Study on the Canine Adenovirus Type 1 (CAdV-1) Infection in Domestic Dogs in Southern Italy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Virus Screening
2.3. Sequence and Phylogenetic Analyses
2.4. Virus Isolation
2.5. Histopathological Examination
3. Results
3.1. Detection of CAdV and Co-Infections
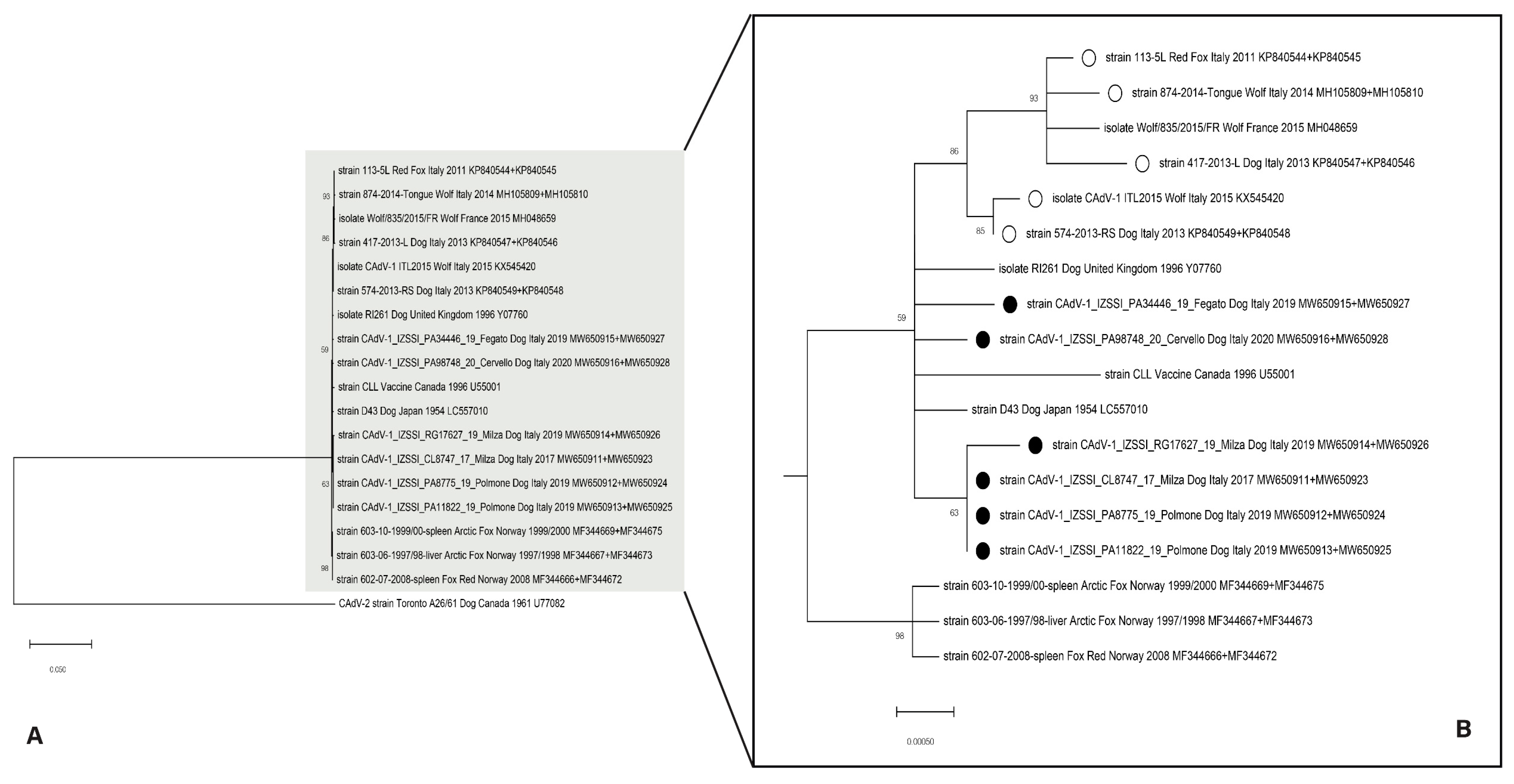
3.2. Sequence Analysis
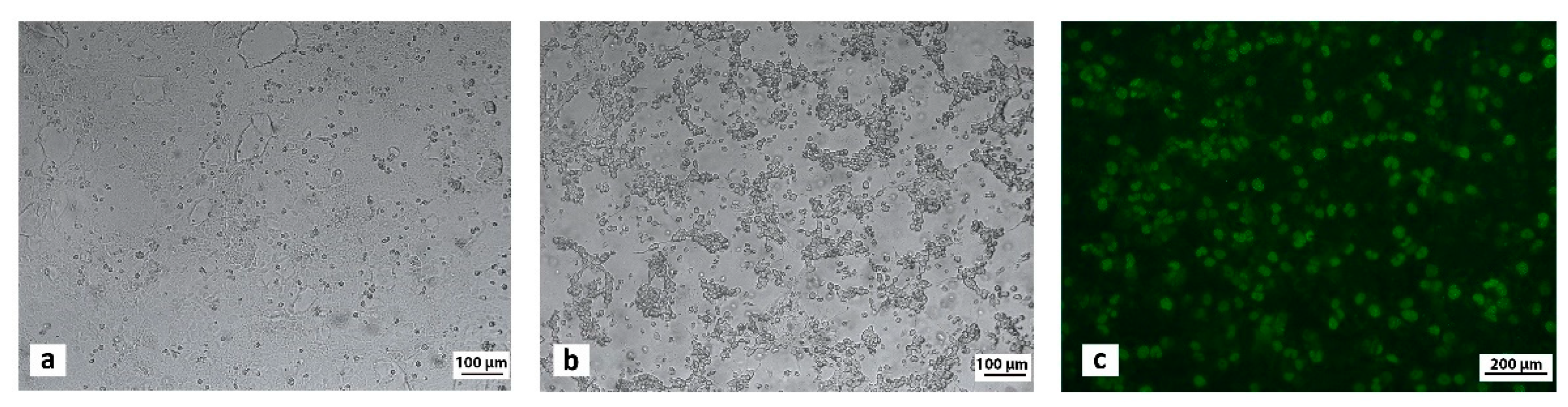
3.3. Virus Isolation
3.4. Histopathological Examination
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rubarth, S. An Acute Virus-Disease with Liver Lesion in Dogs (Hepatitis Contagiosa Canis); a Pathologico-Anatomical and Etiological Investigation. Acta Pathol. Microbiol. Scand. 1947. Available online: https://www.ncbi.nlm.nih.gov/nlmcatalog?cmd=PureSearch&term=0012136R%5Bnlmid%5D (accessed on 20 August 2022).
- Whittem, J.H.; Blood, D.C. Hepatitis Contagiosa Canis (Rubarth) in Australia. Aust. Vet. J. 1949, 25, 166–171. [Google Scholar] [CrossRef]
- Parry, H.B. Viral Hepatitis of Dogs (Rubarth’s Disease). I. Clinical and Pathological Observations on a Spontaneous Epidemic. Vet. Rec. 1950, 62, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Appel, M.J. Canine adenovirus type 1 (infectious canine hepatitis virus). In Virus Infections of Carnivores; Elsevier Science: Amsterdam, The Netherlands, 1987; pp. 29–43. [Google Scholar]
- Adams, M.J.; Lefkowitz, E.J.; King, A.M.Q.; Carstens, E.B. Ratification Vote on Taxonomic Proposals to the International Committee on Taxonomy of Viruses (2014). Arch. Virol. 2014, 159, 2831–2841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bass, E.P.; Gill, M.A.; Beckenhauer, W.H. Evaluation of a Canine Adenovirus Type 2 Strain as a Replacement for Infectious Canine Hepatitis Vaccine. J. Am. Vet. Med. Assoc. 1980, 177, 234–242. [Google Scholar]
- Bergmann, M.; Freisl, M.; Zablotski, Y.; Speck, S.; Truyen, U.; Hartmann, K. Antibody Response to Canine Adenovirus-2 Virus Vaccination in Healthy Adult Dogs. Viruses 2020, 12, 1198. [Google Scholar] [CrossRef]
- Park, N.Y.; Lee, M.C.; Kurkure, N.V.; Cho, H.S. Canine Adenovirus Type 1 Infection of a Eurasian River Otter (Lutra lutra). Vet. Pathol. 2007, 44, 536–539. [Google Scholar] [CrossRef]
- Decaro, N.; Martella, V.; Buonavoglia, C. Canine Adenoviruses and Herpesvirus. Vet. Clin. North. Am. Small Anim. Pract. 2008, 38, 799–814. [Google Scholar] [CrossRef]
- Knowles, S.; Bodenstein, B.L.; Hamon, T.; Saxton, M.W.; Hall, J.S. Infectious Canine Hepatitis in a Brown Bear (Ursus arctos horribilis) from Alaska, USA. J. Wildl. Dis. 2018, 54, 642–645. [Google Scholar] [CrossRef]
