Abstract
Emerging and re-emerging viral diseases pose continuous public health threats, and effective control requires a combination of non-pharmacologic interventions, treatment with antivirals, and prevention with vaccines. The COVID-19 pandemic has demonstrated that the world was least prepared to provide effective treatments. This lack of preparedness has been due, in large part, to a lack of investment in developing a diverse portfolio of antiviral agents, particularly those ready to combat viruses of pandemic potential. Here, we focus on a drug target called macrodomain that is critical for the replication and pathogenesis of alphaviruses and coronaviruses. Some mutations in alphavirus and coronaviral macrodomains are not tolerated for virus replication. In addition, the coronavirus macrodomain suppresses host interferon responses. Therefore, macrodomain inhibitors have the potential to block virus replication and restore the host’s protective interferon response. Viral macrodomains offer an attractive antiviral target for developing direct acting antivirals because they are highly conserved and have a structurally well-defined (druggable) binding pocket. Given that this target is distinct from the existing RNA polymerase and protease targets, a macrodomain inhibitor may complement current approaches, pre-empt the threat of resistance and offer opportunities to develop combination therapies for combating COVID-19 and future viral threats.
1. Introduction
Emerging and re-emerging viral diseases pose continuous public health threats that have increased with globalization, population growth, urbanization, and climate change. These disease-causing viruses range from zoonotic and sexually transmitted viruses such as human immunodeficiency virus (HIV), to vector-borne viruses such as Zika virus and chikungunya virus (CHIKV), to respiratory viruses such as H1N1 influenza virus and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The rapid mutation of these RNA viruses can facilitate infection of new hosts, improve transmission, and increase virulence with sudden spread into new regions. The medical, social, and political consequences of these emerging infections are global but often disproportionately affect resource-poor countries, resulting in unequal social, health system, and economic burdens on these populations.
Effective control requires a combination of non-pharmacologic interventions, treatment with antiviral drugs and antibodies, and prevention with vaccines. Experience with the COVID-19 pandemic and other recently emergent viral infections has exposed a lack of effective treatments capable of protecting humans from the devastating effects of highly deadly and contagious viral pathogens. This gap in preparedness has been due, in large part, to a lack of investment in development of antiviral agents for classes of viruses that were only “potential threats” and, if they did emerge, often caused only acute disease and were quickly contained. When SARS-CoV-2 emerged, this dearth of therapeutics was tragically exposed. Therefore, a multi-pronged investment in development of multiple classes of direct-acting antiviral drugs effective against emergent RNA viruses is crucial to prepare for the next epidemic/pandemic.
Coronaviruses, once thought to be mere contributors to the common cold, have now caused three notable epidemics of life-threatening disease in the last two decades: SARS-CoV in 2002–2003, Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012–2015, and the current pandemic of SARS-CoV-2. All of these viruses are capable of causing respiratory infection that can result in severe disease and even death in a high proportion of those infected [1]. Currently, SARS-CoV-2 is the major public health challenge worldwide, with more than 5 million deaths, accompanied by prolonged destabilizing consequences for the global economy, life-expectancy, and education. The COVID-19-induced global recession resulted in an economic contraction of 3.5% in 2020, with the most pronounced downturn in the poorest countries. Furthermore, disruptions to health care systems have increased deaths from other causes and hindered delivery of routine health care services, such as immunizations. Another class of viruses with pandemic potential are the mosquito-borne alphaviruses. Although previously geographically restricted, these viruses continue to expand into new regions to cause epidemics of rash, arthritis, and encephalitis [2,3]. The past 50 years have seen a dramatic emergence/re-emergence of epidemic arboviral diseases, with 3.6 billion people (nearly half the world population) living in urban areas with exposure to efficient Aedes aegypti mosquito vectors [4]. In addition to incapacitating acute disease, a high proportion of individuals infected with arthritis-causing alphaviruses, such as CHIKV, Sindbis virus (SINV), and Mayaro virus (MAYV), develop chronic joint pain [5,6]. Although infectious virus is cleared promptly after most acute RNA virus infections, including those caused by alphaviruses and coronaviruses, viral RNA often persists, accompanied by ongoing immune stimulation that may contribute to prolonged symptoms [5,7,8,9,10,11,12,13]. Therefore, lack of effective treatment results not only in deaths due to acute disease but also in prolonged disability that could likely be prevented with antiviral treatment [14,15]. Availability of effective antiviral drugs for both coronavirus and alphavirus infections would decrease hospitalizations, deaths, and long-term disability, with benefits for individuals and society. A common feature of these virus families that offers a potential target for antiviral drug development is the highly conserved macrodomain.
2. Macrodomains Represent a Unique Target for Pathogens of Pandemic Potential
Few antivirals are available for treatment of SARS-CoV-2 [16], and no treatments are available for infection with any alphavirus. Antiviral drug development for coronaviruses has mostly focused on nucleoside analogs as inhibitors of RNA polymerase function (e.g., remdesivir or molnupiravir) and peptide analogs that inhibit viral proteases (e.g., nirmatrelvir). Likewise, preliminary evaluation of drugs that inhibit alphavirus replication have most commonly targeted the proteases or polymerase [17,18]. However, there remains a substantial need for developing additional antivirals with novel targets. Identifying distinct targets that require different antiviral mechanisms may complement current approaches, pre-empt the threat of resistance, and offer opportunities to develop combination therapy. Both viral families contain a highly conserved macrodomain that is critical for viral replication and virulence, making it an attractive therapeutic target [19,20]. Macrodomain inhibitors, if successfully developed, could be significant weapons in the future antiviral armamentarium to combat these pathogenic viruses with epidemic potential. Here, we summarize the role of the macrodomain in viral replication and virulence and review the current efforts to develop macrodomain inhibitors as direct-acting antivirals.
