Cellular Organelles Involved in Hepatitis E Virus Infection
Abstract
:1. Introduction
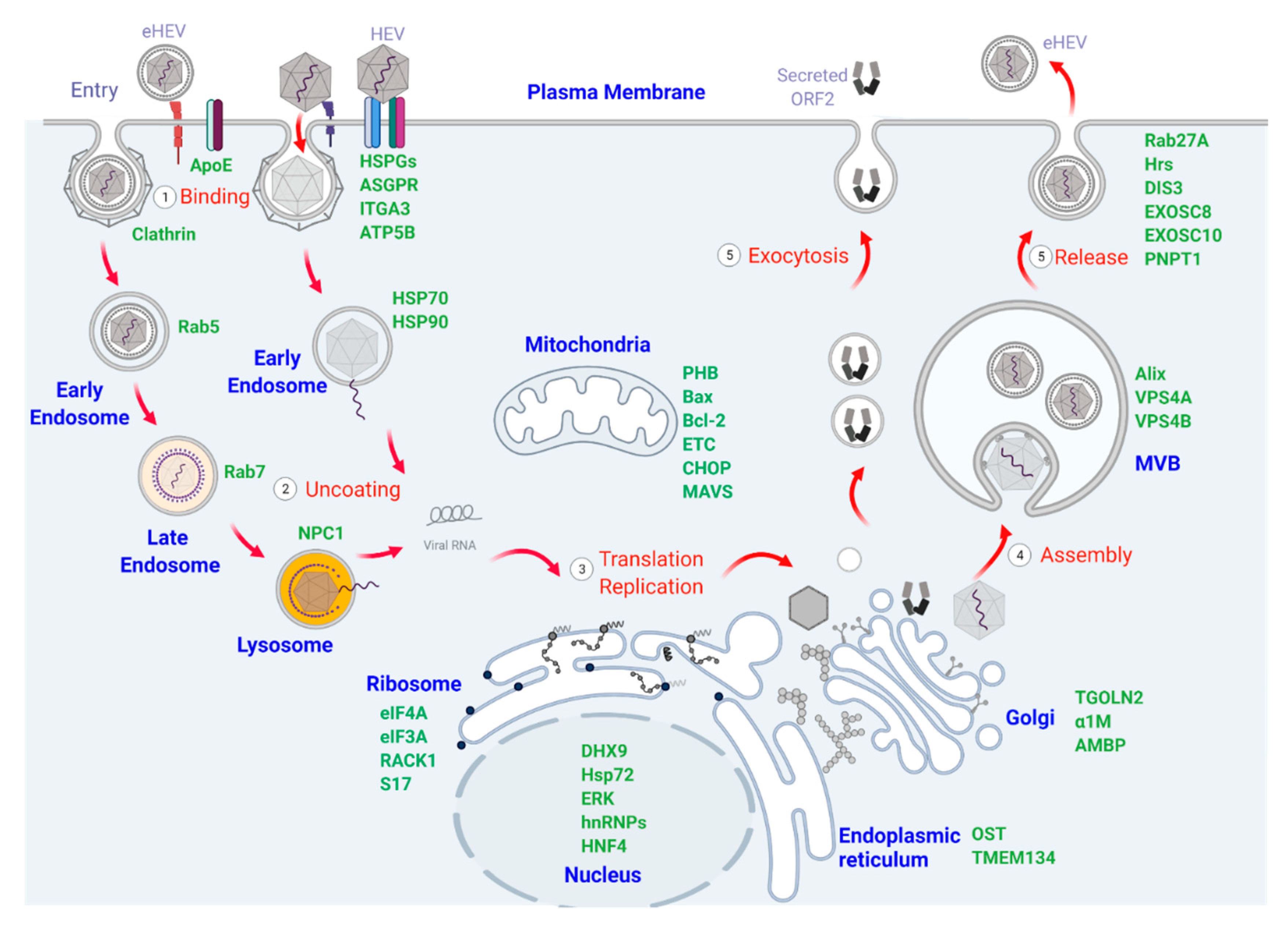
2. Cellular Organelles and Proteins Participating in HEV Binding, Entry, and Uncoating
2.1. Plasma Membrane and Membrane-Associated Proteins That Mediate HEV Binding and Entry
2.2. Endosomal Vesicles and Related Signaling Pathways Involved in HEV Trafficking
3. Cellular Organelles and Proteins Involved in HEV Translation and Replication
3.1. Endoplasmic Reticulum and ER-Resident Proteins Involved in HEV Translation and Replication
3.2. Ribosomes and Associated Factors Necessary for Translation of HEV Proteins
3.3. Mitochondria and Related Signaling Participating in HEV Infection
3.4. Interactions between HEV Components and the Nucleus during Virus Infection
4. Cellular Organelles and Proteins Involved in HEV Assembly and Release
4.1. The Role of the Golgi Apparatus in HEV Assembly
4.2. Multivesicular Bodies (MVB) and Exosomal Pathways in HEV Egress
5. Conclusions and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- WHO. Hepatitis E. Available online: https://www.who.int/news-room/fact-sheets/detail/hepatitis-e (accessed on 27 July 2021).
