Abstract
Non-tuberculous mycobacteria that cannot be identified at the species level represent a challenge for clinical laboratories, as proper species assignment is key to implementing successful treatments or epidemiological studies. We re-identified forty-eight isolates of Ziehl–Neelsen (ZN)-staining-positive “acid-fast bacilli” (AFB), which were isolated in a clinical laboratory and previously identified as Mycobacterium species but were unidentifiable at the species level with the hsp65 PCR restriction fragment length polymorphism analysis (PRA). As most isolates also could not be identified confidently via 16S, hsp65, or rpoB DNA sequencing and a nBLAST search analysis, we employed a phylogenetic method for their identification using the sequences of the 16S rDNA, which resulted in the identification of most AFB and a Mycobacterium species diversity not found before in our laboratory. Most were rare species with only a few clinical reports. Moreover, although selected with the ZN staining as AFB, not all isolates belonged to the genus Mycobacterium, and we report for the first time in Latin America the isolation of Nocardia puris, Tsukamurella pulmosis, and Gordonia sputi from sputum samples of symptomatic patients. We conclude that ZN staining does not differentiate between the genus Mycobacterium and other genera of AFB. Moreover, there is a need for a simple and more accurate tree-based identification method for mycobacterial species. For this purpose, and in development in our lab, is a web-based identification system using a phylogenetic analysis (including all AFB genera) based on 16S rDNA sequences (and in the future multigene datasets) and the closest relatives.
1. Introduction
Non-tuberculous mycobacteria (NTM) are facultative intracellular pathogens that in general are not transmitted from person to person but acquired by susceptible individuals from the environment [,]. Recently, however, it has also been shown that NTM infections can be acquired through transmission, via fomites and aerosols []. Approximately 200 NTM species have been identified, of which approximately 140 are pathogenic to humans and animals [].
Accurately identifying NTM at the species level is essential for providing a correct disease diagnosis, identifying expected antimicrobial susceptibility patterns, and tracing outbreaks for infection source analyses [,,,,]. The Ziehl–Neelsen (ZN) staining method is the first step in identifying mycobacteria, since they show up as “acid-fast bacilli” (AFB), a physical property that gives this bacterium the ability to resist decolorization by acids during staining procedures. Even though acid fastness can be attributed to many different genera of bacteria (the genus Nocardia, for example), in clinical practice it is a fairly unique characteristic of Mycobacterium species. The further identification of AFB at the species level has become complicated because of the steady increase in species in the last 10 years. The phenotypic approaches based on morphological characteristics, growth rates, preferred growth medium, pigmentation, and biochemical tests are time-consuming and nowadays useless [,]. Several commercial molecular identification methods are available but these are relatively expensive and are only used in laboratories with enough resources []. Small regional laboratories have chosen alternative methods for identification, and a popular, fast, cheap technique—described for the first time about 30 years ago in 1993—is the PCR restriction fragment length polymorphism analysis (PRA) method, targeting a fragment of the heat shock protein 65 (hsp65) gene []. This method has been shown to be adequate for identifying the most important species in a convenient way []. For less prevalent mycobacterial species, the technique fails because of the presence of identical or near-identical PRA patterns of other mycobacteria species in the database [,].
The present study aimed to identify 48 unidentified NTM species isolated in a tuberculosis diagnostic laboratory in Caracas, Venezuela. Re-identification was considered important, as these strains were of clinical relevance. Forty-five strains were isolated from patients with respiratory problems or other tuberculosis-like clinical manifestations. Three strains were isolated from environmental samples and were associated with NTM outbreaks []. The laboratory had tried to identify these AFB with the PRA technique, but this technique either failed to assign a definitive species name, the restriction enzyme pattern could not be found in the PRA database [], or no hsp65 PCR signal was obtained and the isolates were reported as Mycobacterium spp. to the physician. Here, we sequenced the 16S rRNA, rpoB, and hsp65 genes of these strains and used a phylogenetic analysis for their identification.