- Balboni, A.; Tryland, M.; Mørk, T.; Killengreen, S.T.; Fuglei, E.; Battilani, M. Unique Genetic Features of Canine Adenovirus Type 1 (CAdV-1) Infecting Red Foxes (Vulpes vulpes) in Northern Norway and Arctic Foxes (Vulpes lagopus) in Svalbard. Vet. Res. Commun. 2019, 43, 67–76. [Google Scholar] [CrossRef] [Green Version]
- Verin, R.; Forzan, M.; Schulze, C.; Rocchigiani, G.; Balboni, A.; Poli, A.; Mazzei, M. Multicentric Molecular and Pathologic Study on Canine Adenovirus Type 1 in Red Foxes (Vulpes vulpes) in Three European Countries. J. Wildl. Dis. 2019, 55, 935. [Google Scholar] [CrossRef] [PubMed]
- Oleaga, A.; Balseiro, A.; Espí, A.; Royo, L.J. Wolf (Canis lupus) as Canine Adenovirus Type 1 (CAdV-1) Sentinel for the Endangered Cantabrian Brown Bear (Ursus arctos arctos). Transbound. Emerg. Dis. 2022, 69, 516–523. [Google Scholar] [CrossRef] [PubMed]
- Balboni, A.; Urbani, L.; Delogu, M.; Musto, C.; Fontana, M.C.; Merialdi, G.; Lucifora, G.; Terrusi, A.; Dondi, F.; Battilani, M. Integrated Use of Molecular Techniques to Detect and Genetically Characterise DNA Viruses in Italian Wolves (Canis lupus italicus). Animals 2021, 11, 2198. [Google Scholar] [CrossRef] [PubMed]
- Ndiana, L.A.; Lanave, G.; Desario, C.; Berjaoui, S.; Alfano, F.; Puglia, I.; Fusco, G.; Colaianni, M.L.; Vincifori, G.; Camarda, A.; et al. Circulation of Diverse Protoparvoviruses in Wild Carnivores, Italy. Transbound Emerg Dis 2020, 68, 2489–2502. [Google Scholar] [CrossRef]
- Balboni, A.; Verin, R.; Morandi, F.; Poli, A.; Prosperi, S.; Battilani, M. Molecular Epidemiology of Canine Adenovirus Type 1 and Type 2 in Free-Ranging Red Foxes (Vulpes vulpes) in Italy. Vet. Microbiol. 2013, 162, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Balboni, A.; Mollace, C.; Giunti, M.; Dondi, F.; Prosperi, S.; Battilani, M. Investigation of the Presence of Canine Adenovirus (CAdV) in Owned Dogs in Northern Italy. Res. Vet. Sci. 2014, 97, 631–636. [Google Scholar] [CrossRef] [PubMed]
- Balboni, A.; Musto, C.; Kaehler, E.; Verin, R.; Caniglia, R.; Fabbri, E.; Carra, E.; Cotti, C.; Battilani, M.; Delogu, M. Genetic Characterization of Canine Adenovirus Type 1 Detected by Real-Time Polymerase Chain Reaction in an Oral Sample of an Italian Wolf (Canis lupus). J. Wildl. Dis. 2019, 55, 737. [Google Scholar] [CrossRef]
- Dowgier, G.; Lahoreau, J.; Lanave, G.; Losurdo, M.; Varello, K.; Lucente, M.S.; Ventriglia, G.; Bozzetta, E.; Martella, V.; Buonavoglia, C.; et al. Sequential Circulation of Canine Adenoviruses 1 and 2 in Captive Wild Carnivores, France. Vet. Microbiol. 2018, 221, 67–73. [Google Scholar] [CrossRef]
