413. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple Sequence Alignment Using ClustalW and ClustalX. Curr. Protoc. Bioinforma. 2003, 1, 1–22. [Google Scholar] [CrossRef]
- Tamura, K. Estimation of the Number of Nucleotide Substitutions When There Are Strong Transition-Transversion and G+C-Content Biases. Mol. Biol. Evol. 1992, 9, 678–687. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information. Yarrowia Lipolytica. Available online: https://www.ncbi.nlm.nih.gov/genome/browse/#!/eukaryotes/194/ (accessed on 25 September 2022).
- Madzak, C. Yarrowia lipolytica Strains and Their Biotechnological Applications: How Natural Biodiversity and Metabolic Engineering Could Contribute to Cell Factories Improvement. J. Fungi 2021, 7, 548. [Google Scholar] [CrossRef] [PubMed]
- Gaikwad, P.; Joshi, S.; Mandlecha, A.; RaviKumar, A. Phylogenomic and Biochemical Analysis Reassesses Temperate Marine Yeast Yarrowia lipolytica NCIM 3590 to Be Yarrowia bubula. Sci. Rep. 2021, 11, 5487. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information. BioSample: SAMN19575489. Available online: https://www.ncbi.nlm.nih.gov/biosample/SAMN19575489 (accessed on 25 September 2022).
- Zinjarde, S.S.; Pant, A.A. Hydrocarbon Degraders from Tropical Marine Environments. Mar. Pollut. Bull. 2002, 44, 118–121. [Google Scholar] [CrossRef]
- Quarterman, J.; Slininger, P.J.; Kurtzman, C.P.; Thompson, S.R.; Dien, B.S. A Survey of Yeast from the Yarrowia Clade for Lipid Production in Dilute Acid Pretreated Lignocellulosic Biomass Hydrolysate. Appl. Microbiol. Biotechnol. 2017, 101, 3319–3334. [Google Scholar] [CrossRef] [PubMed]
- Barth, G.; Weber, H. Genetic Studies on the Yeast Saccharomycopsis lipolytica Inactivation and mutagenesis. Z. Allg. Mikrobiol. 1983, 23, 147–157. [Google Scholar]
- Wojtatowicz, M.; Rymowicz, W.; Kautola, H. Comparison of Different Strains of the Yeast Yarrowia lipolytica for Citric Acid Production from Glucose Hydrol. Appl. Biochem. Biotechnol. 1991, 31, 165–174. [Google Scholar] [CrossRef]
- Zhou, J.; Zhou, H.; Du, G.; Liu, L.; Chen, J. Screening of a Thiamine-Auxotrophic Yeast α-Ketoglutaric Acid Overproduction. Lett. Appl. Microbiol. 2010, 51, 264–271. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. BioSample: SAMN21354843. Available online: https://www.ncbi.nlm.nih.gov/biosample/SAMN21354843 (accessed on 25 September 2022).
- National Center for Biotechnology Information. BioSample: SAMN08667771. Available online: https://www.ncbi.nlm.nih.gov/biosample/SAMN08667771 (accessed on 25 September 2022).
- National Center for Biotechnology Information. BioSample: SAMN08563947. Available online: https://www.ncbi.nlm.nih.gov/biosample/SAMN08563947 (accessed on 25 September 2022).
- National Center for Biotechnology Information. BioSample: SAMN25408340. Available online: https://www.ncbi.nlm.nih.gov/biosample/SAMN25408340 (accessed on 25 September 2022).
- National Center for Biotechnology Information. BioSample: SAMN08667773. Available online: https://www.ncbi.nlm.nih.gov/biosample/SAMN08667773 (accessed on 25 September 2022).