2. Materials and Methods
2.1. Acid-Fast Mycobacterial Strains
The 48 strains came from a strain bank present in the ‘Dr. José María Vargas’ Hospital in Caracas and were isolated from 2014 to 2021 from patients with a presumed tuberculosis infection (42 strains were isolated from sputum, pus, biopsies, pleural liquid, or urine samples). Three strains were isolated from a water sample, food, or cosmetic product; all products were associated with NTM outbreaks [,]. Three strains were isolated from patients, but the type of clinical sample was unknown as these strains were referred to our lab for identification. The strains were isolated on Lowenstein–Jensen (L-J) medium and all were ZN-positive; for that reason, they were classified as mycobacteria. Most of these 48 isolates were rapid growers with the exception of 6 strains that were slow growers. The previous identification was unsuccessfully carried out with the PRA technique, obtaining an uncertain or ambiguous result after digestion with restriction enzymes. Three inoculation loops of growth of the strains on L-J medium were stored in TE buffer (for DNA isolation) and in TSB with 10% glycerol at −20 °C or −70 °C, respectively. See Supplementary Table S1 for more detailed information regarding the origin of the strains, type of clinical sample from which the strains were isolated, year of isolation, and preliminary identification (where available).
2.2. PCR and Sequencing
The DNA was liberated from the 48 strains by boiling the strains in TE buffer for 5 min. After a brief centrifugation step (1 min at 10,000× g), the supernatant was used for the PCR reaction. The PCR amplification of three genes (16S, hsp65, and rpoB) was performed with GoTaq® Green Master Mix from Promega®. The primers for the amplification of the genes can be found in Table 1. The PCR conditions have been described previously []. The amplicons were visualized via electrophoresis in a 1% agarose gel, and when specific products were amplified, these products were sequenced with the Sanger method and analyzed with the ABI 3500xL Genetic Analyzer from Applied Biosystems at the Service Department of Universidad de las Americas, Quito, Ecuador.

Table 1.
Oligonucleotide primers used for the sequencing of parts of the 16S, rpoB, and hsp65 genes of the mycobacteria in this study. The primers used were previously described [,,].
2.3. Identification with a Phylogenetic Analysis
The sequences of the 16S rRNA, rpoB, and hsp65 genes, obtained with forward and reverse primers, were assembled and consensus sequences were edited using MEGA 11. The identity of each sequence was confirmed using BLAST in NCBI resources. For the phylogenetic analysis, a total of 40 sequences deposited in GenBank NCBI from 40 species of Mycobacterium were included as related groups. Tsukamurella tyrosinosolvens, Tsukamurella pulmonis, Nocardia puris, Nocardia cyriacigeorgica, Gordonia sputi, Gordonia iterans, and Corynebacterium jeikeium were selected as outgroups (GenBank Accession numbers can be found in Supplementary Table S2). The DNA sequence matrices of 88 sequences x 497 nucleotides for 16S, 80 sequences x 288 nucleotides for rpoB, and 69 sequences x 389 nucleotides for hsp65 were aligned using MEGA 11 [,] with the ClustalW algorithm with high gap creation and extension penalties of 16.0 and 6.0, respectively, searching for a strong positional homology.
Afterward, a maximum parsimony (MP) analysis was implemented using the heuristic search option with a tree bisection reconnection branch-swapping algorithm with a random stepwise addition of 10 replicates for each search and 100–1000 replications per analysis. The gaps were treated as missing data. The characters were treated as unordered and equally weighted. We also performed a maximum likelihood (ML) and substitution models estimated on MEGA 11 (Supplementary Table S3) using an MP as a guide tree. In both analyses, the robustness of the trees was estimated using parsimony bootstrapping with 500 pseudo-replicates. For classification purposes for complexes and groups, and for the verification of phylogenetic species, each monophyletic clade was evaluated by means of the divergence between and within clades (uncorrected p-distance calculated), as well as between sequences identified in the NCBI and what was obtained in the laboratory.
2.4. Ethical Considerations
The Institutional Review Board of Servicio Autónomo Instituto de Biomedicina Dr. Jacinto Convit in Caracas, Venezuela, approved the study protocol with a waiver of informed consent (IRB SAIB 06-06-2022).