3. The Biochemistry of Macrodomains
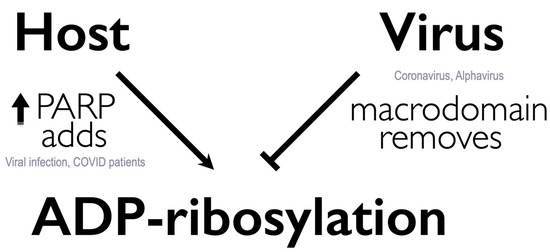
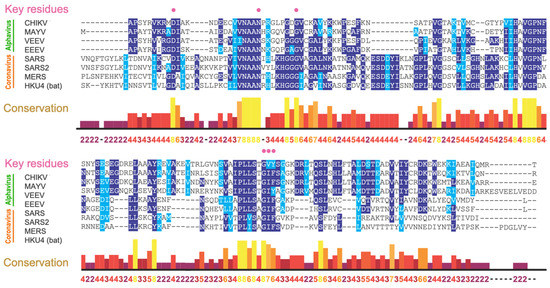
ADP-ribosylation is a post-translational modification catalyzed by ADP-ribosyltransferases (ARTs, also known as PARPs [21]) that transfer an ADP-ribose moiety from NAD+ onto target proteins [22]. The ADP-ribose molecule is transferred as a single unit of mono-ADP-ribose (MAR) or as consecutively attached single units of MAR through glycosidic bonds to preceding ADP-ribose units covalently to form a poly-ADP-ribose (PAR) chain. A macrodomain is a conserved protein fold, existing either as a single protein or embedded within a larger protein, capable of binding to and, in some cases, reversing this modification [19,20,23]. The macrodomain structure consists of a three-layered α/β/α fold and a conserved ADP-ribose binding pocket (Figure 1a,b) [24].
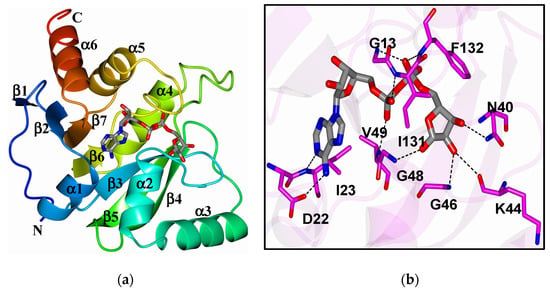
Figure 1.
(a) The structure of SARS-CoV-2 macrodomain complexed with ADP-ribose (6WOJ); (b) hydrogen bonds (dashed lines) between amino acids in the binding pocket and ADP-ribose. Obtained from Alhammad et al., 2020.
These macrodomains are identified in all kingdoms of life [22], including a subset of plus-strand RNA viruses: alphaviruses, coronaviruses, rubella virus, and hepatitis E virus (HEV) [19,20,23]. Viral macrodomains bind to ADP-ribose, its derivatives, and protein-conjugated ADP-ribose. Viral macrodomains are highly conserved in the nonstructural proteins of both alphaviruses and coronaviruses and belong to the MacroD subclass that has ADP-ribosylhydrolase activity [19,25,26,27,28,29,30]. All alphaviruses contain a single macrodomain in the N-terminal portion of nonstructural protein 3 (nsP3) while the highly pathogenic β-coronaviruses SARS-CoV, SARS-CoV-2, and MERS-CoV contain two to three tandem macrodomains in their nsP3, but only the first (Mac1) possesses ADP-ribosylhydrolase activity [31]. The coronavirus and alphavirus macrodomains are primarily MAR-hydrolases, although alphavirus and HEV macrodomains may also have PAR-hydrolase activity, especially when coupled with other proteins [27,32,33]. Therefore, these viral macrodomains, while conserved across different viral families, may have distinct functions.
4. Viral Macrodomains Are Critical for Viral Replication and Disease Pathogenesis
Some mutations in the ADP-ribose binding regions of coronavirus, alphavirus, and HEV macrodomains are not tolerated and viruses cannot replicate, indicating an essential function for this domain [28,34,35,36]. Alphavirus macrodomain mutants without binding or hydrolase activity are not viable due to defects in initiation of infection and viral RNA synthesis [34,35]. In addition, hydrolase activity is critical later in infection for translation of the sub-genomic RNA to produce the viral structural proteins and for disruption of stress granules, which are enriched with translation initiation factors [28,33,34].
Other viruses without macrodomain activity, such as SARS-CoV N1040A, can replicate normally in some tissue culture cells, but these mutants are attenuated in mice [37]. Studies in animals have shown that macrodomain ADP-ribosylhydrolase activity is critical for both coronavirus and alphavirus pathogenesis (e.g., Figure 2a,b) [20,38]. These data indicate that, while there may be virus- and cell-type specific differences in macrodomain function, the macrodomain is universally necessary for virus virulence.
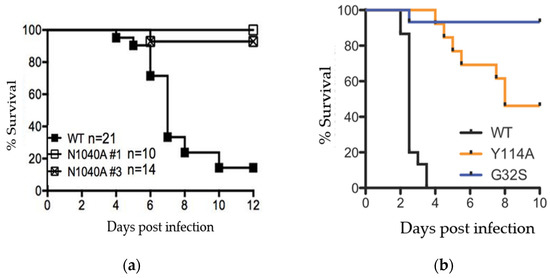
Figure 2.
SARS-CoV and CHIKV macrodomain activity is required for viral pathogenesis. (a) Female Balb/C mice were infected with a lethal dose of SARS-CoV and equivalent amount of 2 separate clones of macrodomain mutant (N1040A) virus and monitored for survival over 12 days. Data from Fehr et al., 2016; (b) 2-day old CD-1 mice (N = 24–28/group) were infected with CHIKV or nsP3 macrodomain mutants Y114A and G32S and monitored for survival over 10 days. Data from McPherson et al., 2017.