- Gaillardin, C.M.; Charoy, V.; Heslot, H. A Study of Copulation, Sporulation and Meiotic Segregation in Candida lipolytica. Arch. Mikrobiol. 1973, 92, 69–83. [Google Scholar] [CrossRef]
- Du, J.; Wang, X.; Luo, H.; Wang, Y.; Liu, X.; Zhou, X. Epidemiological Investigation of Non-Albicans Candida Species Recovered from Mycotic Mastitis of Cows in Yinchuan, Ningxia of China. BMC Vet. Res. 2018, 14, 251. [Google Scholar] [CrossRef]
- Gründer, S.; Mayser, P.; Redmann, T.; Kaleta, E.F. Mycological Examinations on the Fungal Flora of the Chicken Comb. Mycoses 2005, 48, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Leme, F.C.O.; de Barros Negreiros, M.M.; Koga, F.A.; de Moraes Gimenes Bosco, S.; Bagagli, E.; Haddad, V.H. Evaluation of Pathogenic Fungi Occurrence in Traumatogenic Structures of Freshwater Fish. Rev. Soc. Bras. Med. Trop. 2011, 44, 182–185. [Google Scholar] [CrossRef] [PubMed]
- Pitarch, A.; Gil, C.; Blanco, G. Oral Mycoses in Avian Scavengers Exposed to Antibiotics from Livestock Farming. Sci. Total Environ. 2017, 605, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Pollock, C.G.; Rohrbach, B.; Ramsay, E.C. Fungal Dermatitis in Captive Pinnipeds. J. Zoo Wildl. Med. 2000, 31, 374–378. [Google Scholar] [CrossRef]
- Terrell, K.A.; Ballmann, A.E.; Brown, A.; Childers, C.; Knowles, S.; Meredith, A.; Sparks, D. Environmental Contamination and Unusual Snake Mortality in an Urban National Wildlife Refuge. Herpetol. Conserv. Biol. 2020, 15, 652–665. [Google Scholar]
- Irby, R.F.; Kandula, M.; Zadikany, R.; Sandin, R.L.; Greene, J.N. Yarrowia lipolytica as Normal Human Flora. Infect. Dis. Clin. Pract. 2014, 22, 207–209. [Google Scholar] [CrossRef]
- Cohen, M.S.; Isturiz, R.E.; Malech, H.L.; Root, R.K.; Wilfert, C.M.; Gutman, L.; Buckley, R.H. Fungal Infection in Chronic Granulomatous Disease: The Importance of the Phagocyte in Defense against Fungi. Am. J. Med. 1981, 71, 59–66. [Google Scholar] [CrossRef]
- Abbes, S.; Amouri, I.; Trabelsi, H.; Neji, S.; Sellami, H.; Rahmouni, F.; Makni, F.; Rebai, T.; Ayadi, A. Analysis of Virulence Factors and In Vivo Biofilm-Form Ing Capacity of Yarrowia lipolytica Isolated from Patients with Fungemia. Med. Mycol. 2017, 55, 193–202. [Google Scholar] [CrossRef] [Green Version]
- Pawaiya, R.V.S.; Sharma, A.K.; Swarup, D.; Somvanshi, R. Pathology of Mycotic Gastritis in a Wild Indian Freshwater/Marsh Crocodile (Mugger; Crocodylus palustris): A Case Report. Vet. Med. 2011, 56, 135–139. [Google Scholar] [CrossRef] [Green Version]
- Ciccarelli, S.; Valastro, C.; Di Bello, A.; Paci, S.; Caprio, F.; Corrente, M.L.; Trotta, A.; Franchini, D. Diagnosis and Treatment of Pulmonary Disease in Sea Turtles (Caretta caretta). Animals 2020, 10, 1355. [Google Scholar] [CrossRef]
- Petty, L.A.; Gallan, A.J.; Detrick, J.A.; Ridgway, J.P.; Mueller, J.; Pisano, J. Candida dubliniensis Pneumonia: A Case Report and Review of Literature. Mycopathologia 2016, 181, 765–768. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yu, Y.T.; Xu, C.H.; Chen, D.C. Candida Colonization in the Respiratory Tract: What Is the Significance? Front. Med. 2021, 7, 598037. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, C.G.P.; Fitzpatrick, D.A. Pangloss: A Tool for Pan-Genome Analysis of Microbial Eukaryotes. Genes 2019, 10, 521. [Google Scholar] [CrossRef] [PubMed]





| Assembly | Alignment Coverage | Percentage Identity | BioSample | Origin | State | Reference |
|---|---|---|---|---|---|---|
| % | % | |||||
| GCA_023374015.1 | 97.359 | 99.492 | XZ-2019 | Meat | China | [37] |
| GCA_003571385.1 | 97.286 | 99.508 | NCIM 3589 | Marine strain | India | [38] |
