High Rates of Detection and Molecular Characterization of Porcine Adenovirus Serotype 5 (Porcine mastadenovirus C) from Diarrheic Pigs
Abstract
1. Introduction
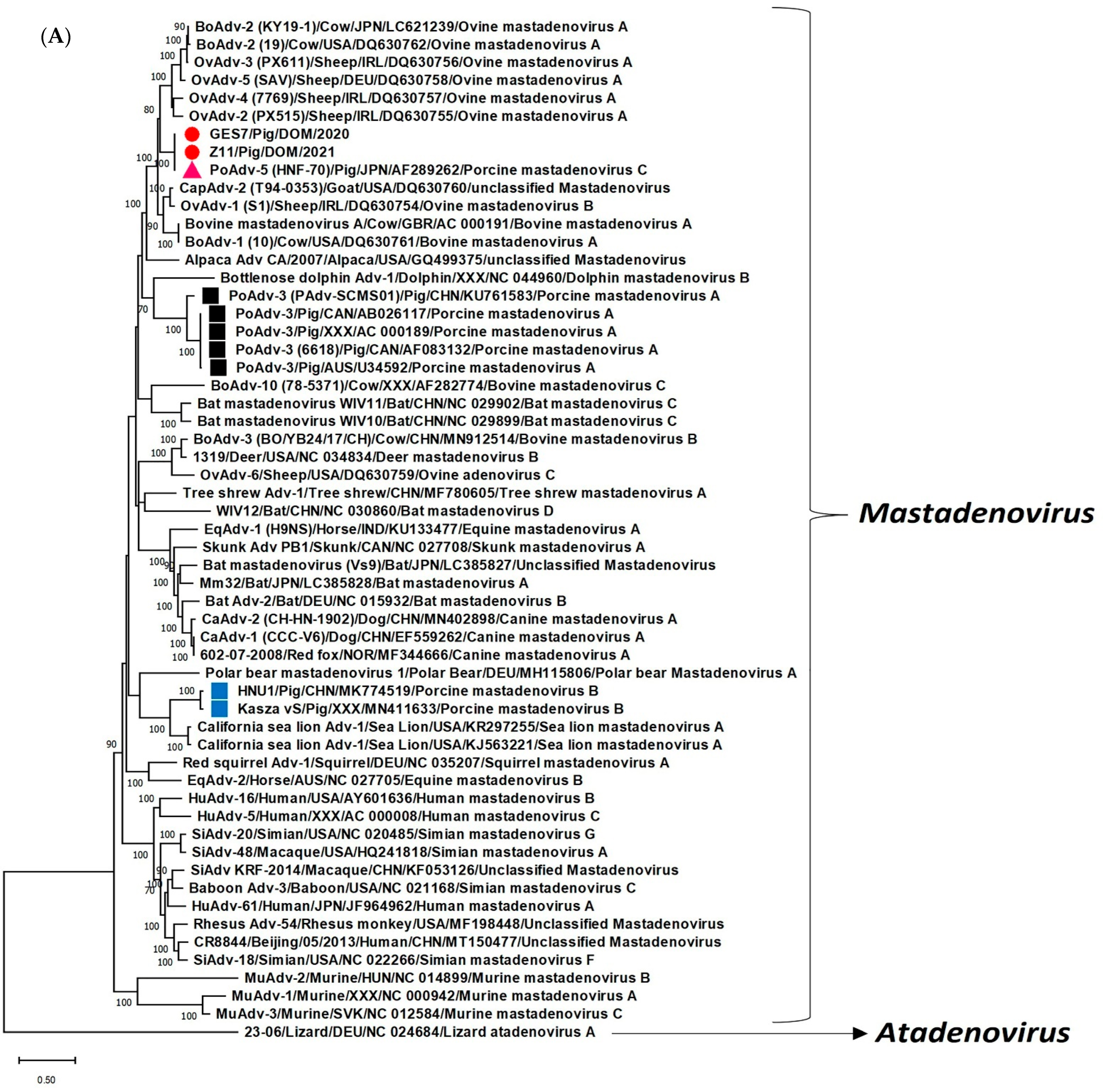
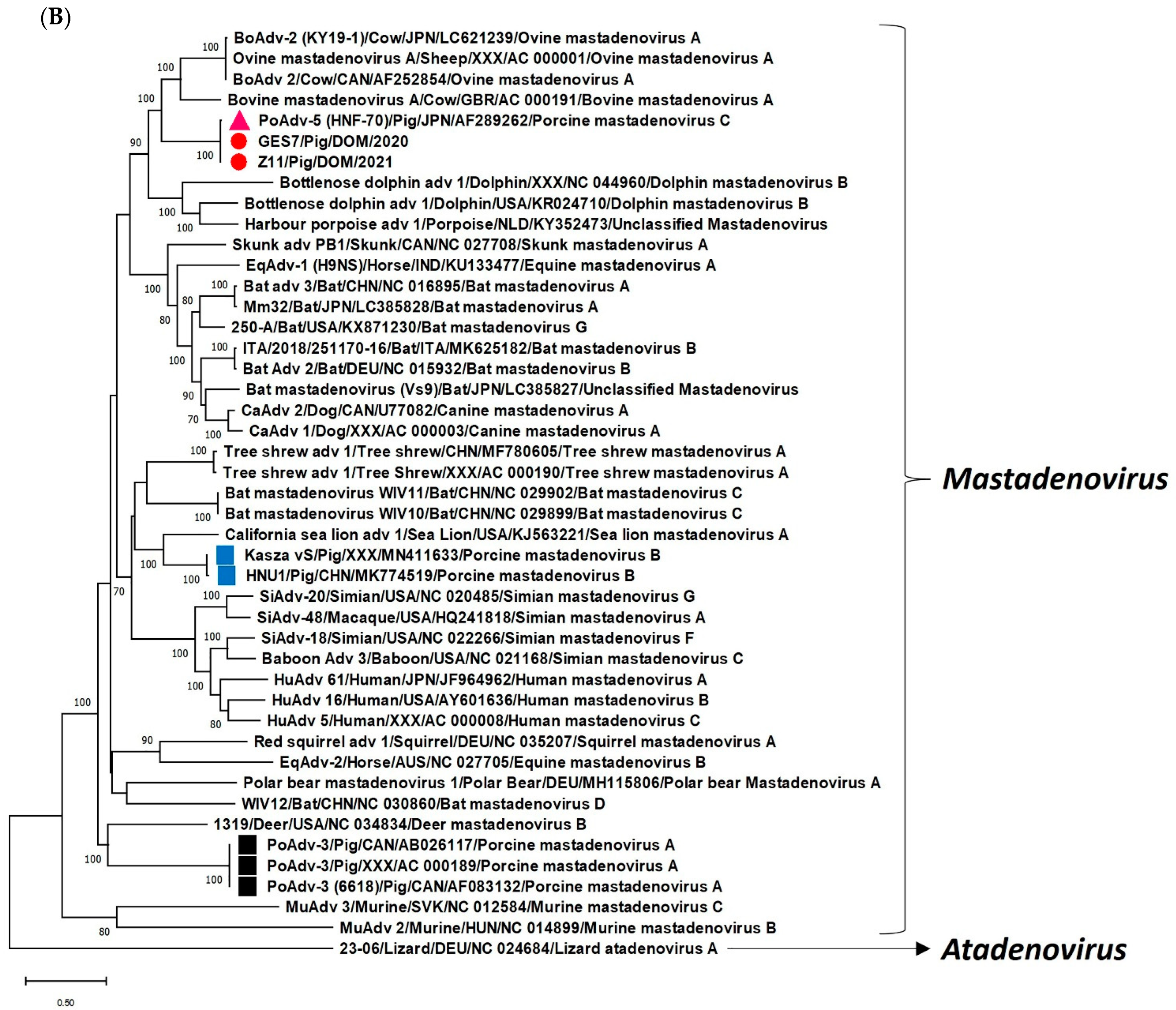
2. Results and Discussion
3. Materials and Methods
3.1. Sampling
3.2. Amplification of Viral Genome
3.3. Nucleotide Sequencing
3.4. Sequence Analysis
3.5. GenBank Accession Numbers
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Benkő, M.; Aoki, K.; Arnberg, N.; Davison, A.J.; Echavarría, M.; Hess, M.; Jones, M.S.; Kaján, G.L.; Kajon, A.E.; Mittal, S.K.; et al. ICTV Virus Taxonomy Profile: Adenoviridae 2022. J. Gen. Virol. 2022, 103, 001721. [Google Scholar] [CrossRef] [PubMed]
- Benfield, D.A.; Hesse, R.A. Adenoviruses. In Diseases of Swine; Wiley: Hoboken, NJ, USA, 2019; pp. 438–442. [Google Scholar] [CrossRef]
- Nagy, M.; Tuboly, T. Porcine Adenoviruses: An Update on Genome Analysis and Vector Development. Acta Vet. Hung. 2000, 48, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Sibley, S.D.; Goldberg, T.L.; Pedersen, J.A. Detection of Known and Novel Adenoviruses in Cattle Wastes via Broad-Spectrum Primers. Appl. Environ. Microbiol. 2011, 77, 5001–5008. [Google Scholar] [CrossRef]
- Jerman, U.; Kolenc, M.; Steyer, A.; Veranič, P.; Prijatelj, M.; Kreft, M. A Novel Strain of Porcine Adenovirus Detected in Urinary Bladder Urothelial Cell Culture. Viruses 2014, 6, 2505–2518. [Google Scholar] [CrossRef]