The coronavirus macrodomain is also required for full repression of the interferon (IFN) response during infection. Mouse hepatitis virus (MHV) and SARS-CoV macrodomain mutant viruses increase innate immune responses following infection [37,39]. Furthermore, it was demonstrated in a co-infection model that this antiviral response helped protect mice from a lethal SARS-CoV infection [37]. Importantly, an early IFN response after infection is critical for its protective effects. Early administration of IFN-I or IFN-III is protective in mouse models of SARS-CoV and MERS-CoV infection, but administration of IFN in the later stages of infection is not [40,41]. Unlike the wild-type virus which suppresses IFN response, macrodomain mutant virus infection elicits IFN induction in the very early stages of infection [37,39]. Thus, macrodomain inhibitors have the potential to restore the host’s protective early and robust IFN response in addition to blocking virus replication.
8. High-Throughput Assays for Compound Screening
Not only is the macrodomain a suitable target both virologically and biochemically, but high-throughput screening assays are now available to quickly identify hit compounds. These assays can efficiently screen for compounds that can inhibit macrodomain-ADP-ribose binding or macrodomain ADP-ribosylhydrolase activity. One of the first macrodomain high-throughput screening (HTS) assays, developed by Schuller et al., is an ADP-ribose displacement assay using AlphaScreen technology [69]. It was initially described in a screen used to identify inhibitors of the 2nd macrodomain of PARP14 and has since been used for the SARS-CoV-2 macrodomain [67]. AlphaScreen is a bead-based, non-radioactive Amplified Luminescent Proximity Homogenous Assay, where a “donor” bead converts ambient oxygen to singlet oxygen, which interacts with an “acceptor” bead generating chemiluminescence at 370 nm and in turn activates additional fluorophores in the bead with emission at 520-620 nm. To give off a signal, the two beads must be in close proximity to each other or the singlet oxygen will go undetected. To make this assay suitable to measure macrodomain-ADP-ribose binding, a peptide was developed that has a biotin molecule attached to one lysine and a non-hydrolysable ADP-ribose on another lysine. Histidine-tagged macrodomains that are bound to this peptide will interact with a donor streptavidin bead and an Ni2+ acceptor bead, which will then give off the light signal (Figure 6a). Another ADP-ribose binding assay recently developed by Sowa et al. is a FRET-based assay [70]. In this assay, a YFP-tagged Gαi protein-based peptide is ADP-ribosylated by pertussis toxin at a cysteine residue, which cannot be hydrolyzed by viral macrodomains. This protein is then incubated with a CFP-labeled macrodomain, which will then give off a FRET signal upon binding. Using this assay, the authors identified suramin as a non-specific inhibitor of multiple macrodomain-containing proteins which, interestingly, was previously shown to inhibit alphavirus replication in cell culture [71].
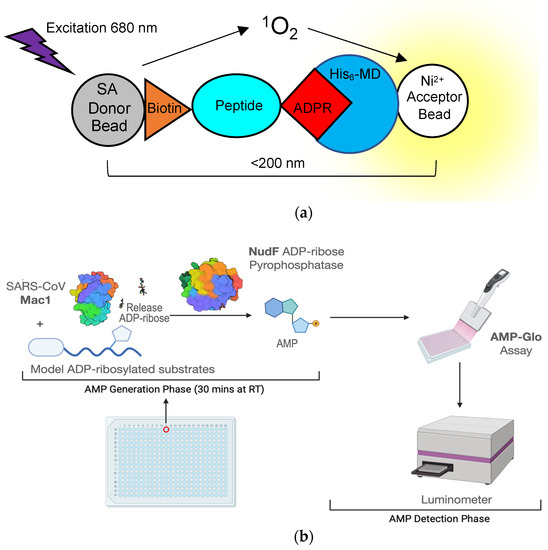
Figure 6.
Assays developed to use high-throughput screening for macrodomain inhibitors. (a) Cartoon diagram depicting a bead-based AlphaScreen assay for measuring macrodomain interaction with an ADP-ribosylated peptide. SA—streptavidin; Ni2+—Nickel; His6-MD—Histidine-tagged macrodomain; ADPR—ADP-ribose; (b) Schematics of ADPr-Glo assay (see text for more details).
In addition, the binding of ADP-ribose or chemical compounds to the macrodomain can be indirectly assessed by differential scanning fluorimetry (DSF). DSF involves the use of a fluorescent dye (e.g., SYPRO Orange) with affinity for hydrophobic portions of proteins, which are exposed as proteins unfold. Binding of ADP-ribose to the macrodomain reduced the increase in fluorescence upon heat denaturation, indicating an increase in the melting temperature, and thus the stability of the SARS-CoV-2 macrodomain when bound to ADP-ribose [32,58]. Virdi et al. used this assay to screen ~2500 compounds for their ability to alter the melting temperature of the macrodomain. This screen identified nucleotides, steroids, antibiotics, and benzimidazoles as potential macrodomain binders. Molecular docking experiments suggested that some of these compounds may interact in the ADP-ribose binding pocket of the SARS-CoV-2 macrodomain [58]. However, many compounds influence the observed protein melting temperature by either quenching fluorescence, increasing fluorescence, or interacting with the reporter dye when mixed with SYPRO Orange, and may therefore be scored as a false positive or false negative in a DSF screen [58].