- Li, P.; Liu, J.; Li, Y.; Su, J.; Ma, Z.; Bramer, W.M.; Cao, W.; de Man, R.A.; Peppelenbosch, M.P.; Pan, Q. The global epidemiology of hepatitis E virus infection: A systematic review and meta-analysis. Liver Int. 2020, 40, 1516–1528. [Google Scholar] [CrossRef] [Green Version]
- Primadharsini, P.P.; Nagashima, S.; Okamoto, H. Mechanism of Cross-Species Transmission, Adaptive Evolution and Pathogenesis of Hepatitis E Virus. Viruses 2021, 13, 909. [Google Scholar] [CrossRef] [PubMed]
- Treagus, S.; Wright, C.; Baker-Austin, C.; Longdon, B.; Lowther, J. The Foodborne Transmission of Hepatitis E Virus to Humans. Food Environ. Virol. 2021, 13, 127–145. [Google Scholar] [CrossRef] [PubMed]
- Teshale, E.H.; Grytdal, S.P.; Howard, C.; Barry, V.; Kamili, S.; Drobeniuc, J.; Hill, V.R.; Okware, S.; Hu, D.J.; Holmberg, S.D. Evidence of person-to-person transmission of hepatitis E virus during a large outbreak in Northern Uganda. Clin. Infect. Dis. 2010, 50, 1006–1010. [Google Scholar] [CrossRef]
- Clayson, E.T.; Innis, B.L.; Myint, K.S.; Narupiti, S.; Vaughn, D.W.; Giri, S.; Ranabhat, P.; Shrestha, M.P. Detection of hepatitis E virus infections among domestic swine in the Kathmandu Valley of Nepal. Am. J. Trop. Med. Hyg. 1995, 53, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Neumann, S.; Hackl, S.S.; Piepenschneider, M.; Vina-Rodriguez, A.; Dremsek, P.; Ulrich, R.G.; Groschup, M.H.; Eiden, M. Serologic and Molecular Survey of Hepatitis E Virus in German Deer Populations. J. Wildl. Dis. 2016, 52, 106–113. [Google Scholar] [CrossRef]
- Geng, Y.; Zhao, C.; Song, A.; Wang, J.; Zhang, X.; Harrison, T.J.; Zhou, Y.; Wang, W.; Wang, Y. The serological prevalence and genetic diversity of hepatitis E virus in farmed rabbits in China. Infect. Genet. Evol. 2011, 11, 476–482. [Google Scholar] [CrossRef]
- Yamamoto, H.; Suzuki, J.; Matsuda, A.; Ishida, T.; Ami, Y.; Suzaki, Y.; Adachi, I.; Wakita, T.; Takeda, N.; Li, T.C. Hepatitis E virus outbreak in monkey facility, Japan. Emerg. Infect. Dis. 2012, 18, 2032–2034. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Li, Y.; Yu, W.; Jing, S.; Wang, J.; Long, F.; He, Z.; Yang, C.; Bi, Y.; Cao, W.; et al. Excretion of infectious hepatitis E virus into milk in cows imposes high risks of zoonosis. Hepatology 2016, 64, 350–359. [Google Scholar] [CrossRef] [Green Version]
- Long, F.; Yu, W.; Yang, C.; Wang, J.; Li, Y.; Li, Y.; Huang, F. High prevalence of hepatitis E virus infection in goats. J. Med. Virol. 2017, 89, 1981–1987. [Google Scholar] [CrossRef]
- Di Pasquale, S.; De Santis, P.; La Rosa, G.; Di Domenico, K.; Iaconelli, M.; Micarelli, G.; Martini, E.; Bilei, S.; De Medici, D.; Suffredini, E. Quantification and genetic diversity of Hepatitis E virus in wild boar (Sus scrofa) hunted for domestic consumption in Central Italy. Food Microbiol. 2019, 82, 194–201. [Google Scholar] [CrossRef]
- Meng, X.J. Expanding Host Range and Cross-Species Infection of Hepatitis E Virus. PLoS Pathog. 2016, 12, e1005695. [Google Scholar] [CrossRef] [PubMed]
- Rasche, A.; Saqib, M.; Liljander, A.M.; Bornstein, S.; Zohaib, A.; Renneker, S.; Steinhagen, K.; Wernery, R.; Younan, M.; Gluecks, I.; et al. Hepatitis E Virus Infection in Dromedaries, North and East Africa, United Arab Emirates, and Pakistan, 1983–2015. Emerg. Infect. Dis. 2016, 22, 1249–1252. [Google Scholar] [CrossRef]
- Wang, L.; Teng, J.L.L.; Lau, S.K.P.; Sridhar, S.; Fu, H.; Gong, W.; Li, M.; Xu, Q.; He, Y.; Zhuang, H.; et al. Transmission of a Novel Genotype of Hepatitis E Virus from Bactrian Camels to Cynomolgus Macaques. J. Virol. 2019, 93, e02014-18. [Google Scholar] [CrossRef] [Green Version]
- Dalton, H.R.; Seghatchian, J. Hepatitis E virus: Emerging from the shadows in developed countries. Transfus. Apher. Sci. 2016, 55, 271–274. [Google Scholar] [CrossRef] [PubMed]