- García Marín, J.F.; Royo, L.J.; Oleaga, A.; Gayo, E.; Alarcia, O.; Pinto, D.; Martínez, I.Z.; González, P.; Balsera, R.; Marcos, J.L.; et al. Canine Adenovirus Type 1 (CAdV-1) in Free-Ranging European Brown Bear (Ursus arctos arctos): A Threat for Cantabrian Population? Transbound. Emerg. Dis. 2018, 65, 2049–2056. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Ochiai, K.; Itakura, C. Dual Infection with Canine Distemper Virus and Infectious Canine Hepatitis Virus (Canine Adenovirus Type 1) in a Dog. J. Vet. Med. Sci. 1993, 55, 699–701. [Google Scholar] [CrossRef] [Green Version]
- Müller, C.; Sieber-Ruckstuhl, N.; Decaro, N.; Keller, S.; Quante, S.; Tschuor, F.; Wenger, M.; Reusch, C. Hepatitis contagiosa canis Infektion bei 4 Hunden in der Schweiz. Schweiz. Arch. Tierheilk. 2010, 152, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Alves, C.D.B.T.; Granados, O.F.O.; da F. Budaszewski, R.; Streck, A.F.; Weber, M.N.; Cibulski, S.P.; Pinto, L.D.; Ikuta, N.; Canal, C.W. Identification of Enteric Viruses Circulating in a Dog Population with Low Vaccine Coverage. Braz. J. Microbiol. 2018, 49, 790–794. [Google Scholar] [CrossRef] [PubMed]
- Headley, S.A.; Oliveira, T.E.S.; Pereira, A.H.T.; Moreira, J.R.; Michelazzo, M.M.Z.; Pires, B.G.; Marutani, V.H.B.; Xavier, A.A.C.; Di Santis, G.W.; Garcia, J.L.; et al. Canine Morbillivirus (Canine Distemper Virus) with Concomitant Canine Adenovirus, Canine Parvovirus-2, and Neospora Caninum in Puppies: A Retrospective Immunohistochemical Study. Sci. Rep. 2018, 8, 13477. [Google Scholar] [CrossRef]
- Hornsey, S.J.; Philibert, H.; Godson, D.L.; Snead, E.C.R. Canine Adenovirus Type 1 Causing Neurological Signs in a 5-Week-Old Puppy. BMC Vet. Res. 2019, 15, 418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pratelli, A.; Martella, V.; Elia, G.; Tempesta, M.; Guarda, F.; Capucchio, M.T.; Carmichael, L.E.; Buonavoglia, C. Severe Enteric Disease in an Animal Shelter Associated with Dual Infections by Canine Adenovirus Type 1 and Canine Coronavirus. J. Vet. Med. B 2001, 48, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Decaro, N.; Campolo, M.; Elia, G.; Buonavoglia, D.; Colaianni, M.L.; Lorusso, A.; Mari, V.; Buonavoglia, C. Infectious Canine Hepatitis: An “Old” Disease Reemerging in Italy. Res. Vet. Sci. 2007, 83, 269–273. [Google Scholar] [CrossRef]
- Balboni, A.; Dondi, F.; Agnoli, C.; Verin, R.; Gruarin, M.; Morini, M.; Battilani, M. Novel Sequence Variants of Viral Hexon and Fibre Genes in Two Dogs with Canine Adenovirus Type 1-Associated Disease. Vet. J. 2017, 223, 73–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pintore, M.D.; Corbellini, D.; Chieppa, M.N.; Vallino Costassa, E.; Florio, C.L.; Varello, K.; Bozzetta, E.; Adriano, D.; Decaro, N.; Casalone, C.; et al. Canine Adenovirus Type 1 and Pasteurella Pneumotropica Co-infection in a Puppy. Vet. Ital. 2016, 52, 57–62. [Google Scholar] [CrossRef]