- Liu, S.-J.; Wang, Q.; Li, T.-T.; Zhang, S.-H.; Li, J.-Y.; Wu, L.-J.; Qiu, Y.; Ge, X.-Y. Characterization of the First Genome of Porcine Mastadenovirus B (HNU1 Strain) and Implications on Its Lymphoid and Special Origin. Virol. Sin. 2020, 35, 528–537. [Google Scholar] [CrossRef]
- Horak, S.; Leedom Larsen, K. Porcine Adenovirus; Iowa State University: Ames, IA, USA, 2016; Available online: http://www.cfsph.iastate.edu/pdf/shic-factsheet-porcine-adenovirus (accessed on 28 July 2022).
- Tuboly, T.; Nagy, É. Construction and Characterization of Recombinant Porcine Adenovirus Serotype 5 Expressing the Transmissible Gastroenteritis Virus Spike Gene. J. Gen. Virol. 2001, 82, 183–190. [Google Scholar] [CrossRef]
- Kumthip, K.; Khamrin, P.; Kongkaew, A.; Vachirachewin, R.; Malasao, R.; Ushijima, H.; Maneekarn, N. Molecular Epidemiology and Characterization of Porcine Adenoviruses in Pigs with Diarrhea in Thailand. Infect. Genet. Evol. 2019, 67, 73–77. [Google Scholar] [CrossRef]
- Elazhary, M.A.; Dea, S.; Mittal, K.R.; Higgins, R. Prevalence of Antibodies to Swine Influenza Virus, Porcine Adenovirus Type 4 and Haemophilus Pleuropneumoniae in Quebec Pig Farms with Respiratory Problems. Can. Vet. J. 1985, 26, 190–192. [Google Scholar]
- Abid, H.; Holscher, M.; Byerly, C. An Outbreak of Adenovirus Enteritis in Piglets. Vet Med. Sm. An. Clin. 1984, 79, 105. [Google Scholar]
- Nagy, M.; Nagy, É.; Tuboly, T. The Complete Nucleotide Sequence of Porcine Adenovirus Serotype 5. J. Gen. Virol. 2001, 82, 525–529. [Google Scholar] [CrossRef]