Measurements of macrodomain ADP-ribosylhydrolase activity have historically relied on gel-based autoradiography and Western blot assays that are not practical for screening large numbers of compounds [26,29]. More recently, our group developed a novel assay that measures ADP-ribosylhydrolase activity in a high-throughput, luminescence-based format called ADPr-Glo (Figure 6b) [54]: First, ADP-ribose is released from a defined MARylated substrate by the macrodomain ADP-ribosylhydrolase of interest. Second, the phosphodiesterase NudF cleaves the released ADP-ribose into phosphoribose and AMP. Finally, AMP is converted to luminescence with the commercially available AMP-Glo kit. This method takes advantage of the substrate selectivity of NudF, which cleaves free ADP-ribose but has no activity toward protein-conjugated ADP-ribose [72]. Therefore, the luminescence signal is controlled by the rate of the ADP-ribosylhydrolase. ADPr-Glo can be performed in 384-well plates with reaction volumes as low as 5 µL, greatly minimizing time and costs compared to traditional gel-based activity assays [28,32]. We established ADPr-Glo conditions for inhibitor screening and multiple macrodomains, including SARS-CoV-2 Mac1, MERS-CoV Mac1, and their closest human homolog, MacroD2. In a pilot screen of the 3233 pharmacologically active compounds, we identified dasatinib and dihydralazine as ADP-ribosylhydrolase inhibitors for both human and viral macrodomains tested. Importantly, dasatinib inhibits SARS-CoV-2 and MERS-CoV Mac1 but not the closest human homolog, MacroD2. The selectivity demonstrates it is possible to discover drugs that specifically inhibit viral macrodomains. Although cytotoxic when used at µM concentration, dasatinib has antiviral activities against SARS-CoV and MERS-CoV through an unknown mechanism [73]. These proof-of-concept data pave the way for screening large compound libraries to identify improved macrodomain inhibitors and explore their potential as antiviral therapies for SARS-CoV-2 and future viral threats [54].
Finally, Russo et al. also developed an immunofluorescence-based assay to assess the activity of SARS-CoV-2 Mac1 activity in cells [74]. Activation of IFN responses, by treating cells with IFN-γ or the double-stranded RNA mimic, poly(I:C), robustly induces ADP-ribosylation in cells, which can be removed by ectopic expression of wild type, but not catalytically inactive mutant, Mac1. Although the initial screen of a limited set of compounds did not yield any hits, this assay may potentially be used in the future for testing the target engagement of potential macrodomain inhibitors in cells.
9. Conclusions
Viral macrodomains offer an attractive antiviral target because they are highly conserved and have structurally well-defined (druggable) binding pockets. Because the viral macrodomain is mechanistically distinct from more common antiviral targets (e.g., viral polymerases or proteases), a macrodomain inhibitor would facilitate development of combination therapies for optimal treatment (as successfully developed for HIV). Besides inhibiting viral replication, macrodomain inhibitors may also boost immune responses that contribute to the recovery from disease [75].
Furthermore, coronaviruses and alphaviruses are prominent veterinary pathogens, so macrodomain inhibitors may also be useful for treatment of animals. While the current focus is on targeting macrodomains for antiviral therapy, drug development efforts may identify compounds that can inhibit human macrodomains as well. These inhibitors may have important uses in other human diseases such as cancer, metabolic disorders, and inflammatory diseases. In addition, first-generation macrodomain inhibitors may also serve as tools to probe pathways regulated by ADP-ribosylation that may be attractive novel targets for development of therapeutic interventions.
Funding
This research was funded by COVID-19 PreClinical Research Discovery Fund from Johns Hopkins University (A.K.L.L.) and discretionary funds from Johns Hopkins Bloomberg School of Public Health (A.K.L.L.). Macrodomain biology investigations have been funded by a Johns Hopkins Catalyst Award (A.K.L.L.), pilot grants from the Johns Hopkins University School of Medicine’s Sherrilyn and Ken Fisher Center for Environmental Infectious Disease (D.E.G. and A.K.L.L.), and NIH grants R56AI137264 (D.E.G. and A.K.L.L.), R01GM104135 (A.K.L.L.), R35GM138029 (A.R.F.), K22AI134993-01 (A.R.F.), P20GM113117 (A.R.F.), and startup funds from the University of Kansas (A.R.F).
Acknowledgments
Conflicts of Interest
D.E.G. is on advisory boards for Takeda Pharmaceuticals, GlaxoSmithKline, and GreenLight Biosciences. The other authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Wang, Y.; Grunewald, M.; Perlman, S. Coronaviruses: An Updated Overview of Their Replication and Pathogenesis. Methods Mol. Biol. 2020, 2203, 1–29. [Google Scholar] [CrossRef]