- Kamar, N.; Pischke, S. Acute and Persistent Hepatitis E Virus Genotype 3 and 4 Infection: Clinical Features, Pathogenesis, and Treatment. Cold Spring Harb. Perspect. Med. 2019, 9, a031872. [Google Scholar] [CrossRef] [PubMed]
- Todt, D.; Meister, T.L.; Steinmann, E. Hepatitis E virus treatment and ribavirin therapy: Viral mechanisms of nonresponse. Curr. Opin. Virol. 2018, 32, 80–87. [Google Scholar] [CrossRef]
- Wands, J.R. Non-A, non-B hepatitis. Hepatology 1983, 3, 764–766. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Tanaka, T.; Takahashi, H.; Hoshino, Y.; Nagashima, S.; Jirintai, F.; Mizuo, H.; Yazaki, Y.; Takagi, T.; Azuma, M.; et al. Hepatitis E Virus (HEV) strains in serum samples can replicate efficiently in cultured cells despite the coexistence of HEV antibodies: Characterization of HEV virions in blood circulation. J. Clin. Microbiol. 2010, 48, 1112–1125. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, M.; Yamada, K.; Hoshino, Y.; Takahashi, H.; Ichiyama, K.; Tanaka, T.; Okamoto, H. Monoclonal antibodies raised against the ORF3 protein of hepatitis E virus (HEV) can capture HEV particles in culture supernatant and serum but not those in feces. Arch. Virol. 2008, 153, 1703–1713. [Google Scholar] [CrossRef]
- Balayan, M.S.; Andjaparidze, A.G.; Savinskaya, S.S.; Ketiladze, E.S.; Braginsky, D.M.; Savinov, A.P.; Poleschuk, V.F. Evidence for a virus in non-A, non-B hepatitis transmitted via the fecal-oral route. Intervirology 1983, 20, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Bouvier, N.M.; Palese, P. The biology of influenza viruses. Vaccine 2008, 26 (Suppl. 4), D49–D53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dearborn, A.D.; Marcotrigiano, J. Hepatitis C Virus Structure: Defined by What It Is Not. Cold Spring Harb. Perspect. Med. 2020, 10, a036822. [Google Scholar] [CrossRef]
- Sirohi, D.; Kuhn, R.J. Zika Virus Structure, Maturation, and Receptors. J. Infect. Dis. 2017, 216, S935–S944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
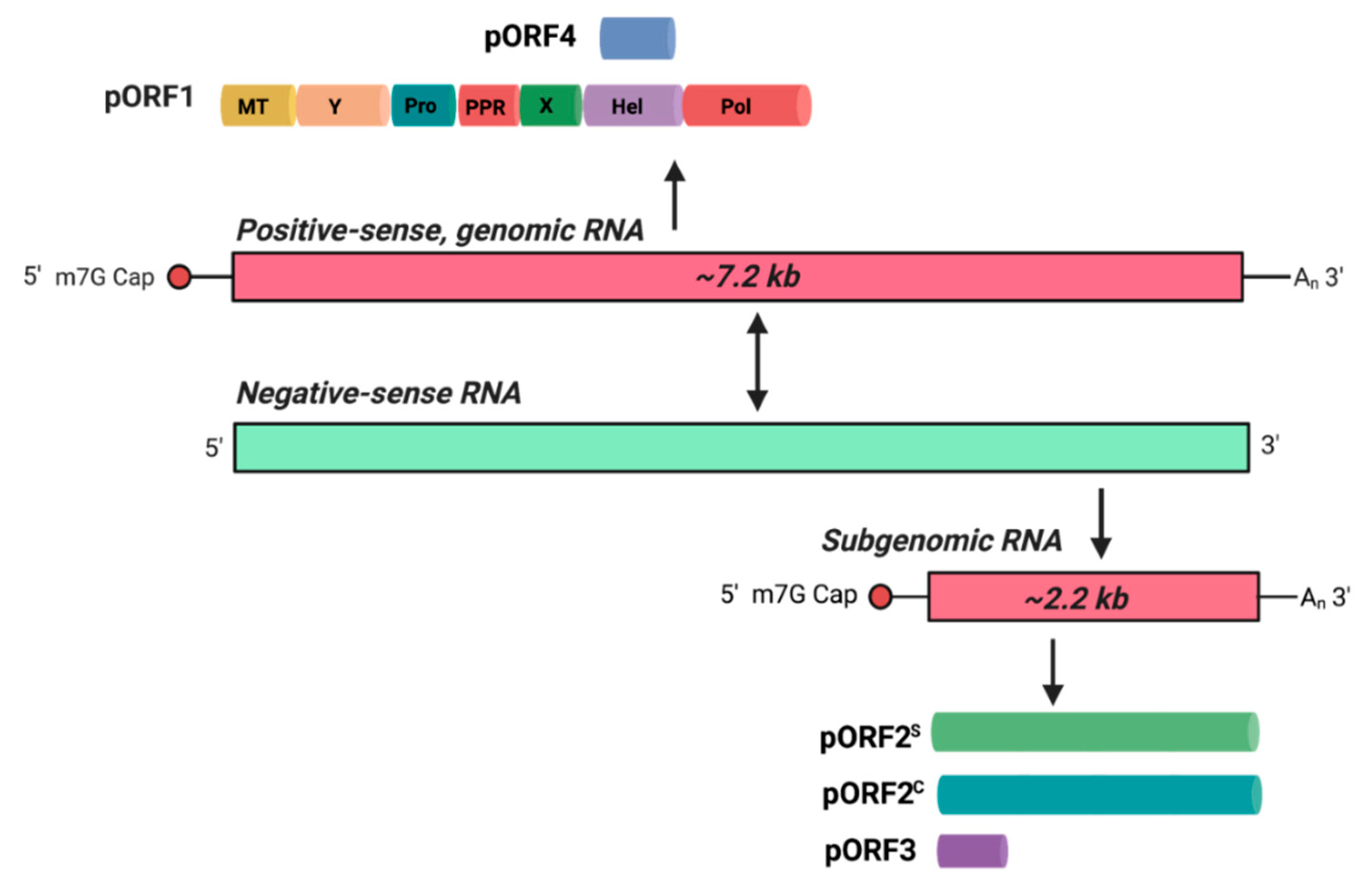
- Kenney, S.P.; Meng, X.J. Hepatitis E Virus Genome Structure and Replication Strategy. Cold Spring Harb. Perspect. Med. 2019, 9, a031724. [Google Scholar] [CrossRef]
- Sehgal, D.; Thomas, S.; Chakraborty, M.; Jameel, S. Expression and processing of the Hepatitis E virus ORF1 nonstructural polyprotein. Virol. J. 2006, 3, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, X.; Ying, D.; Lhomme, S.; Tang, Z.; Walker, C.M.; Xia, N.; Zheng, Z.; Feng, Z. Origin, antigenicity, and function of a secreted form of ORF2 in hepatitis E virus infection. Proc. Natl. Acad. Sci. USA 2018, 115, 4773–4778. [Google Scholar] [CrossRef] [Green Version]
- Gouttenoire, J.; Pollan, A.; Abrami, L.; Oechslin, N.; Mauron, J.; Matter, M.; Oppliger, J.; Szkolnicka, D.; Thi, V.L.D.; van der Goot, F.G.; et al. Palmitoylation mediates membrane association of hepatitis E virus ORF3 protein and is required for infectious particle secretion. PLoS Pathog. 2018, 14, e1007471. [Google Scholar] [CrossRef]
- Glitscher, M.; Hildt, E. Hepatitis E virus egress and beyond—The manifold roles of the viral ORF3 protein. Cell. Microbiol. 2021, e13379. [Google Scholar] [CrossRef]
- Ding, Q.; Heller, B.; Capuccino, J.M.; Song, B.; Nimgaonkar, I.; Hrebikova, G.; Contreras, J.E.; Ploss, A. Hepatitis E virus ORF3 is a functional ion channel required for release of infectious particles. Proc. Natl. Acad. Sci. USA 2017, 114, 1147–1152. [Google Scholar] [CrossRef] [Green Version]
- Nair, V.P.; Anang, S.; Subramani, C.; Madhvi, A.; Bakshi, K.; Srivastava, A.; Shalimar; Nayak, B.; Kumar, C.T.R.; Surjit, M. Endoplasmic Reticulum Stress Induced Synthesis of a Novel Viral Factor Mediates Efficient Replication of Genotype-1 Hepatitis E Virus. PLoS Pathog. 2016, 12, e1005521. [Google Scholar] [CrossRef]