3. Results
3.1. PCR and Sequencing
All 48 strains yielded a sequence of the 16S rRNA and rpoB genes and only 43 strains yielded an hsp65 sequence, as five strains did not amplify with the hsp65 primers TB11 and TB12 (LTM4391, LTM626213, LTN3490, LTL3092, and LTQ5825). The sequences were introduced in GenBank for preliminary identification. The nucleotide sequence data of these strains are available in the GenBank database and the accession numbers can be found in Supplementary Table S4. Nevertheless, none of the sequences or combinations of sequences could definitively identify the strains, so most ended up with an ambiguous taxonomic name with multiple independent records that shared the same or nearly the same gene sequence similarity. That was why all species were subjected to a phylogenetic analysis.
3.2. Sequence Analysis Using GenBank and a Phylogenetic Analysis
The strains were further analyzed using a phylogenetic approach. Based on the 16S sequences of the unidentified strains, 33 references strains of mycobacteria and 7 outgroup strains were chosen, and the sequences of the 16S gene (40 sequences), rpoB gene (32 sequences), and hsp65 gene (26 sequences) of these strains were downloaded to be used in the matrix analysis. See Supplementary Table S2 for the GenBank access numbers of the sequences from the reference species and outgroup strains. The sequences were aligned in three matrices according to the genes. Three phylogenetic trees were obtained using the phylogeny approach. The 16S tree demonstrated a consistent topology and identified the Mycobacterium species (Supplementary Figure S1). This tree was selected for species identification. The other two trees (not shown), the rpoB and hsp65 trees, showed highly variable phylogenetic information and were not further used for the purpose of this article.
The monophyletic associations in the 16S rRNA tree clustered the strains into 16 groups and 5 complexes, with a divergence percentage of 1.328% in the groups (see Supplementary Table S3). If the divergence values of each species in relation to the samples were between 0 and 1.328%, they were considered as species under the monophyletic association. If the values slightly exceeded this range, they were considered as NR (near) the corresponding species, and if the associations were not close to any species, they were considered non-identifiable.
Supplementary Table S1 shows the results obtained for each strain along with the correct species identification of the pathogens. Here, 83.33% (40/48) of the total strains were identified or closely related to a species with this phylogenetic approach. The 16S rRNA gene analysis only allowed the identification of 68.75% (33/48) of the strains. The tree strains were also identified via rpoB (sample LTP1387 as near M. senegalense and M. farcinogenes and LTQ2392–LTS855 as M. conceptionense). The hsp65 gene allowed us to identify LTR2603, LTS244, and LTO3295 as M. fortuitum and LTN4726 as M. abscessus. Remarkably, five “acid-fast” strains were not identified with sequencing as Mycobacterium species but were identified as belonging to the genera Tsukamurella, Nocardia, and Gordonia. See Figure 1 and Figure 2 for examples of the identification using a phylogenetic approach for some of the strains.
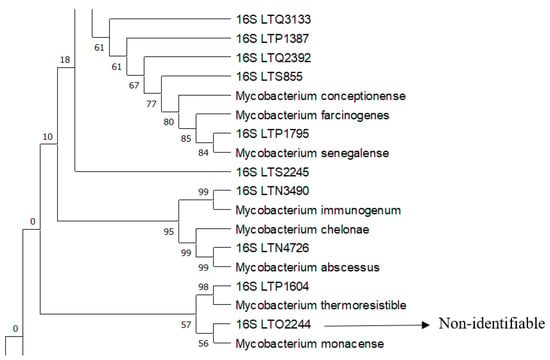
Figure 1.
A segment of the 16S rRNA tree showing complex 2 and the identification of the laboratory strains indicated with 16S using a phylogenetic approach with sequences of reference strains. Phylogenetic construction was performed via ML and T92+G+I using an MP as a guide tree, with the robustness of the bootstrapping set at 500 pseudo-replicates.