- Weaver, S.C.; Winegar, R.; Manger, I.D.; Forrester, N.L. Alphaviruses: Population genetics and determinants of emergence. Antivir. Res. 2012, 94, 242–257. [Google Scholar] [CrossRef] [Green Version]
- Zeller, H.; Van Bortel, W.; Sudre, B. Chikungunya: Its History in Africa and Asia and Its Spread to New Regions in 2013–2014. J. Infect. Dis. 2016, 214, S436–S440. [Google Scholar] [CrossRef] [PubMed]
- Girard, M.; Nelson, C.B.; Picot, V.; Gubler, D.J. Arboviruses: A global public health threat. Vaccine 2020, 38, 3989–3994. [Google Scholar] [CrossRef] [PubMed]
- Hoarau, J.-J.; Bandjee, M.-C.J.; Trotot, P.K.; Das, T.; Li-Pat-Yuen, G.; Dassa, B.; Denizot, M.; Guichard, E.; Ribera, A.; Henni, T.; et al. Persistent Chronic Inflammation and Infection by Chikungunya Arthritogenic Alphavirus in Spite of a Robust Host Immune Response. J. Immunol. 2010, 184, 5914–5927. [Google Scholar] [CrossRef] [Green Version]
- Borgherini, G.; Poubeau, P.; Jossaume, A.; Gouix, A.; Cotte, L.; Michault, A.; Arvin-Berod, C.; Paganin, F. Persistent Arthralgia Associated with Chikungunya Virus: A Study of 88 Adult Patients on Reunion Island. Clin. Infect. Dis. 2008, 47, 469–475. [Google Scholar] [CrossRef] [Green Version]
- Ceulemans, L.J.; Khan, M.; Yoo, S.-J.; Zapiec, B.; Van Gerven, L.; Van Slambrouck, J.; Vanstapel, A.; Van Raemdonck, D.; Vos, R.; Wauters, E.; et al. Persistence of SARS-CoV-2 RNA in lung tissue after mild COVID-19. Lancet Respir. Med. 2021, 9, e78–e79. [Google Scholar] [CrossRef]
- Gaspar-Rodríguez, A.; Padilla-González, A.; Rivera-Toledo, E. Coronavirus persistence in human respiratory tract and cell culture: An overview. Braz. J. Infect. Dis. 2021, 25, 101632. [Google Scholar] [CrossRef]
- Lin, W.-H.W.; Kouyos, R.; Adams, R.J.; Grenfell, B.T.; Griffin, D.E. Prolonged persistence of measles virus RNA is characteristic of primary infection dynamics. Proc. Natl. Acad. Sci. USA 2012, 109, 14989–14994. [Google Scholar] [CrossRef] [Green Version]
- Metcalf, T.U.; Griffin, D.E. Alphavirus-Induced Encephalomyelitis: Antibody-Secreting Cells and Viral Clearance from the Nervous System. J. Virol. 2011, 85, 11490–11501. [Google Scholar] [CrossRef] [Green Version]
- Nelson, A.N.; Lin, W.-H.W.; Shivakoti, R.; Putnam, N.E.; Mangus, L.M.; Adams, R.J.; Hauer, D.; Baxter, V.K.; Griffin, D.E. Association of persistent wild-type measles virus RNA with long-term humoral immunity in rhesus macaques. JCI Insight 2020, 5, e134992. [Google Scholar] [CrossRef] [Green Version]
- Yang, B.; Fan, J.; Huang, J.; Guo, E.; Fu, Y.; Liu, S.; Xiao, R.; Liu, C.; Lu, F.; Qin, T.; et al. Clinical and molecular characteristics of COVID-19 patients with persistent SARS-CoV-2 infection. Nat. Commun. 2021, 12, 3501. [Google Scholar] [CrossRef]
- Levine, B.; Hardwick, J.M.; Griffin, D.E. Persistence of alphaviruses in vertebrate hosts. Trends Microbiol. 1994, 2, 25–28. [Google Scholar] [CrossRef]
- De Andrade, D.C.; Jean, S.; Clavelou, P.; Dallel, R.; Bouhassira, D. Chronic pain associated with the Chikungunya Fever: Long lasting burden of an acute illness. BMC Infect. Dis. 2010, 10, 31. [Google Scholar] [CrossRef]
- Soumahoro, M.-K.; Gérardin, P.; Boelle, P.-Y.; Perrau, J.; Fianu, A.; Pouchot, J.; Malvy, D.; Flahault, A.; Favier, F.; Hanslik, T. Impact of Chikungunya Virus Infection on Health Status and Quality of Life: A Retrospective Cohort Study. PLoS ONE 2009, 4, e7800. [Google Scholar] [CrossRef] [PubMed]
- Shyr, Z.A.; Gorshkov, K.; Chen, C.Z.; Zheng, W. Drug Discovery Strategies for SARS-CoV-2. J. Pharmacol. Exp. Ther. 2020, 375, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Battisti, V.; Urban, E.; Langer, T. Antivirals against the Chikungunya Virus. Viruses 2021, 13, 1307. [Google Scholar] [CrossRef]
- Kaur, P.; Chu, J.J.H. Chikungunya virus: An update on antiviral development and challenges. Drug Discov. Today 2013, 18, 969–983. [Google Scholar] [CrossRef]
- Fehr, A.R.; Jankevicius, G.; Ahel, I.; Perlman, S. Viral Macrodomains: Unique Mediators of Viral Replication and Pathogenesis. Trends Microbiol. 2018, 26, 598–610. [Google Scholar] [CrossRef]
- Leung, A.K.L.; McPherson, R.L.; Griffin, D.E. Macrodomain ADP-ribosylhydrolase and the pathogenesis of infectious diseases. PLoS Pathog. 2018, 14, e1006864. [Google Scholar] [CrossRef]
- Lüscher, B.; Ahel, I.; Altmeyer, M.; Ashworth, A.; Bai, P.; Chang, P.; Cohen, M.; Corda, D.; Dantzer, F.; Daugherty, M.D.; et al. ADP-ribosyltransferases, an update on function and nomenclature. FEBS J. 2021. Available online: https://doi.org/10.1111/febs.16142 (accessed on 7 January 2022). [CrossRef] [PubMed]