- Guu, T.S.; Liu, Z.; Ye, Q.; Mata, D.A.; Li, K.; Yin, C.; Zhang, J.; Tao, Y.J. Structure of the hepatitis E virus-like particle suggests mechanisms for virus assembly and receptor binding. Proc. Natl. Acad. Sci. USA 2009, 106, 12992–12997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, X.; Ambardekar, C.; Lu, Y.; Feng, Z. Distinct Entry Mechanisms for Nonenveloped and Quasi-Enveloped Hepatitis E Viruses. J. Virol. 2016, 90, 4232–4242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graff, J.; Torian, U.; Nguyen, H.; Emerson, S.U. A bicistronic subgenomic mRNA encodes both the ORF2 and ORF3 proteins of hepatitis E virus. J. Virol. 2006, 80, 5919–5926. [Google Scholar] [CrossRef] [Green Version]
- Tyagi, S.; Korkaya, H.; Zafrullah, M.; Jameel, S.; Lal, S.K. The phosphorylated form of the ORF3 protein of hepatitis E virus interacts with its non-glycosylated form of the major capsid protein, ORF2. J. Biol. Chem. 2002, 277, 22759–22767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagashima, S.; Takahashi, M.; Kobayashi, T.; Tanggis; Nishizawa, T.; Nishiyama, T.; Primadharsini, P.P.; Okamoto, H. Characterization of the Quasi-Enveloped Hepatitis E Virus Particles Released by the Cellular Exosomal Pathway. J. Virol. 2017, 91, e00822-17. [Google Scholar] [CrossRef] [Green Version]
- Bagchi, P.; Banerjee, I.; Martin-Acebes, M.A. Editorial: Cell Organelle Exploitation by Viruses During Infection. Front. Microbiol. 2021, 12, 675152. [Google Scholar] [CrossRef]
- Fu, R.M.; Decker, C.C.; Thi, V.L.D. Cell Culture Models for Hepatitis E Virus. Viruses 2019, 11, 608. [Google Scholar] [CrossRef] [Green Version]
- van Deventer, S.; Arp, A.B.; van Spriel, A.B. Dynamic Plasma Membrane Organization: A Complex Symphony. Trends Cell Biol. 2021, 31, 119–129. [Google Scholar] [CrossRef]
- Burckhardt, C.J.; Greber, U.F. Virus movements on the plasma membrane support infection and transmission between cells. PLoS Pathog. 2009, 5, e1000621. [Google Scholar] [CrossRef]
- Kalia, M.; Chandra, V.; Rahman, S.A.; Sehgal, D.; Jameel, S. Heparan sulfate proteoglycans are required for cellular binding of the hepatitis E virus ORF2 capsid protein and for viral infection. J. Virol. 2009, 83, 12714–12724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, D.; Meng, X.J. Molecular biology and replication of hepatitis E virus. Emerg. Microbes Infect. 2012, 1, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Tian, Y.; Wen, Z.; Zhang, F.; Qi, Y.; Huang, W.; Zhang, H.; Wang, Y. Asialoglycoprotein receptor facilitates infection of PLC/PRF/5 cells by HEV through interaction with ORF2. J. Med. Virol. 2016, 88, 2186–2195. [Google Scholar] [CrossRef] [PubMed]
- Shiota, T.; Li, T.C.; Nishimura, Y.; Yoshizaki, S.; Sugiyama, R.; Shimojima, M.; Saijo, M.; Shimizu, H.; Suzuki, R.; Wakita, T.; et al. Integrin alpha3 is involved in non-enveloped hepatitis E virus infection. Virology 2019, 536, 119–124. [Google Scholar] [CrossRef]
- Christianson, H.C.; Belting, M. Heparan sulfate proteoglycan as a cell-surface endocytosis receptor. Matrix Biol. 2014, 35, 51–55. [Google Scholar] [CrossRef]
- Boulant, S.; Stanifer, M.; Lozach, P.Y. Dynamics of virus-receptor interactions in virus binding, signaling, and endocytosis. Viruses 2015, 7, 2794–2815. [Google Scholar] [CrossRef] [Green Version]
- Rogee, S.; Le Gall, M.; Chafey, P.; Bouquet, J.; Cordonnier, N.; Frederici, C.; Pavio, N. Quantitative proteomics identifies host factors modulated during acute hepatitis E virus infection in the swine model. J. Virol. 2015, 89, 129–143. [Google Scholar] [CrossRef] [Green Version]