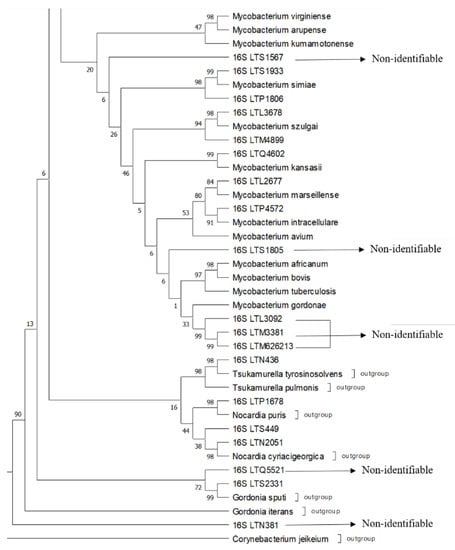
Figure 2.
A segment of the 16S rRNA tree showing complexes 4 and 5, outgroups of the laboratory strains indicated for identification with 16S using a phylogenetic approach with sequences of reference strains and outgroup strains. The phylogenetic construction was performed using ML and T92+G+I using an MP as a guide tree, with the robustness of the bootstrapping set at 500 pseudo-replicates.
4. Discussion
The identification with the sequences of a partial 16S, hsp 65, or rpoB gene or with a phylogenetic approach showed a variety of acid-fast species isolated from patients in a clinical laboratory. Most species were previously not properly identified or could not be identified with the PRA technique, a commonly used technique in many clinical laboratories. Additionally, the 16S rRNA sequencing alone or the sequencing of the hsp65 and rpoB genes and search for a BLAST similarity in GenBank could not assign a species name to most strains. Therefore, the identifications were made with a phylogenetic analysis of the 16S gene. The phylogenetic analysis improved the identification of the strains by placing them close to a group or complex and provided us with the identification based on their evolutionary relationship.
The identification of our isolates showed that many species are hardly mentioned in the medical literature and are relatively unknown as infectious agents for humans. Examples include the species M. porcinum, M. simiae, M. thermoresistibile, M. neoaurum, and M. szulgai [,]. Some strains (Supplementary Table S1 and Figure 1 and Figure 2) were unidentifiable because the percentages of divergence were considerably higher than 1.328%, so these could be new mycobacterial species or variants of already known species, or the references sequences were not considered in the phylogenetic analysis. This needs further investigation.
Furthermore, although we expected to identify only mycobacteria because only AFB were used, other species of the genera Tsukamurella, Nocardia, and Gordonia were found. These genera also have mycolic acid in their cell wall, like mycobacterial species [], which explains their acid-fast properties. In the literature, these bacteria are called “partially or weakly acid-fast”. However, in our laboratory, this “partial acid-fast” nature is hard to differentiate from bacteria with real “acid-fast” characteristics. When viewed under the microscope, short acid-fast red rods were seen by the laboratory technician, who has more than 10 years of experience. Moreover, when we became aware of “weak acid-fast” organisms in our collection of strains, we grew the strains again from the glycerol preparations stored at minus 70 °C. Once again, only acid-fast rods were seen under the microscope. The incorrect identification of the strains in this study could have had grave repercussions for the patients. All isolates came from patients with clinical signs and symptoms that can be confused with tuberculosis. We do not know with which antibiotics these patients were treated with, but incorrect identification can lead to inappropriate antibiotic prescriptions and does not permit species-specific treatment. A correct identification is also important for epidemiological purposes (transmission and infection source studies) and can lead to an underestimation of the importance of the virulence of some species.