- Purpari, G.; Mira, F.; Di Bella, S.; Di Pietro, S.; Giudice, E.; Guercio, A. Investigation on Circulation in Dogs from Sicily (Italy) by Biomolecular Assay. Acta Vet. 2018, 68, 80–94. [Google Scholar] [CrossRef] [Green Version]
- Hu, R.L.; Huang, G.; Qiu, W.; Zhong, Z.H.; Xia, X.Z.; Yin, Z. Detection and Differentiation of CAV-1 and CAV-2 by Polymerase Chain Reaction. Vet. Res. Commun. 2001, 25, 77–84. [Google Scholar] [CrossRef]
- Touihri, L.; Bouzid, I.; Daoud, R.; Desario, C.; El Goulli, A.F.; Decaro, N.; Ghorbel, A.; Buonavoglia, C.; Bahloul, C. Molecular Characterization of Canine Parvovirus-2 Variants Circulating in Tunisia. Virus Genes 2009, 38, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Visser, I.K.G.; Mamaev, L.; Goatley, L.; van Bressem, M.-F.; Osterhaus, A.D.M.E. Dolphin and Porpoise Morbilliviruses Are Genetically Distinct from Phocine Distemper Virus. Virology 1993, 193, 1010–1012. [Google Scholar] [CrossRef] [PubMed]
- Pratelli, A.; Martella, V.; Elia, G.; Decaro, N.; Aliberti, A.; Buonavoglia, D.; Tempesta, M.; Buonavoglia, C. Variation of the Sequence in the Gene Encoding for Transmembrane Protein M of Canine Coronavirus (CCV). Mol. Cell. Probes 2001, 15, 229–233. [Google Scholar] [CrossRef]
- Freeman, M.M.; Kerin, T.; Hull, J.; McCaustland, K.; Gentsch, J. Enhancement of Detection and Quantification of Rotavirus in Stool Using a Modified Real-Time RT-PCR Assay. J. Med. Virol. 2008, 80, 1489–1496. [Google Scholar] [CrossRef] [PubMed]
- Ogbu, K.I.; Mira, F.; Purpari, G.; Nwosuh, C.; Loria, G.R.; Schirò, G.; Chiaramonte, G.; Tion, M.T.; Di Bella, S.; Ventriglia, G.; et al. Nearly Full-length Genome Characterization of Canine Parvovirus Strains Circulating in Nigeria. Transbound. Emerg. Dis. 2020, 67, 635–647. [Google Scholar] [CrossRef] [Green Version]
- Pratelli, A.; Decaro, N.; Tinelli, A.; Martella, V.; Elia, G.; Tempesta, M.; Cirone, F.; Buonavoglia, C. Two Genotypes of Canine Coronavirus Simultaneously Detected in the Fecal Samples of Dogs with Diarrhea. J. Clin. Microbiol. 2004, 42, 1797–1799. [Google Scholar] [CrossRef] [Green Version]
- Erles, K.; Brownlie, J. Sequence Analysis of Divergent Canine Coronavirus Strains Present in a UK Dog Population. Virus Res. 2009, 141, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Li, W.; Hu, W.; Xu, X.; Kan, Y.; Yao, L.; Bi, Y.; Xie, Q. Novel Genotype Definition and the First Epidemiological Investigation of Canine Adenovirus Type 2 in Dogs in Central China. Front. Vet. Sci. 2020, 7, 534. [Google Scholar] [CrossRef]