- Davison, A.J.; Benkő, M.; Harrach, B. Genetic Content and Evolution of Adenoviruses. J. Gen. Virol. 2003, 84, 2895–2908. [Google Scholar] [CrossRef] [PubMed]
- Reddy, P.S.; Idamakanti, N.; Song, J.-Y.; Lee, J.-B.; Hyun, B.-H.; Park, J.-H.; Cha, S.-H.; Bae, Y.-T.; Tikoo, S.K.; Babiuk, L.A. Nucleotide Sequence and Transcription Map of Porcine Adenovirus Type 3. Virology 1998, 251, 414–426. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hirahara, T.; Yasuhara, H.; Matsui, O.; Yamanaka, M.; Tanaka, M.; Fukuyama, S.; Izumida, A.; Yoshiki, K.; Kodama, K.; Nakai, M.; et al. Isolation of Porcine Adenovirus from the Respiratory Tract of Pigs in Japan. JPN J. Vet. Sci. 1990, 52, 407–409. [Google Scholar] [CrossRef] [PubMed]
- Kadoi, K.; Inoue, Y.; Ikeda, T.; Kamata, H.; Yukawa, M.; Iwabuchi, M.; Inaba, Y. Isolation of Porcine Adenovirus as a Candidate of 5th Serotype. J. Basic Microbiol. 1995, 35, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; Hewitt, J.; Greening, G.E. Viral Multiplex Quantitative PCR Assays for Tracking Sources of Fecal Contamination. Appl. Environ. Microbiol. 2010, 76, 1388–1394. [Google Scholar] [CrossRef] [PubMed]
- Hause, B.; Duff, J.; Scheidt, A.; Anderson, G. Virus Detection Using Metagenomic Sequencing of Swine Nasal and Rectal Swabs. J. Swine Health Prod. 2016, 24, 304–308. Available online: https://www.aasv.org/jshap/issues/v24n6/v24n6p304.pdf (accessed on 28 July 2022).