- Gong, Y.; Cun, W. The Role of ApoE in HCV Infection and Comorbidity. Int. J. Mol. Sci. 2019, 20, 2037. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Yesupriya, A.; Chang, M.H.; Teshale, E.; Teo, C.G. Apolipoprotein E and protection against hepatitis E viral infection in American non-Hispanic blacks. Hepatology 2015, 62, 1346–1352. [Google Scholar] [CrossRef] [Green Version]
- Weller, R.; Todt, D.; Engelmann, M.; Friesland, M.; Wedemeyer, H.; Pietschmann, T.; Steinmann, E. Apolipoprotein E polymorphisms and their protective effect on hepatitis E virus replication. Hepatology 2016, 64, 2274–2276. [Google Scholar] [CrossRef]
- Zheng, Z.Z.; Miao, J.; Zhao, M.; Tang, M.; Yeo, A.E.; Yu, H.; Zhang, J.; Xia, N.S. Role of heat-shock protein 90 in hepatitis E virus capsid trafficking. J. Gen. Virol. 2010, 91, 1728–1736. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, Z.; Holla, P.; Ahmad, I.; Jameel, S. The ATP synthase subunit β (ATP5B) is an entry factor for the hepatitis E virus. bioRxiv 2016, 060434. [Google Scholar] [CrossRef] [Green Version]
- Neefjes, J.; Jongsma, M.M.L.; Berlin, I. Stop or Go? Endosome Positioning in the Establishment of Compartment Architecture, Dynamics, and Function. Trends Cell Biol. 2017, 27, 580–594. [Google Scholar] [CrossRef]
- DeTulleo, L.; Kirchhausen, T. The clathrin endocytic pathway in viral infection. EMBO J. 1998, 17, 4585–4593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, H.; Shi, W.; Freund, L.B. Mechanics of receptor-mediated endocytosis. Proc. Natl. Acad. Sci. USA 2005, 102, 9469–9474. [Google Scholar] [CrossRef] [Green Version]
- Costafreda, M.I.; Abbasi, A.; Lu, H.; Kaplan, G. Exosome mimicry by a HAVCR1—NPC1 pathway of endosomal fusion mediates hepatitis A virus infection. Nat. Microbiol. 2020, 5, 1096–1106. [Google Scholar] [CrossRef]
- Sun, S.; Tang, X.; Guo, Y.; Hu, J. Endoplasmic reticulum composition and form: Proteins in and out. Curr. Opin. Cell Biol. 2021, 71, 1–6. [Google Scholar] [CrossRef]
- Chen, S.; Novick, P.; Ferro-Novick, S. ER structure and function. Curr. Opin. Cell Biol. 2013, 25, 428–433. [Google Scholar] [CrossRef] [Green Version]
- Eymieux, S.; Blanchard, E.; Uzbekov, R.; Hourioux, C.; Roingeard, P. Annulate lamellae and intracellular pathogens. Cell. Microbiol. 2021, 23, e13328. [Google Scholar] [CrossRef] [PubMed]
- Ravindran, M.S.; Bagchi, P.; Cunningham, C.N.; Tsai, B. Opportunistic intruders: How viruses orchestrate ER functions to infect cells. Nat. Rev. Microbiol. 2016, 14, 407–420. [Google Scholar] [CrossRef] [PubMed]
- Inoue, T.; Tsai, B. How viruses use the endoplasmic reticulum for entry, replication, and assembly. Cold Spring Harb. Perspect. Biol. 2013, 5, a013250. [Google Scholar] [CrossRef]
- Romero-Brey, I.; Bartenschlager, R. Endoplasmic Reticulum: The Favorite Intracellular Niche for Viral Replication and Assembly. Viruses 2016, 8, 160. [Google Scholar] [CrossRef] [Green Version]
- Perttila, J.; Spuul, P.; Ahola, T. Early secretory pathway localization and lack of processing for hepatitis E virus replication protein pORF1. J. Gen. Virol. 2013, 94, 807–816. [Google Scholar] [CrossRef]
- John, L.; Thomas, S.; Herchenroder, O.; Putzer, B.M.; Schaefer, S. Hepatitis E virus ORF2 protein activates the pro-apoptotic gene CHOP and anti-apoptotic heat shock proteins. PLoS ONE 2011, 6, e25378. [Google Scholar] [CrossRef] [PubMed]
- Rehman, S.; Kapur, N.; Durgapal, H.; Panda, S.K. Subcellular localization of hepatitis E virus (HEV) replicase. Virology 2008, 370, 77–92. [Google Scholar] [CrossRef] [Green Version]