- Hall, T.A. Bioedit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program For Windows 95/98/ Nt. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar] [CrossRef]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A Greedy Algorithm for Aligning DNA Sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Pizzurro, F.; Marcacci, M.; Zaccaria, G.; Orsini, M.; Cito, F.; Rosamilia, A.; Di Renzo, L.; Malatesta, D.; Di Sabatino, D.; Lorusso, A. Genome Sequence of Canine Adenovirus Type 1 Isolated from a Wolf (Canis lupus) in Southern Italy. Genome Announc. 2017, 5, e00225-17. [Google Scholar] [CrossRef] [PubMed]
- Cardillo, L.; Piegari, G.; Iovane, V.; Viscardi, M.; Alfano, F.; Cerrone, A.; Pagnini, U.; Montagnaro, S.; Galiero, G.; Pisanelli, G.; et al. Lifestyle as Risk Factor for Infectious Causes of Death in Young Dogs: A Retrospective Study in Southern Italy (2015–2017). Vet. Med. Int. 2020, 2020, 1–10. [Google Scholar] [CrossRef]
- Piegari, G.; Cardillo, L.; Alfano, F.; Vangone, L.; Iovane, V.; Fusco, G. Pathological, Bacteriological and Virological Findings in Sudden and Unexpected Deaths in Young Dogs. Animals 2020, 10, 1134. [Google Scholar] [CrossRef] [PubMed]
- Decaro, N.; Desario, C.; Elia, G.; Campolo, M.; Lorusso, A.; Mari, V.; Martella, V.; Buonavoglia, C. Occurrence of Severe Gastroenteritis in Pups after Canine Parvovirus Vaccine Administration: A Clinical and Laboratory Diagnostic Dilemma. Vaccine 2007, 25, 1161–1166. [Google Scholar] [CrossRef] [PubMed]
- Duijvestijn, M.; Mughini-Gras, L.; Schuurman, N.; Schijf, W.; Wagenaar, J.A.; Egberink, H. Enteropathogen Infections in Canine Puppies: (Co-)Occurrence, Clinical Relevance and Risk Factors. Vet. Microbiol. 2016, 195, 115–122. [Google Scholar] [CrossRef] [Green Version]
- Dowgier, G.; Lorusso, E.; Decaro, N.; Desario, C.; Mari, V.; Lucente, M.S.; Lanave, G.; Buonavoglia, C.; Elia, G. A Molecular Survey for Selected Viral Enteropathogens Revealed a Limited Role of Canine Circovirus in the Development of Canine Acute Gastroenteritis. Vet. Microbiol. 2017, 204, 54–58. [Google Scholar] [CrossRef]
- Caudell, D.; Confer, A.W.; Fulton, R.W.; Berry, A.; Saliki, J.T.; Fent, G.M.; Ritchey, J.W. Diagnosis of Infectious Canine Hepatitis Virus (CAV-1) Infection in Puppies with Encephalopathy. J. Vet. Diagn. Invest. 2005, 17, 58–61. [Google Scholar] [CrossRef] [Green Version]
- Sykes, J.E.; Greene, C.E. Infectious Diseases of the Dog and Cat, 4th ed.; Elsevier Health Sciences: St. Louis, MO, USA, 2013. [Google Scholar]
- Fujimoto, Y. Studies on Infectious Canine Hepatitis I: Histopathological Studies on Experimental Cases. Jpn. J. Vet. Res. 1957, 5, 51–70. [Google Scholar] [CrossRef]