The reason we identified these bacteria as acid-fast and consequently as mycobacteria could possibly be found in the composition of our Ziehl–Neelsen stain or decolorizing solution. We used a more concentrated 1% carbol–fuchsine solution (a mixture of phenol and basic fuchsin) in the staining procedures as recommended by the Stop TB Partnership [], while other ZN staining protocols use a 0.3% solution of carbol–fuchsine [,]. It is well established that a higher concentration of fuchsine produces a better staining of the mycobacterial cells [,]. Additionally, the phenol concentrations and acid–alcohol solutions vary in the Ziehl–Neelsen staining protocols [,,]. Moreover, the efficiency of the Ziehl–Neelsen method for destaining acid-fast bacteria depends on the composition of the solutions. We used 3% HCL in 95% ethanol as a destaining solution, but 6% HCL has also been recommended or 20–25% sulfuric acid [,,]. The staining and destaining of “partial or weak” AFB with the different solutions has never been evaluated. Despite its application for well over a century, the ZN technique remains non-standardized, and we believe that standardization is urgently required and should incorporate the partially acid-fast genera, which include Nocardia, Tsukamurella, and Gordonia. These genera are uncommon pathogens and are prone to being misdiagnosed as mycobacteria []. An example in our clinical laboratory was strain LTN436, a Tsukamurella pulmonis strain, which was isolated from a sputum sample after a decontamination step and grown on L-J medium. The patient was misdiagnosed as infected with a Mycobacterium spp. Such an error may be common among lung infections caused by Tsukamurella species because the disease manifestation shares a striking similarity with mycobacterial infections []. Similarly, strain LTS2331, with a preliminary identification as a Nocardia spp., was eventually identified as Gordonia sputi. Only in recent years have Gordonia species been recognized as emerging pathogens []. Globally, from 1996 to 2015, only 16 cases of infections by Gordonia sputi were reported, and these were associated with infections from catheters or with lung disease in immunocompromised patients []. Gordonia spp. are environmental bacteria whose implication in human disease seems to be increasing []. Finally, two isolates were respectively identified as Nocardia puris and Nocardia cyriacigeorgica. The genus Nocardia comprises opportunistic pathogens that cause a variety of clinical diseases in immunocompromised people, mostly pulmonary infections []. Nocardia cyriacigeorgica has been isolated before from lung infections in patients in Venezuela []. However, the isolation and identification of Nocardia puris as well as the other “weak” acid-fast microorganisms—Tsukamurella pulmonis and Gordonia sputi—are the first reports of human infection in Latin America.
5. Conclusions
A high variety of mycobacterial strains were identified, with many species rarely described as pathogens in the medical literature. Moreover, acid-fast microorganisms of other genera, also rarely mentioned as pathogens, were found, and three species were identified that have never been reported before in Latin America, namely Nocardia puris, Tsukamurella pulmosis, and Gordonia sputi. We also found that several of the mycobacteria species of this study could not be identified with a “one gene only” sequence mainly due to the great diversity of new species and groups of species and the absence of reference sequences in GenBank, but that the interpretation of the 16S sequences data with a phylogenetic approach in general permits the identification of genus and species groups based on the closest evolutionary relatives. For the clinical laboratory, a simpler straightforward method for the identification of acid-fast microorganisms is needed. This method could be based on the sequencing of a particular gene or a computational phylogenetic algorithm to identify the acid-fast microorganism. For this purpose, and in development in our lab, is an online web-based identification system that will use a phylogenetic analysis (including all AFB genera) based on 16S rDNA sequences, phylogenetic trees, and close relatives. This database will be constantly updated with genetic information concerning the mycobacterial species and will permit the identification based on the relationships with its closest relatives. Several online databases that track the pathogen identification and evolution processes in real time are already available, including the platform Nextstrain (available online: https://nextstrain.org/; accessed on 20 September 2022),which tracks the pathogenic evolution of several viruses and Yersinia pestis, or the Pango Network (available online: https://www.pango.network/; accessed on 20 September 2022, developed for identifying SARS-CoV-2 genetic lineages of epidemiological relevance.
6. Limitations of This Study
We used no clinical data and did not perform follow-up checks on the patients. Most isolates were from many years ago and telephone contact was lost. Thus, we were unable to determine what effect the incorrect identification of these AFB had on the disease outcomes.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pathogens11101159/s1, Figure S1: Phylogenetic tree of 16S rRNA sequences of the Mycobacterium reference sequences, outgroup strains and the Mycobacterium spp. Strains; Table S1: Origin and identification of acid-fast bacteria, identified as Mycobacteria with the ZN staining; Table S2: GenBank accession numbers of Mycobacterium species sequences used in the phylogenetic analysis; Table S3: DNA Models test, divergence percentages, and number of identifications for each gene; Table S4: 16S rRNA sequences of the 48 strains that were identified with ZN staining as belonging to the genus Mycobacteria.