| GCA_003367945.1 | 97.209 | 99.495 | YB-566 | N.R. | USA | [39] |
| GCA_900537225.1 | 97.165 | 99.489 | H222 | Soil | Germany | [40] |
| GCA_003367845.1 | 97.149 | 99.493 | YB-567 | Gluten settling water | USA | [39] |
| GCA_900087985.1 | 97.113 | 99.497 | A101 | Carwash effluent | Poland | [41] |
| GCA_000613145.2 | 96.974 | 99.445 | WSH-Z06 | Oil-polluted soil | China | [42] |
| GCA_020826875.1 | 96.876 | 99.511 | CGMCC7326 | Honeycomb | China | [43] |
| GCA_003367925.1 | 96.561 | 99.445 | YB-419 | Maize fiber tailings | USA | [39] |
| GCA_003054365.1 | 96.121 | 99.414 | IBT 446 | Feta cheese | Denmark | [44] |
| GCA_003243785.1 | 95.874 | 99.493 | CBA6003 | Kimchi | South Korea | [45] |
| GCA_023272835.1 | 95.401 | 99.475 | 22301-5 | Waste waters | France | [46] |
| GCA_003367965.1 | 95.315 | 99.350 | YB-420 | Milled corn | USA | [39] |
| GCA_003054305.1 | 95.174 | 99.439 | H222 (CLIB 80) | Soil | Germany | [47] |
| GCA_003367865.1 | 86.806 | 99.557 | YB-392 | Gluten settler | USA | [39] |
| GCA_012654145.1 | 86.162 | 99.568 | W29 | Waste waters | France | [48] |
| Animal | Number of Y. lipolytica Positive/Number of Animals Tested | Origin of Isolation | Country | Lesions | Reference |
|---|---|---|---|---|---|
| Harbor seal, Phoca vitulina | 2/16 | 1 swab and 1 skin scraping of skin | Tennessee, USA | Fungal dermatitis in facial-periorbital in one animal, not reported in the other | [53] |
| Fish (corvina or freshwater silver croaker, Plagioscion squamosissimus, piranha, Serrasalmus maculatus, dogfish, Acestrothychus lacustris, mandijuba or mandiamarelo catfish, Pimelodus maculatus, tilapia, Tilapia spp., and wolf fish, Hoplias malabaricus) | N.R. | Traumatogenic structures (stings, rays of fins, and sharp teeth) | Brazil | N.A. | [51] |
| Cinereous vultures, Gyps fulvus | 3/6 nestlings | Scraped oral mucosa | Spain | Oral lesions | [52] |
| Cow | 5 /484 cows with mastitis | Milk | China | Mastitis | [49] |
| Black-masked Racer, Coluber constrictor latrunculus | 1/3 | Liver | Louisiana, USA | Hepatic necrosis | [54] |
| Green sea turtle, Chelonia mydas | 9/270 | Granulomas from lung and liver | Brazil | Mild to moderate granulomatous pneumonia and severe hepatitis | [14] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iurescia, M.; Santini, A.; Montagnani, M.; Diaconu, E.L.; Stravino, F.; Agnelli, D.; Vergari, E.; Fichi, G.; Eleni, C. First Isolation of Yarrowia lipolytica in a Granulomatous Pneumonia of a Spectacled Caiman, Caiman crocodilus Linnaeus, 1758. Pathogens 2022, 11, 1255. https://doi.org/10.3390/pathogens11111255
Iurescia M, Santini A, Montagnani M, Diaconu EL, Stravino F, Agnelli D, Vergari E, Fichi G, Eleni C. First Isolation of Yarrowia lipolytica in a Granulomatous Pneumonia of a Spectacled Caiman, Caiman crocodilus Linnaeus, 1758. Pathogens. 2022; 11(11):1255. https://doi.org/10.3390/pathogens11111255
Chicago/Turabian StyleIurescia, Manuela, Andrea Santini, Marco Montagnani, Elena Lavinia Diaconu, Fiorentino Stravino, Devid Agnelli, Emanuela Vergari, Gianluca Fichi, and Claudia Eleni. 2022. "First Isolation of Yarrowia lipolytica in a Granulomatous Pneumonia of a Spectacled Caiman, Caiman crocodilus Linnaeus, 1758" Pathogens 11, no. 11: 1255. https://doi.org/10.3390/pathogens11111255
APA StyleIurescia, M., Santini, A., Montagnani, M., Diaconu, E. L., Stravino, F., Agnelli, D., Vergari, E., Fichi, G., & Eleni, C. (2022). First Isolation of Yarrowia lipolytica in a Granulomatous Pneumonia of a Spectacled Caiman, Caiman crocodilus Linnaeus, 1758. Pathogens, 11(11), 1255. https://doi.org/10.3390/pathogens11111255