- Balboni, A.; Dondi, F.; Prosperi, S.; Battilani, M. Development of a SYBR Green Real-Time PCR Assay with Melting Curve Analysis for Simultaneous Detection and Differentiation of Canine Adenovirus Type 1 and Type 2. J. Virol. Methods 2015, 222, 34–40. [Google Scholar] [CrossRef]
- Canuti, M.; Fry, K.; Cluff, H.D.; Mira, F.; Fenton, H.; Lang, A.S. Co-Circulation of Five Species of Dog Parvoviruses and Canine Adenovirus Type 1 among Gray Wolves (Canis lupus) in Northern Canada. Transbound. Emerg. Dis. 2022, 69, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Ochi, Y.; Konishi, S.; Takizawa, T.; Ikegami, T.; Yamamoto, S.; Ishida, K.; Sato, A. Studies on Infectious Hepatitis in the Dog (I). Zentralbl. Veterinärmed. 2010, 3, 55–62. [Google Scholar] [CrossRef]
- Mira, F.; Purpari, G.; Di Bella, S.; Vicari, D.; Schirò, G.; Di Marco, P.; Macaluso, G.; Battilani, M.; Guercio, A. Update on Canine Distemper Virus (CDV) Strains of Arctic-like Lineage Detected in Dogs in Italy. Vet. Ital. 2018, 54, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Mira, F.; Purpari, G.; Di Bella, S.; Colaianni, M.L.; Schirò, G.; Chiaramonte, G.; Gucciardi, F.; Pisano, P.; Lastra, A.; Decaro, N.; et al. Spreading of Canine Parvovirus Type 2c Mutants of Asian Origin in Southern Italy. Transbound. Emerg. Dis. 2019, 66, 2297–2304. [Google Scholar] [CrossRef]
- Mira, F.; Purpari, G.; Lorusso, E.; Di Bella, S.; Gucciardi, F.; Desario, C.; Macaluso, G.; Decaro, N.; Guercio, A. Introduction of Asian Canine Parvovirus in Europe through Dog Importation. Transbound. Emerg. Dis. 2018, 65, 16–21. [Google Scholar] [CrossRef]
- Walker, D.; Abbondati, E.; Cox, A.L.; Mitchell, G.B.; Pizzi, R.; Sharp, C.P.; Philbey, A.W. Infectious canine hepatitis in red foxes (Vulpes vulpes) in wildlife rescue centres in the UK. Vet. Rec. 2016, 178, 421. [Google Scholar] [CrossRef]
- De Jonge, B.; Van Brantegem, L.; Chiers, K. Infectious canine hepatitis, not only in the textbooks: A brief review and three case reports. Vlaams. Diergeneeskd. Tijdschr. 2020, 89, 284–291. [Google Scholar] [CrossRef]
- Ford, R.B.; Larson, L.J.; McClure, K.D.; Schultz, R.D.; Welborn, L.V. 2017 AAHA Canine Vaccination Guidelines. J. Am. Anim. Hosp. Assoc. 2017, 53, 243–251. [Google Scholar] [CrossRef]
- Day, M.J.; Horzinek, M.C.; Schultz, R.D.; Squires, R.A. Vaccination Guidelines Group (VGG) of the World Small Animal Veterinary Association (WSAVA). WSAVA Guidelines for the vaccination of dogs and cats. J. Small Anim. Pract. 2016, 57, E1–E45. [Google Scholar] [CrossRef] [Green Version]