Author Contributions
Conceptualization, Y.L., G.E., F.E.C.-A., D.S. and J.H.d.W.; methodology, Y.L., G.E., F.E.C.-A., D.S., S.G.-F., Y.R., C.B.-C., J.C.N. and J.H.d.W.; formal analysis, Y.L., G.E., S.G.-F., Y.R., C.B.-C., J.C.N. and J.H.d.W.; data curation, Y.L., G.E., J.C.N. and J.H.d.W.; writing, Y.L., G.E. and J.C.N.; writing—review and editing, Y.L., G.E., F.E.C.-A., D.S., S.G.-F., Y.R., C.B.-C., J.C.N. and J.H.d.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research and the APC were funded by the Universidad de Las Américas, UDLA (Servicio Beta-Caseína), Quito, Ecuador.
Institutional Review Board Statement
The Institutional Review Board of Servicio Autónomo Instituto de Biomedicina Dr. Jacinto Convit in Caracas, Venezuela, approved the study protocol with a waiver for informed consent (IRB SAIB 06-06-2022).
Informed Consent Statement
No written informed consent for publication was obtained from the participating patients, since the patients cannot be identified in this study and only mycobacterial isolates were used, with no personal information or clinical data form part of the report.
Data Availability Statement
All sequence data are published in GenBank.
Acknowledgments
We thank the personnel at the research laboratories at Universidad de las Américas in Quito, Ecuador, and the Tuberculosis Laboratory in Caracas, Venezuela.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Turenne, C.Y. Nontuberculous mycobacteria: Insights on taxonomy and evolution. Infect. Genet. Evol. 2019, 72, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Misch, E.A.; Saddler, C.; Davis, J.M. Skin and Soft Tissue Infections Due to Nontuberculous Mycobacteria. Curr. Infect. Dis. Rep. 2018, 20, 6. [Google Scholar] [CrossRef]
- Bryant, J.M.; Grogono, D.M.; Rodriguez-Rincon, D.; Everall, I.; Brown, K.P.; Moreno, P.; Verma, D.; Hill, E.; Drijkoningen, J.; Gilligan, P.; et al. Emergence and spread of a human-transmissible multidrug-resistant nontuberculous Mycobacterium. Science 2016, 354, 751–757. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Kinjo, T.; Motooka, D.; Nabeya, D.; Jung, N.; Uechi, K.; Horii, T.; Iida, T.; Fujita, J.; Nakamura, S. Comprehensive subspecies identification of 175 nontuberculous mycobacteria species based on 7547 genomic profiles. Emerg. Microbes Infect. 2019, 8, 1043–1053. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, I.; Tiberi, S.; Farooqi, J.; Jabeen, K.; Yeboah-Manu, D.; Migliori, G.B.; Hasan, R. Non-tuberculous mycobacterial infections—A neglected and emerging problem. Int. J. Infect. Dis. 2020, 92, S46–S50. [Google Scholar] [CrossRef]
- Forbes, B.A. Mycobacterial Taxonomy. J. Clin. Microbiol. 2017, 55, 380–383. [Google Scholar] [CrossRef] [PubMed]
- Tortoli, E.; Fedrizzi, T.; Meehan, C.J.; Trovato, A.; Grottola, A.; Giacobazzi, E.; Serpini, G.F.; Tagliazucchi, S.; Fabio, A.; Bettua, C.; et al. The new phylogeny of the genus Mycobacterium: The old and the news. Infect. Genet. Evol. 2017, 56, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Jardín, O.D.M.; Hernández-Pérez, R.; Corrales, H.; Cardoso-Leao, S.; de Waard, J.H. Seguimiento de un brote de infección en tejido blando causado por Mycobacterium abscessus posterior a la mesoterapia en Venezuela. Enferm. Infecc. Y Microbiol. Clínica 2010, 28, 596–601. (In Spanish) [Google Scholar] [CrossRef]
- Torres-Coy, J.A.; A Rodríguez-Castillo, B.; Pérez-Alfonzo, R.; DE Waard, J.H. Source investigation of two outbreaks of skin and soft tissue infection by Mycobacterium abscessus subsp. abscessus in Venezuela. Epidemiol. Infect. 2015, 144, 1117–1120. [Google Scholar] [CrossRef]