- Palazzo, L.; Mikolčević, P.; Mikoč, A.; Ahel, I. ADP-ribosylation signalling and human disease. Open Biol. 2019, 9, 190041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rack, J.G.M.; Perina, D.; Ahel, I. Macrodomains: Structure, Function, Evolution, and Catalytic Activities. Annu. Rev. Biochem. 2016, 85, 431–454. [Google Scholar] [CrossRef]
- Karras, G.; Kustatscher, G.; Buhecha, H.R.; Allen, M.D.; Pugieux, C.; Sait, F.; Bycroft, M.; Ladurner, A.G. The macro domain is an ADP-ribose binding module. EMBO J. 2005, 24, 1911–1920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eckei, L.; Krieg, S.; Bütepage, M.; Lehmann, A.; Gross, A.; Lippok, B.; Grimm, A.R.; Kümmerer, B.M.; Rossetti, G.; Lüscher, B.; et al. The conserved macrodomains of the non-structural proteins of Chikungunya virus and other pathogenic positive strand RNA viruses function as mono-ADP-ribosylhydrolases. Sci. Rep. 2017, 7, 41746. [Google Scholar] [CrossRef] [Green Version]
- Jankevicius, G.; Hassler, M.; Golia, B.; Rybin, V.; Zacharias, M.; Timinszky, G.; Ladurner, A.G. A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nat. Struct. Mol. Biol. 2013, 20, 508–514. [Google Scholar] [CrossRef]
- Li, C.; Debing, Y.; Jankevicius, G.; Neyts, J.; Ahel, I.; Coutard, B.; Canard, B. Viral Macro Domains Reverse Protein ADP-Ribosylation. J. Virol. 2016, 90, 8478–8486. [Google Scholar] [CrossRef] [Green Version]
- McPherson, R.L.; Abraham, R.; Sreekumar, E.; Ong, S.-E.; Cheng, S.-J.; Baxter, V.K.; Kistemaker, H.A.V.; Filippov, D.V.; Griffin, D.E.; Leung, A.K.L. ADP-ribosylhydrolase activity of Chikungunya virus macrodomain is critical for virus replication and virulence. Proc. Natl. Acad. Sci. USA 2017, 114, 1666–1671. [Google Scholar] [CrossRef] [Green Version]
- Rosenthal, F.; Feijs, K.L.; Frugier, E.; Bonalli, M.; Forst, A.H.; Imhof, R.; Winkler, H.C.; Fischer, D.; Caflisch, A.; Hassa, P.O.; et al. Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nat. Struct. Mol. Biol. 2013, 20, 502–507. [Google Scholar] [CrossRef]
- Daniels, C.M.; Ong, S.-E.; Leung, A.K. Phosphoproteomic Approach to Characterize Protein Mono- and Poly(ADP-ribosyl)ation Sites from Cells. J. Proteome Res. 2014, 13, 3510–3522. [Google Scholar] [CrossRef] [Green Version]
- Neuman, B.W. Bioinformatics and functional analyses of coronavirus nonstructural proteins involved in the formation of replicative organelles. Antivir. Res. 2016, 135, 97–107. [Google Scholar] [CrossRef]
- Alhammad, Y.M.O.; Kashipathy, M.M.; Roy, A.; Gagné, J.-P.; McDonald, P.; Gao, P.; Nonfoux, L.; Battaile, K.P.; Johnson, D.K.; Holmstrom, E.D.; et al. The SARS-CoV-2 Conserved Macrodomain Is a Mono-ADP-Ribosylhydrolase. J. Virol. 2021, 95, e01969-20. [Google Scholar] [CrossRef]
- Jayabalan, A.K.; Adivarahan, S.; Koppula, A.; Abraham, R.; Batish, M.; Zenklusen, D.; Griffin, D.E.; Leung, A.K.L. Stress granule formation, disassembly, and composition are regulated by alphavirus ADP-ribosylhydrolase activity. Proc. Natl. Acad. Sci. USA 2021, 118, e2021719118. [Google Scholar] [CrossRef]
- Abraham, R.; Hauer, D.; McPherson, R.L.; Utt, A.; Kirby, I.T.; Cohen, M.S.; Merits, A.; Leung, A.K.L.; Griffin, D.E. ADP-ribosyl–binding and hydrolase activities of the alphavirus nsP3 macrodomain are critical for initiation of virus replication. Proc. Natl. Acad. Sci. USA 2018, 115, E10457–E10466. [Google Scholar] [CrossRef] [Green Version]
- Abraham, R.; McPherson, R.L.; Dasovich, M.; Badiee, M.; Leung, A.K.L.; Griffin, D.E. Both ADP-Ribosyl-Binding and Hydrolase Activities of the Alphavirus nsP3 Macrodomain Affect Neurovirulence in Mice. mBio 2020, 11, e03253-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voth, L.S.; O’Connor, J.J.; Kerr, C.M.; Doerger, E.; Schwarting, N.; Sperstad, P.; Johnson, D.K.; Fehr, A.R. Unique Mutations in the Murine Hepatitis Virus Macrodomain Differentially Attenuate Virus Replication, Indicating Multiple Roles for the Macrodomain in Coronavirus Replication. J. Virol. 2021, 95, e0076621. [Google Scholar] [CrossRef]
- Fehr, A.R.; Channappanavar, R.; Jankevicius, G.; Fett, C.; Zhao, J.; Athmer, J.; Meyerholz, D.K.; Ahel, I.; Perlman, S. The Conserved Coronavirus Macrodomain Promotes Virulence and Suppresses the Innate Immune Response during Severe Acute Respiratory Syndrome Coronavirus Infection. mBio 2016, 7, e01721-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alhammad, Y.M.O.; Fehr, A.R. The Viral Macrodomain Counters Host Antiviral ADP-Ribosylation. Viruses 2020, 12, 384. [Google Scholar] [CrossRef] [Green Version]
- Grunewald, M.E.; Chen, Y.; Kuny, C.; Maejima, T.; Lease, R.; Ferraris, D.; Aikawa, M.; Sullivan, C.S.; Perlman, S.; Fehr, A.R. The coronavirus macrodomain is required to prevent PARP-mediated inhibition of virus replication and enhancement of IFN expression. PLoS Pathog. 2019, 15, e1007756. [Google Scholar] [CrossRef] [Green Version]