- Dall’Ara, P. Vaccini e vaccinazioni core del cane. In Vaccini e Vaccinazioni degli Animali da Compagnia; EDRA: Milano, Italy, 2020; pp. 153–174. [Google Scholar]
- Di Sabatino, D.; Di Francesco, G.; Zaccaria, G.; Malatesta, D.; Brugnola, L.; Marcacci, M.; Portanti, O.; De Massis, F.; Savini, G.; Teodori, L.; et al. Lethal Distemper in Badgers (Meles meles) Following Epidemic in Dogs and Wolves. Infect. Genet. Evol. 2016, 46, 130–137. [Google Scholar] [CrossRef]



| Dog id. | Date of Sampling | Age | Breed | Origin | Viral Co-Infections | CAdV-1 Strain | GenBank Accession Number | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Penton | Hexon | E3 | Fiber | |||||||
| 1 | 19-Sep-2017 | 2 m | Mixed | Stray | CPV-2c | CAdV-1_IZSSI_CL8747_17_Milza | MZ313227 | MW650911 | MW650917 | MW650923 |
| 2 | 01-Apr-2019 | 2 m | Mixed | Stray | CPV-2a | CAdV-1_IZSSI_PA8775_19_Polmone | MZ313228 | MW650912 | MW650918 | MW650924 |
| 3 | 06-May-2019 | 12 m | Mixed | Stray | CPV-2c, CCoV-IIa | CAdV-1_IZSSI_PA11822_19_Polmone | MZ313229 | MW650913 | MW650919 | MW650925 |
| 4 | 09-Oct-2019 | 4 m | Mixed | Stray | CPV-2b | CAdV-1_IZSSI_RG17627_19_Milza | MZ313230 | MW650914 | MW650920 | MW650926 |
| 5 | 12-Dec-2019 | 11 m | Mixed | Stray | CCoV-IIa | CAdV-1_IZSSI_PA34446_19_Fegato | MZ313231 | MW650915 | MW650921 | MW650927 |
| 6 | 17-Nov-2020 | 2 m | Mixed | Stray | no | CAdV-1_IZSSI_PA98748_20_Cervello | MZ313232 | MW650916 | MW650922 | MW650928 |
| Nucleotide and (Amino Acid) Positions | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Penton a | Hexon b | E3 c | U-exon d | Fiber e | |||||||||||||||||
| Strain/Isolate | Host | Country | Year | 414 (138) | 486 (162) | 545 (182) | 1128 (376) | 1163 (388) | 1860 (620) | 2055 (685) | 2238 (746) | 406 (136) | 427 (143) | 80 (27) | 69 (23) | 330 (110) | 513 (171) | 765 (255) | 1162 (388) | 1460 (487) | 1608 (536) |
| CAdV-1 ITL2015 1,2 | Wolf | Italy | 2015 | G (Ala) | T (Leu) | C (Thr) | A (Gly) | G (Ser) | T (Pro) | T (Gly) | C (Phe) | A (Ser) | T (Leu) | C (Thr) | A (Pro) | C (Asp) | T (Pro) | A (Ala) | A (Ile) | C (Ala) | C (Phe) |
| D43 1 | Dog | Japan | 1954 | C (Thr) | A (Gly) | A (Asn) |
T (Pro) |
C (Gly) |
C (Phe) |
A (Ser) |
C (Leu) |
C (Thr) |
C (Pro) |
A (Glu) |
T (Pro) |
A (Ala) |
A (Ile) |
C (Ala) |
C (Phe) | ||
| CAdV-1_IZSSI_CL8747_17_Milza | Dog | Italy | 2017 | A (Asn) | C (-) | C (-) | T (Ile) | C (-) | A (Glu) | T (Val) | T (-) | ||||||||||
| CAdV-1_IZSSI_PA8775_19_Polmone | Dog | Italy | 2019 | A (Asn) | C (-) | C (-) | C (-) | A (Glu) | T (Val) | T (-) | |||||||||||
| CAdV-1_IZSSI_PA11822_19_Polmone | Dog | Italy | 2019 | A (Asn) | C (-) | C (-) | C (-) | A (Glu) | T (Val) | T (-) | |||||||||||