- Jameel, S.; Zafrullah, M.; Ozdener, M.H.; Panda, S.K. Expression in animal cells and characterization of the hepatitis E virus structural proteins. J. Virol. 1996, 70, 207–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Y.; Huang, W.; Yang, J.; Wen, Z.; Geng, Y.; Zhao, C.; Zhang, H.; Wang, Y. Systematic identification of hepatitis E virus ORF2 interactome reveals that TMEM134 engages in ORF2-mediated NF-kappaB pathway. Virus Res. 2017, 228, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Li, S.; Yang, C.; Wei, M.; Song, C.; Zheng, Z.; Gu, Y.; Du, H.; Zhang, J.; Xia, N. Homology model and potential virus-capsid binding site of a putative HEV receptor Grp78. J. Mol. Model. 2011, 17, 987–995. [Google Scholar] [CrossRef]
- Surjit, M.; Jameel, S.; Lal, S.K. Cytoplasmic localization of the ORF2 protein of hepatitis E virus is dependent on its ability to undergo retrotranslocation from the endoplasmic reticulum. J. Virol. 2007, 81, 3339–3345. [Google Scholar] [CrossRef] [Green Version]
- Zafrullah, M.; Ozdener, M.H.; Kumar, R.; Panda, S.K.; Jameel, S. Mutational analysis of glycosylation, membrane translocation, and cell surface expression of the hepatitis E virus ORF2 protein. J. Virol. 1999, 73, 4074–4082. [Google Scholar] [CrossRef] [Green Version]
- Montpellier, C.; Wychowski, C.; Sayed, I.M.; Meunier, J.C.; Saliou, J.M.; Ankavay, M.; Bull, A.; Pillez, A.; Abravanel, F.; Helle, F.; et al. Hepatitis E Virus Lifecycle and Identification of 3 Forms of the ORF2 Capsid Protein. Gastroenterology 2018, 154, 211–223.e8. [Google Scholar] [CrossRef]
- Parvez, M.K. Mutational analysis of hepatitis E virus ORF1 "Y-domain": Effects on RNA replication and virion infectivity. World J. Gastroenterol. 2017, 23, 590–602. [Google Scholar] [CrossRef] [PubMed]
- Lowther, J.; Naismith, J.H.; Dunn, T.M.; Campopiano, D.J. Structural, mechanistic and regulatory studies of serine palmitoyltransferase. Biochem. Soc. Trans. 2012, 40, 547–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ingolia, N.T.; Hussmann, J.A.; Weissman, J.S. Ribosome Profiling: Global Views of Translation. Cold Spring Harb. Perspect. Biol. 2019, 11, a032698. [Google Scholar] [CrossRef] [PubMed]
- Li, S. Regulation of Ribosomal Proteins on Viral Infection. Cells 2019, 8, 508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramani, C.; Nair, V.P.; Anang, S.; Mandal, S.D.; Pareek, M.; Kaushik, N.; Srivastava, A.; Saha, S.; Shalimar; Nayak, B.; et al. Host-Virus Protein Interaction Network Reveals the Involvement of Multiple Host Processes in the Life Cycle of Hepatitis E Virus. mSystems 2018, 3, e00135-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guca, E.; Hashem, Y. Major structural rearrangements of the canonical eukaryotic translation initiation complex. Curr. Opin. Struct. Biol. 2018, 53, 151–158. [Google Scholar] [CrossRef]
- Todt, D.; Moeller, N.; Praditya, D.; Kinast, V.; Friesland, M.; Engelmann, M.; Verhoye, L.; Sayed, I.M.; Behrendt, P.; Thi, V.L.D.; et al. The natural compound silvestrol inhibits hepatitis E virus (HEV) replication in vitro and in vivo. Antiviral. Res. 2018, 157, 151–158. [Google Scholar] [CrossRef]
- Glitscher, M.; Himmelsbach, K.; Woytinek, K.; Johne, R.; Reuter, A.; Spiric, J.; Schwaben, L.; Grunweller, A.; Hildt, E. Inhibition of Hepatitis E Virus Spread by the Natural Compound Silvestrol. Viruses 2018, 10, 301. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.S.; Tabata, K.; Twu, W.I.; Rahman, M.S.; Kim, H.S.; Yu, J.B.; Jee, M.H.; Bartenschlager, R.; Jang, S.K. RACK1 mediates rewiring of intracellular networks induced by hepatitis C virus infection. PLoS Pathog. 2019, 15, e1008021. [Google Scholar] [CrossRef] [Green Version]
- LaFontaine, E.; Miller, C.M.; Permaul, N.; Martin, E.T.; Fuchs, G. Ribosomal protein RACK1 enhances translation of poliovirus and other viral IRESs. Virology 2020, 545, 53–62. [Google Scholar] [CrossRef]
- Dobrikov, M.I.; Dobrikova, E.Y.; Gromeier, M. Ribosomal RACK1:Protein Kinase C betaII Modulates Intramolecular Interactions between Unstructured Regions of Eukaryotic Initiation Factor 4G (eIF4G) That Control eIF4E and eIF3 Binding. Mol. Cell. Biol. 2018, 38, e00306-18. [Google Scholar] [CrossRef] [Green Version]