- Tortoli, E. Standard operating procedure for optimal identification of mycobacteria using 16S rRNA gene sequences. Stand. Genom. Sci. 2010, 3, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.S.; Lo, B.; Son, J. Phylogenomics and Comparative Genomic Studies Robustly Support Division of the Genus Mycobacterium into an Emended Genus Mycobacterium and Four Novel Genera. Front. Microbiol. 2018, 9, 67. [Google Scholar] [CrossRef] [PubMed]
- Terán, R.; de Waard, J.H. Recent advances in the laboratory diagnosis of tuberculosis. J. Int. Fed. Clin. Chem. Lab. Med. 2015, 26, 295–309. [Google Scholar]
- Telenti, A.; Marchesi, F.; Balz, M.; Bally, F.; Böttger, E.C.; Bodmer, T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 1993, 31, 175–178. [Google Scholar] [CrossRef]
- Chimara, E.; Ferrazoli, L.; Ueky, S.Y.M.; Martins, M.C.; Durham, A.M.; Arbeit, R.D.; Leão, S.C. Reliable identification of mycobacterial species by PCR-restriction enzyme analysis (PRA)-hsp65 in a reference laboratory and elaboration of a sequence-based extended algorithm of PRA-hsp65patterns. BMC Microbiol. 2008, 8, 48. [Google Scholar] [CrossRef] [PubMed]
- Tortoli, E. Impact of Genotypic Studies on Mycobacterial Taxonomy: The New Mycobacteria of the 1990s. Clin. Microbiol. Rev. 2003, 16, 319–354. [Google Scholar] [CrossRef]
- Prodhom, G.; Vincent, V.; Pfyffer, G.E.; Telenti, A. Centre Hospitalier Universitaire Vaudoi. PRASITE: Identification of Mycobacteria. 2007. Available online: http://app.chuv.ch/prasite/index.htm (accessed on 2 September 2022).
- Cooksey, R.C.; De Waard, J.H.; Yakrus, M.A.; Rivera, I.; Chopite, M.; Toney, S.R.; Morlock, G.P.; Butler, W.R. Mycobacterium cosmeticum sp. nov., a novel rapidly growing species isolated from a cosmetic infection and from a nail salon. Int. J. Syst. Evol. Microbiol. 2004, 54, 2385–2391. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Park, H.-J.; Cho, S.-N.; Bai, G.-H.; Kim, S.-J. Species Identification of Mycobacteria by PCR-Restriction Fragment Length Polymorphism of the rpoB Gene. J. Clin. Microbiol. 2000, 38, 2966–2971. [Google Scholar] [CrossRef] [PubMed]
- Brunello, F.; Ligozzi, M.; Cristelli, E.; Bonora, S.; Tortoli, E.; Fontana, R. Identification of 54 Mycobacterial Species by PCR-Restriction Fragment Length Polymorphism Analysis of the hsp65 Gene. J. Clin. Microbiol. 2001, 39, 2799–2806. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef]
- Piquero, J.; Casals, J.P.; Higuera, E.L.; Yakrus, M.; Sikes, D.; De Waard, J.H. Iatrogenic Mycobacterium simiae Skin Infection in an Immunocompetent Patient. Emerg. Infect. Dis. 2004, 10, 969–970. [Google Scholar] [CrossRef]
- Torres-Coy, J.A.; Carrera, C.; Rodríguez-Castillo, B.A.; Ramírez-Murga, R.; Ortiz-Cáceres, W.; Pérez-Alfonzo, R.; de Waard, J.H. Mycobacterium szulgai: An unusual cause of skin and soft tissue infection after breast augmentation. Int. J. Dermatol. 2017, 56, e122–e124. [Google Scholar] [CrossRef] [PubMed]
- Safaei, S.; Fatahi-Bafghi, M.; Pouresmaeil, O. Role of Tsukamurella species in human infections: First literature review. New Microbes New Infect. 2017, 22, 6–12. [Google Scholar] [CrossRef]
- Lumb, R.; Van Deun, A.; Bastian, I.; Fitz-Gerald, M. The Handbook, Laboratory Diagnosis of Tuberculosis by Sputum Microscopy; SA Pathology, Australia. 2013. Available online: https://stoptb.org/wg/Gli/assets/documents/TB%20MICROSCOPY%20HANDBOOK_FINAL.pdf (accessed on 13 August 2022).