| CAdV-1_IZSSI_RG17627_19_Milza | Dog | Italy | 2019 | A (Asn) | A (Asn) | C (-) | C (-) | G (Gly) | C (-) | T (Ile) | C (-) | A (Glu) | T (Val) | T (-) | |||||||
| CAdV-1_IZSSI_PA34446_19_Fegato | Dog | Italy | 2019 | A (-) | C (-) | A (Asn) | C (-) | T (-) | C (-) | C (-) | A (Glu) | G (-) | G (Val) | ||||||||
| CAdV-1_IZSSI_PA98748_20_Cervello | Dog | Italy | 2020 | G (-) | A (Asn) | C (-) | C (-) | C (-) | A (Glu) | C (-) | |||||||||||
| Fox/466/2017/ITA 2 | Fox | Italy | 2017 | n.a. 3 |
G (Ser) |
T (-) | n.a. 3 |
C (-) |
C (Asp) | ||||||||||||
| 874/2014 2 | Wolf | Italy | 2014 | n.a. 3 |
G (Ser) |
T (-) |
C (-) |
C (Thr) |
A (Glu) | ||||||||||||
| 574-2013-RS 2 | Dog | Italy | 2013 | n.a. 3 |
G (Ser) |
T (-) |
C (-) |
C (-) |
C (Asp) | ||||||||||||
| 417-2013-L 2 | Dog | Italy | 2013 | n.a. 3 |
G (Ser) |
T (-) |
C (-) |
C (Thr) |
A (Glu) | ||||||||||||
| 113-5L 2 | Fox | Italy | 2013 | n.a. 3 |
G (Ser) |
T (-) |
C (-) |
C (Thr) |
A (Glu) | ||||||||||||
| 300-2012-RS 2 | Dog | Italy | 2012 | n.a. 3 | n.a. 3 | C (-) | n.a. 3 | ||||||||||||||
| 09-13F 2 | Fox | Italy | 2011 | n.a. 3 | n.a. 3 | C (-) | n.a. 3 | ||||||||||||||
| 313-2010-Lparaffin 2 | Dog | Italy | 2010 | n.a. 3 | n.a. 3 | C (-) | n.a. 3 | ||||||||||||||
| 384-1966-FFPEL 2 | Dog | Italy | 1966 | n.a. 3 | n.a. 3 | C (-) | n.a. 3 | ||||||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mira, F.; Puleio, R.; Schirò, G.; Condorelli, L.; Di Bella, S.; Chiaramonte, G.; Purpari, G.; Cannella, V.; Balboni, A.; Randazzo, V.; et al. Study on the Canine Adenovirus Type 1 (CAdV-1) Infection in Domestic Dogs in Southern Italy. Pathogens 2022, 11, 1254. https://doi.org/10.3390/pathogens11111254
Mira F, Puleio R, Schirò G, Condorelli L, Di Bella S, Chiaramonte G, Purpari G, Cannella V, Balboni A, Randazzo V, et al. Study on the Canine Adenovirus Type 1 (CAdV-1) Infection in Domestic Dogs in Southern Italy. Pathogens. 2022; 11(11):1254. https://doi.org/10.3390/pathogens11111254
Chicago/Turabian StyleMira, Francesco, Roberto Puleio, Giorgia Schirò, Lucia Condorelli, Santina Di Bella, Gabriele Chiaramonte, Giuseppa Purpari, Vincenza Cannella, Andrea Balboni, Vincenzo Randazzo, and et al. 2022. "Study on the Canine Adenovirus Type 1 (CAdV-1) Infection in Domestic Dogs in Southern Italy" Pathogens 11, no. 11: 1254. https://doi.org/10.3390/pathogens11111254
APA StyleMira, F., Puleio, R., Schirò, G., Condorelli, L., Di Bella, S., Chiaramonte, G., Purpari, G., Cannella, V., Balboni, A., Randazzo, V., Antoci, F., Vicari, D., & Guercio, A. (2022). Study on the Canine Adenovirus Type 1 (CAdV-1) Infection in Domestic Dogs in Southern Italy. Pathogens, 11(11), 1254. https://doi.org/10.3390/pathogens11111254