- Kenney, S.P.; Meng, X.J. The lysine residues within the human ribosomal protein S17 sequence naturally inserted into the viral nonstructural protein of a unique strain of hepatitis E virus are important for enhanced virus replication. J. Virol. 2015, 89, 3793–3803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shukla, P.; Nguyen, H.T.; Torian, U.; Engle, R.E.; Faulk, K.; Dalton, H.R.; Bendall, R.P.; Keane, F.E.; Purcell, R.H.; Emerson, S.U. Cross-species infections of cultured cells by hepatitis E virus and discovery of an infectious virus-host recombinant. Proc. Natl. Acad. Sci. USA 2011, 108, 2438–2443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, H.T.; Torian, U.; Faulk, K.; Mather, K.; Engle, R.E.; Thompson, E.; Bonkovsky, H.L.; Emerson, S.U. A naturally occurring human/hepatitis E recombinant virus predominates in serum but not in faeces of a chronic hepatitis E patient and has a growth advantage in cell culture. J. Gen. Virol. 2012, 93, 526–530. [Google Scholar] [CrossRef] [PubMed]
- Vasileiou, P.V.S.; Evangelou, K.; Vlasis, K.; Fildisis, G.; Panayiotidis, M.I.; Chronopoulos, E.; Passias, P.G.; Kouloukoussa, M.; Gorgoulis, V.G.; Havaki, S. Mitochondrial Homeostasis and Cellular Senescence. Cells 2019, 8, 686. [Google Scholar] [CrossRef] [Green Version]
- Lai, J.H.; Luo, S.F.; Ho, L.J. Operation of mitochondrial machinery in viral infection-induced immune responses. Biochem. Pharmacol. 2018, 156, 348–356. [Google Scholar] [CrossRef]
- Neumann, S.; El Maadidi, S.; Faletti, L.; Haun, F.; Labib, S.; Schejtman, A.; Maurer, U.; Borner, C. How do viruses control mitochondria-mediated apoptosis? Virus Res. 2015, 209, 45–55. [Google Scholar] [CrossRef]
- Silwal, P.; Kim, J.K.; Kim, Y.J.; Jo, E.K. Mitochondrial Reactive Oxygen Species: Double-Edged Weapon in Host Defense and Pathological Inflammation During Infection. Front. Immunol. 2020, 11, 1649. [Google Scholar] [CrossRef] [PubMed]
- Soomro, M.H.; Shi, R.; She, R.; Yang, Y.; Hu, F.; Li, H. Antigen detection and apoptosis in Mongolian gerbil’s kidney experimentally intraperitoneally infected by swine hepatitis E virus. Virus Res. 2016, 213, 343–352. [Google Scholar] [CrossRef]
- Liu, B.; Chen, Y.; Zhao, L.; Zhang, M.; Ren, X.; Zhang, Y.; Zhang, B.; Fan, M.; Zhao, Q.; Zhou, E.M. Identification and pathogenicity of a novel genotype avian hepatitis E virus from silkie fowl (gallus gallus). Vet. Microbiol. 2020, 245, 108688. [Google Scholar] [CrossRef]
- Yang, Y.; Shi, R.; Soomro, M.H.; Hu, F.; Du, F.; She, R. Hepatitis E Virus Induces Hepatocyte Apoptosis via Mitochondrial Pathway in Mongolian Gerbils. Front. Microbiol. 2018, 9, 460. [Google Scholar] [CrossRef]
- Qu, C.; Zhang, S.; Wang, W.; Li, M.; Wang, Y.; van der Heijde-Mulder, M.; Shokrollahi, E.; Hakim, M.S.; Raat, N.J.H.; Peppelenbosch, M.P.; et al. Mitochondrial electron transport chain complex III sustains hepatitis E virus replication and represents an antiviral target. FASEB J. 2019, 33, 1008–1019. [Google Scholar] [CrossRef]
- Gotoh, T.; Terada, K.; Oyadomari, S.; Mori, M. hsp70-DnaJ chaperone pair prevents nitric oxide- and CHOP-induced apoptosis by inhibiting translocation of Bax to mitochondria. Cell Death Differ. 2004, 11, 390–402. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, C.; Horner, S.M. MAVS Coordination of Antiviral Innate Immunity. J. Virol. 2015, 89, 6974–6977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Liang, Y.; Qu, L.; Chen, Z.; Yi, M.; Li, K.; Lemon, S.M. Disruption of innate immunity due to mitochondrial targeting of a picornaviral protease precursor. Proc. Natl. Acad. Sci. USA 2007, 104, 7253–7258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.D.; Sun, L.; Seth, R.B.; Pineda, G.; Chen, Z.J. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc. Natl. Acad. Sci. USA 2005, 102, 17717–17722. [Google Scholar] [CrossRef] [Green Version]