- International Union Against Tuberculosis and Lung Disease. Technical Guide for Sputum Examination for Tuberculosis by Direct Microscopy. 1978. Available online: https://tbrieder.org/publications/books_english/microscopy.pdf (accessed on 13 August 2022).
- Pan American Health Organization. Handbook for the Bacteriological Diagnosis of Tuberculosis. Part I: Smear Microscopy. 2019. Available online: https://www.paho.org/en/documents/handbook-bacteriological-diagnosis-tuberculosis-part-i-smear-microscopy-update-2018 (accessed on 25 April 2022).
- Selvakumar, N.; Rahman, F.; Rajasekaran, S.; Narayanan, P.R.; Frieden, T.R. Inefficiency of 0.3% Carbol Fuchsin in Ziehl-Neelsen Staining for Detecting Acid-Fast Bacilli. J. Clin. Microbiol. 2002, 40, 3041–3043. [Google Scholar] [CrossRef] [PubMed]
- Selvakumar, N.; Sekar, M.G.; Rahman, F.; Syamsunder, A.; Duraipandian, M.; Wares, F.; Narayanan, P.R. Comparison of variants of carbol-fuchsin solution in Ziehl-Neelsen for detection of acid-fast bacilli. Int. J. Tuberc. Lung Dis. 2005, 9, 226–229. [Google Scholar]
- Angra, P.; Ridderhof, J.; Smithwick, R. Comparison of Two Different Strengths of Carbol Fuchsin in Ziehl-Neelsen Staining for Detecting Acid-Fast Bacilli. J. Clin. Microbiol. 2003, 41, 3459. [Google Scholar] [CrossRef]
- Gross, M. Rapid Staining of Acid-Fast Bacteria*. Am. J. Clin. Pathol. 1952, 22, 1034–1035. [Google Scholar] [CrossRef]
- Murray, S.J.; Barrett, A.; Magee, J.G.; Freeman, R. Optimisation of acid fast smears for the direct detection of mycobacteria in clinical samples. J. Clin. Pathol. 2003, 56, 613–615. [Google Scholar] [CrossRef]
- Yang, L.; Cao, Y.; Dan, Z.; Wang, Z.; Wang, X. Community-acquired Tsukamurella pneumonia in a young immunocompetent adult: A case misdiagnosed as pulmonary tuberculosis and literature review. Postgrad. Med. 2017, 129, 563–566. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Yu, Y.; Chen, M.; Wang, C.; Kang, Y.; Li, H.; Lou, J. Bacteremia due to Gordonia polyisoprenivorans: Case report and review of literature. BMC Infect. Dis. 2017, 17, 419. [Google Scholar] [CrossRef][Green Version]
- Fang, W.; Li, J.; Cui, H.-S.; Jin, X.; Zhai, J.; Dai, Y.; Li, Y. First identification of Gordonia sputi in a post-traumatic endophthalmitis patient—A case report and literatures review. BMC Ophthalmol. 2017, 17, 190. [Google Scholar] [CrossRef] [PubMed]
- Renvoise, A.; Harle, J.-R.; Raoult, D.; Roux, V. Gordonia sputi Bacteremia. Emerg. Infect. Dis. 2009, 15, 1535–1537. [Google Scholar] [CrossRef] [PubMed]
- Hamdi, A.M.; Fida, M.; Deml, S.M.; Abu Saleh, O.M.; Wengenack, N.L. Retrospective Analysis of Antimicrobial Susceptibility Profiles of Nocardia Species from a Tertiary Hospital and Reference Laboratory, 2011 to 2017. Antimicrob. Agents Chemother. 2020, 64, e01868-19. [Google Scholar] [CrossRef] [PubMed]
- Uzcátegui-Negrón, M.; Serrano, J.; Boiron, P.; Rodriguez-Nava, V.; Couble, A.; Moniée, D.; Herrera, K.S.; Sandoval, H.; Reviákina, V.; Panizo, M.; et al. Reclassification by molecular methods of actinobacteria strains isolated from clinical cases in Venezuela. J. Mycol. Médicale 2011, 21, 100–105. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).