- Channappanavar, R.; Fehr, A.; Vijay, R.; Mack, M.; Zhao, J.; Meyerholz, D.; Perlman, S. Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice. Cell Host Microbe 2016, 19, 181–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Channappanavar, R.; Fehr, A.R.; Zheng, J.; Wohlford-Lenane, C.; Abrahante, J.E.; Mack, M.; Sompallae, R.; McCray, P.B., Jr.; Meyerholz, D.K.; Perlman, S. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Investig. 2019, 129, 3625–3639. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.E.; Hughes, J.; Gu, Q.; Behdenna, A.; Singer, J.B.; Dennis, T.; Orton, R.J.; Varela, M.; Gifford, R.J.; Wilson, S.J.; et al. Fundamental properties of the mammalian innate immune system revealed by multispecies comparison of type I interferon responses. PLoS Biol. 2017, 15, e2004086. [Google Scholar] [CrossRef] [PubMed]
- Mavian, C.; Ramirez-Mata, A.S.; Dollar, J.J.; Nolan, D.J.; Cash, M.; White, K.; Rich, S.N.; Magalis, B.R.; Marini, S.; Prosperi, M.C.F.; et al. Brain tissue transcriptomic analysis of SIV-infected macaques identifies several altered metabolic pathways linked to neuropathogenesis and poly (ADP-ribose) polymerases (PARPs) as potential therapeutic targets. J. NeuroVirol. 2021, 27, 101–115. [Google Scholar] [CrossRef] [PubMed]
- Heer, C.D.; Sanderson, D.J.; Voth, L.S.; Alhammad, Y.M.O.; Schmidt, M.S.; Trammell, S.A.J.; Perlman, S.; Cohen, M.S.; Fehr, A.R.; Brenner, C. Coronavirus infection and PARP expression dysregulate the NAD metabolome: An actionable component of innate immunity. J. Biol. Chem. 2020, 295, 17986–17996. [Google Scholar] [CrossRef]
- Fehr, A.R.; Singh, S.A.; Kerr, C.M.; Mukai, S.; Higashi, H.; Aikawa, M. The impact of PARPs and ADP-ribosylation on inflammation and host–pathogen interactions. Genes Dev. 2020, 34, 341–359. [Google Scholar] [CrossRef]
- Atasheva, S.; Akhrymuk, M.; Frolova, E.I.; Frolov, I. New PARP Gene with an Anti-Alphavirus Function. J. Virol. 2012, 86, 8147–8160. [Google Scholar] [CrossRef] [Green Version]
- Atasheva, S.; Frolova, E.I.; Frolov, I. Interferon-Stimulated Poly(ADP-Ribose) Polymerases Are Potent Inhibitors of Cellular Translation and Virus Replication. J. Virol. 2014, 88, 2116–2130. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Zhao, H.; Liu, P.; Li, C.; Quanquin, N.; Ji, X.; Sun, N.; Du, P.; Qin, C.-F.; Lu, N.; et al. PARP12 suppresses Zika virus infection through PARP-dependent degradation of NS1 and NS3 viral proteins. Sci. Signal. 2018, 11, eaas9332. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Shi, Y.; Li, S.; Liu, J.; Zu, S.; Xu, X.; Gao, M.; Sun, N.; Pan, C.; Peng, L.; et al. ADP-ribosyltransferase PARP11 suppresses Zika virus in synergy with PARP12. Cell Biosci. 2021, 11, 116. [Google Scholar] [CrossRef]
- Lee, S.; Lee, Y.-S.; Choi, Y.; Son, A.; Park, Y.; Lee, K.-M.; Kim, J.; Kim, J.-S.; Kim, V.N. The SARS-CoV-2 RNA interactome. Mol. Cell 2021, 81, 2838–2850.e2836. [Google Scholar] [CrossRef]
- Guo, T.; Zuo, Y.; Qian, L.; Liu, J.; Yuan, Y.; Xu, K.; Miao, Y.; Feng, Q.; Chen, X.; Jin, L.; et al. ADP-ribosyltransferase PARP11 modulates the interferon antiviral response by mono-ADP-ribosylating the ubiquitin E3 ligase β-TrCP. Nat. Microbiol. 2019, 4, 1872–1884. [Google Scholar] [CrossRef]
- Yamada, T.; Horimoto, H.; Kameyama, T.; Hayakawa, S.; Yamato, H.; Dazai, M.; Takada, A.; Kida, H.; Bott, D.; Zhou, A.C.; et al. Constitutive aryl hydrocarbon receptor signaling constrains type I interferon–mediated antiviral innate defense. Nat. Immunol. 2016, 17, 687–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, E.; Griffin, D.E. The nsP3 macro domain is important for Sindbis virus replication in neurons and neurovirulence in mice. Virology 2009, 388, 305–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dasovich, M.; Zhuo, J.; Goodman, J.A.; Thomas, A.G.; McPherson, R.L.; Jayabalan, A.K.; Busa, V.F.; Cheng, S.-J.; Murphy, B.A.; Redinger, K.R.; et al. High-Throughput Activity Assay for Screening Inhibitors of the SARS-CoV-2 Mac1 Macrodomain. ACS Chem. Biol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Malet, H.; Coutard, B.; Jamal, S.; Dutartre, H.; Papageorgiou, N.; Neuvonen, M.; Ahola, T.; Forrester, N.; Gould, E.A.; Lafitte, D.; et al. The Crystal Structures of Chikungunya and Venezuelan Equine Encephalitis Virus nsP3 Macro Domains Define a Conserved Adenosine Binding Pocket. J. Virol. 2009, 83, 6534–6545. [Google Scholar] [CrossRef] [Green Version]
- Rack, J.G.M.; Palazzo, L.; Ahel, I. (ADP-ribosyl)hydrolases: Structure, function, and biology. Genes Dev. 2020, 34, 263–284. [Google Scholar] [CrossRef]
- Frick, D.N.; Virdi, R.S.; Vuksanovic, N.; Dahal, N.; Silvaggi, N.R. Molecular Basis for ADP-Ribose Binding to the Mac1 Domain of SARS-CoV-2 nsp3. Biochemistry 2020, 59, 2608–2615. [Google Scholar] [CrossRef]
- Virdi, R.S.; Bavisotto, R.V.; Hopper, N.C.; Vuksanovic, N.; Melkonian, T.R.; Silvaggi, N.R.; Frick, D.N. Discovery of Drug-Like Ligands for the Mac1 Domain of SARS-CoV-2 Nsp3. SLAS Discov. 2020, 25, 1162–1170. [Google Scholar] [CrossRef]