- Hundesa, A.; Maluquer de Motes, C.; Bofill-Mas, S.; Albinana-Gimenez, N.; Girones, R. Identification of Human and Animal Adenoviruses and Polyomaviruses for Determination of Sources of Fecal Contamination in the Environment. Appl. Environ. Microbiol. 2006, 72, 7886–7893. [Google Scholar] [CrossRef]
- Shih, Y.-J.; Tao, C.-W.; Tsai, H.-C.; Huang, W.-C.; Huang, T.-Y.; Chen, J.-S.; Chiu, Y.-C.; Hsu, T.-K.; Hsu, B.-M. First Detection of Enteric Adenoviruses Genotype 41 in Recreation Spring Areas of Taiwan. Environ. Sci. Pollut. Res. 2017, 24, 18392–18399. [Google Scholar] [CrossRef]
- Chen, J.-S.; Tsai, H.-C.; Nagarajan, V.; Hsu, B.-M. Adenovirus in Fishery Harbours and Identification of Contamination Sources. Water Res. 2022, 219, 118538. [Google Scholar] [CrossRef]
- Nagarajan, V.; Chen, J.; Hsu, B.; Hsu, G.; Wang, J.; Hussain, B. Prevalence, Distribution, and Genotypes of Adenovirus and Norovirus in the Puzi River and Its Tributaries and the Surrounding Areas in Taiwan. Geohealth 2021, 5, e2021GH000465. [Google Scholar] [CrossRef]
- Gainor, K.; Becker, A.A.M.J.; Malik, Y.S.; Ghosh, S. First Report on Detection and Molecular Characterization of Adenoviruses in the Small Indian Mongoose (Urva Auropunctata). Viruses 2021, 13, 2194. [Google Scholar] [CrossRef] [PubMed]
- Wellehan, J.F.X.; Johnson, A.J.; Harrach, B.; Benko, M.; Pessier, A.P.; Johnson, C.M.; Garner, M.M.; Childress, A.; Jacobson, E.R. Detection and Analysis of Six Lizard Adenoviruses by Consensus Primer PCR Provides Further Evidence of a Reptilian Origin for the Atadenoviruses. J. Virol. 2004, 78, 13366–13369. [Google Scholar] [CrossRef] [PubMed]
- Kleymann, A.; Soto, E.; Illanes, O.; Malik, Y.S.; Fuentealba, C.; Ghosh, S. High Rates of Detection and Complete Genomic Analysis of Porcine Circovirus 2 (PCV2) in the Lesser Antilles Island of St. Kitts: Identification of PCV2b-PCV2d Recombinants. Transbound. Emerg. Dis. 2020, 67, 2282–2289. [Google Scholar] [CrossRef] [PubMed]
- Navarro, R.; Aung, M.S.; Cruz, K.; Ketzis, J.; Gallagher, C.A.; Beierschmitt, A.; Malik, Y.S.; Kobayashi, N.; Ghosh, S. Whole Genome Analysis Provides Evidence for Porcine-to-Simian Interspecies Transmission of Rotavirus-A. Infect. Genet. Evol. 2017, 49, 21–31. [Google Scholar] [CrossRef]
- Russell, W.C. Adenoviruses: Update on Structure and Function. J. Gen. Virol. 2009, 90, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Hoeben, R.C.; Uil, T.G. Adenovirus DNA Replication. Cold Spring Harb. Perspect. Biol. 2013, 5, a013003. [Google Scholar] [CrossRef]
- Segalés, J.; Sibila, M. Revisiting Porcine Circovirus Disease Diagnostic Criteria in the Current Porcine Circovirus 2 Epidemiological Context. Vet. Sci. 2022, 9, 110. [Google Scholar] [CrossRef]
- Nagy, M.; Nagy, É.; Tuboly, T. Sequence Analysis of Porcine Adenovirus Serotype 5 Fibre Gene: Evidence for Recombination. Virus Genes 2002, 24, 181–185. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