- Babar, Z.; Khan, M.; Zahra, M.; Anwar, M.; Noor, K.; Hashmi, H.F.; Suleman, M.; Waseem, M.; Shah, A.; Ali, S.; et al. Drug similarity and structure-based screening of medicinal compounds to target macrodomain-I from SARS-CoV-2 to rescue the host immune system: A molecular dynamics study. J. Biomol. Struct. Dyn. 2020, 40, 523–537. [Google Scholar] [CrossRef]
- Bajusz, D.; Wade, W.S.; Satała, G.; Bojarski, A.J.; Ilaš, J.; Ebner, J.; Grebien, F.; Papp, H.; Jakab, F.; Douangamath, A.; et al. Exploring protein hotspots by optimized fragment pharmacophores. Nat. Commun. 2021, 12, 3201. [Google Scholar] [CrossRef]
- Debnath, P.; Debnath, B.; Bhaumik, S.; Debnath, S. In Silico Identification of Potential Inhibitors of ADP-Ribose Phosphatase of SARS-CoV-2 nsP3 by Combining E-Pharmacophore- and Receptor-Based Virtual Screening of Database. ChemistrySelect 2020, 5, 9388–9398. [Google Scholar] [CrossRef]
- Jung, L.S.; Gund, T.M.; Narayan, M. Comparison of Binding Site of Remdesivir and Its Metabolites with NSP12-NSP7-NSP8, and NSP3 of SARS-CoV-2 Virus and Alternative Potential Drugs for COVID-19 Treatment. Protein J. 2020, 39, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Ni, X.; Schröder, M.; Olieric, V.; Sharpe, M.E.; Hernandez-Olmos, V.; Proschak, E.; Merk, D.; Knapp, S.; Chaikuad, A. Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain. ACS Med. Chem. Lett. 2021, 12, 603–609. [Google Scholar] [CrossRef]
- Selvaraj, C.; Dinesh, D.C.; Panwar, U.; Boura, E.; Singh, S.K. High-Throughput Screening and Quantum Mechanics for Identifying Potent Inhibitors Against Mac1 Domain of SARS-CoV-2 Nsp3. IEEE/ACM Trans. Comput. Biol. Bioinform. 2021, 18, 1262–1270. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Kushwaha, P.P.; Prajapati, K.S.; Shuaib, M.; Gupta, S.; Kumar, S. Identification of FDA approved drugs and nucleoside analogues as potential SARS-CoV-2 A1pp domain inhibitor: An in silico study. Comput. Biol. Med. 2021, 130, 104185. [Google Scholar] [CrossRef]
- Zhang, S.; Garzan, A.; Haese, N.; Bostwick, R.; Martinez-Gzegozewska, Y.; Rasmussen, L.; Streblow, D.N.; Haise, M.T.; Pathak, A.K.; Augelli-Szafran, C.E.; et al. Pyrimidone inhibitors targeting Chikungunya Virus nsP3 macrodomain by fragment-based drug design. PLoS ONE 2021, 16, e0245013. [Google Scholar] [CrossRef]
- Schuller, M.; Correy, G.J.; Gahbauer, S.; Fearon, D.; Wu, T.; Díaz, R.E.; Young, I.D.; Martins, L.C.; Smith, D.H.; Schulze-Gahmen, U.; et al. Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking. Sci. Adv. 2021, 7, eabf8711. [Google Scholar] [CrossRef]
- Rack, J.G.M.; Zorzini, V.; Zhu, Z.; Schuller, M.; Ahel, D.; Ahel, I. Viral macrodomains: A structural and evolutionary assessment of the pharmacological potential. Open Biol. 2020, 10, 200237. [Google Scholar] [CrossRef]
- Schuller, M.; Riedel, K.; Gibbs-Seymour, I.; Uth, K.; Sieg, C.; Gehring, A.P.; Ahel, I.; Bracher, F.; Kessler, B.M.; Elkins, J.M.; et al. Discovery of a Selective Allosteric Inhibitor Targeting Macrodomain 2 of Polyadenosine-Diphosphate-Ribose Polymerase 14. ACS Chem. Biol. 2017, 12, 2866–2874. [Google Scholar] [CrossRef] [PubMed]
- Sowa, S.T.; Galera-Prat, A.; Wazir, S.; Alanen, H.I.; Maksimainen, M.M.; Lehtiö, L. A molecular toolbox for ADP-ribosyl binding proteins. Cell Rep. Methods 2021, 1, 100121. [Google Scholar] [CrossRef]
- Albulescu, I.C.; White-Scholten, L.; Tas, A.; Hoornweg, T.E.; Ferla, S.; Kovacikova, K.; Smit, J.M.; Brancale, A.; Snijder, E.J.; van Hemert, M.J. Suramin Inhibits Chikungunya Virus Replication by Interacting with Virions and Blocking the Early Steps of Infection. Viruses 2020, 12, 314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daniels, C.; Thirawatananond, P.; Ong, S.-E.; Gabelli, S.B.; Leung, A.K.L. Nudix hydrolases degrade protein-conjugated ADP-ribose. Sci. Rep. 2015, 5, 18271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dyall, J.; Coleman, C.M.; Hart, B.J.; Venkataraman, T.; Holbrook, M.R.; Kindrachuk, J.; Johnson, R.F.; Olinger, G.G., Jr.; Jahrling, P.B.; Laidlaw, M.; et al. Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob. Agents Chemother. 2014, 58, 4885–4893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russo, L.C.; Tomasin, R.; Araújo Matos, I.; Manucci, A.C.; Sowa, S.T.; Dale, K.; Caldecott, K.W.; Lehtiö, L.; Schechtman, D.; Meotti, F.C.; et al. The SARS-CoV-2 Nsp3 macrodomain reverses PARP9/DTX3L-dependent ADP-ribosylation induced by interferon signalling. J. Biol. Chem. 2021, 297, 101041. [Google Scholar] [CrossRef] [PubMed]
- Lazear, H.M.; Schoggins, J.W.; Diamond, M.S. Shared and Distinct Functions of Type I and Type III Interferons. Immunity 2019, 50, 907–923. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).