| Animal/Sample Number | Age Group/Category of Animal 1 | Location of the Pig Farm | Year of Sample Collection | Coinfection with | |
|---|---|---|---|---|---|
| Porcine circovirus 2 2 | Rotavirus A 3 | ||||
| DE10 | Weaner | Pedro Brand 4 | 2020 | Positive | Negative |
| DE22 | Weaner | Cabrera 4 | 2020 | Positive | Negative |
| DE92 | Weaner | Pedro Brand | 2020 | Positive | Negative |
| ENG2 | Grower | Cabrera | 2020 | Positive | Negative |
| ENG4 | Grower | Cabrera | 2020 | Positive | Negative |
| ENG5 | Grower | Cabrera | 2020 | Positive | Negative |
| ENG7 | Grower | Cabrera | 2020 | Negative | Negative |
| ENG14 | Grower | Cabrera | 2020 | Positive | Negative |
| ENG21 | Grower | Cabrera | 2020 | Positive | Negative |
| ENG52 | Grower | Cabrera | 2020 | Positive | Negative |
| GE4 | Farrow 5 /Pregnant | Pedro Brand | 2020 | Negative | Negative |
| GE12 | Farrow/Pregnant | Pedro Brand | 2020 | Negative | Negative |
| GES7 | Farrow/Pregnant | Pedro Brand | 2020 | Positive | Negative |
| GES15 | Farrow/Pregnant | Pedro Brand | 2020 | Positive | Negative |
| M8 | Piglet | Cabrera | 2020 | Positive | Negative |
| MA1 | Piglet | Cabrera | 2020 | Negative | Negative |
| MA5 | Piglet | Cabrera | 2020 | Positive | Negative |
| MA9 | Piglet | Cabrera | 2020 | Positive | Negative |
| N8 | Grower | Cabrera | 2020 | Positive | Negative |
| VE8 | Boar | Cabrera | 2020 | Positive | Negative |
| VE22 | Boar | Cabrera | 2020 | Positive | Negative |
| D17 | Weaner | Villa Mella 4 | 2021 | Positive | Negative |
| PP3 | Dry Sow | Villa Mella | 2021 | Negative | Negative |
| PP8 | Dry Sow | Villa Mella | 2021 | Negative | Negative |
| Z11 | Grower | Villa Mella | 2021 | Positive | Negative |
| Z13 | Grower | Villa Mella | 2021 | Positive | Negative |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gainor, K.; Fortuna, Y.C.; Alakkaparambil, A.S.; González, W.; Malik, Y.S.; Ghosh, S. High Rates of Detection and Molecular Characterization of Porcine Adenovirus Serotype 5 (Porcine mastadenovirus C) from Diarrheic Pigs. Pathogens 2022, 11, 1210. https://doi.org/10.3390/pathogens11101210
Gainor K, Fortuna YC, Alakkaparambil AS, González W, Malik YS, Ghosh S. High Rates of Detection and Molecular Characterization of Porcine Adenovirus Serotype 5 (Porcine mastadenovirus C) from Diarrheic Pigs. Pathogens. 2022; 11(10):1210. https://doi.org/10.3390/pathogens11101210
Chicago/Turabian StyleGainor, Kerry, Yussaira Castillo Fortuna, Angeline Steny Alakkaparambil, Wendy González, Yashpal Singh Malik, and Souvik Ghosh. 2022. "High Rates of Detection and Molecular Characterization of Porcine Adenovirus Serotype 5 (Porcine mastadenovirus C) from Diarrheic Pigs" Pathogens 11, no. 10: 1210. https://doi.org/10.3390/pathogens11101210
APA StyleGainor, K., Fortuna, Y. C., Alakkaparambil, A. S., González, W., Malik, Y. S., & Ghosh, S. (2022). High Rates of Detection and Molecular Characterization of Porcine Adenovirus Serotype 5 (Porcine mastadenovirus C) from Diarrheic Pigs. Pathogens, 11(10), 1210. https://doi.org/10.3390/pathogens11101210