- Yin, X.; Li, X.; Ambardekar, C.; Hu, Z.; Lhomme, S.; Feng, Z. Hepatitis E virus persists in the presence of a type III interferon response. PLoS Pathog. 2017, 13, e1006417. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Zhou, X.; Wang, W.; Wang, Y.; Yin, Y.; Laan, L.J.; Sprengers, D.; Metselaar, H.J.; Peppelenbosch, M.P.; Pan, Q. IFN regulatory factor 1 restricts hepatitis E virus replication by activating STAT1 to induce antiviral IFN-stimulated genes. FASEB J. 2016, 30, 3352–3367. [Google Scholar] [CrossRef] [Green Version]
- Dong, C.; Zafrullah, M.; Mixson-Hayden, T.; Dai, X.; Liang, J.; Meng, J.; Kamili, S. Suppression of interferon-alpha signaling by hepatitis E virus. Hepatology 2012, 55, 1324–1332. [Google Scholar] [CrossRef]
- Wissing, M.H.; Bruggemann, Y.; Steinmann, E.; Todt, D. Virus—Host Cell Interplay during Hepatitis E Virus Infection. Trends Microbiol. 2021, 29, 309–319. [Google Scholar] [CrossRef]
- Lenggenhager, D.; Gouttenoire, J.; Malehmir, M.; Bawohl, M.; Honcharova-Biletska, H.; Kreutzer, S.; Semela, D.; Neuweiler, J.; Hurlimann, S.; Aepli, P.; et al. Visualization of hepatitis E virus RNA and proteins in the human liver. J. Hepatol. 2017, 67, 471–479. [Google Scholar] [CrossRef]
- Paingankar, M.S.; Arankalle, V.A. Identification and characterization of cellular proteins interacting with Hepatitis E virus untranslated regions. Virus Res. 2015, 208, 98–109. [Google Scholar] [CrossRef]
- Korkaya, H.; Jameel, S.; Gupta, D.; Tyagi, S.; Kumar, R.; Zafrullah, M.; Mazumdar, M.; Lal, S.K.; Xiaofang, L.; Sehgal, D.; et al. The ORF3 protein of hepatitis E virus binds to Src homology 3 domains and activates MAPK. J. Biol. Chem. 2001, 276, 42389–42400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pingale, K.D.; Kanade, G.D.; Karpe, Y.A. Heterogeneous Nuclear Ribonucleoproteins Participate in Hepatitis E Virus Replication. J. Mol. Biol. 2020, 432, 2369–2387. [Google Scholar] [CrossRef]
- Gonzalez, F.J. Regulation of hepatocyte nuclear factor 4 alpha-mediated transcription. Drug Metab. Pharmacokinet. 2008, 23, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Chandra, V.; Holla, P.; Ghosh, D.; Chakrabarti, D.; Padigaru, M.; Jameel, S. The hepatitis E virus ORF3 protein regulates the expression of liver-specific genes by modulating localization of hepatocyte nuclear factor 4. PLoS ONE 2011, 6, e22412. [Google Scholar] [CrossRef] [Green Version]
- Yamashita, T.; Mori, Y.; Miyazaki, N.; Cheng, R.H.; Yoshimura, M.; Unno, H.; Shima, R.; Moriishi, K.; Tsukihara, T.; Li, T.C.; et al. Biological and immunological characteristics of hepatitis E virus-like particles based on the crystal structure. Proc. Natl. Acad. Sci. USA 2009, 106, 12986–12991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.C.; Yamakawa, Y.; Suzuki, K.; Tatsumi, M.; Razak, M.A.; Uchida, T.; Takeda, N.; Miyamura, T. Expression and self-assembly of empty virus-like particles of hepatitis E virus. J. Virol. 1997, 71, 7207–7213. [Google Scholar] [CrossRef] [Green Version]
- Xing, L.; Kato, K.; Li, T.; Takeda, N.; Miyamura, T.; Hammar, L.; Cheng, R.H. Recombinant hepatitis E capsid protein self-assembles into a dual-domain T = 1 particle presenting native virus epitopes. Virology 1999, 265, 35–45. [Google Scholar] [CrossRef] [Green Version]
- Li, T.C.; Takeda, N.; Miyamura, T.; Matsuura, Y.; Wang, J.C.; Engvall, H.; Hammar, L.; Xing, L.; Cheng, R.H. Essential elements of the capsid protein for self-assembly into empty virus-like particles of hepatitis E virus. J. Virol. 2005, 79, 12999–13006. [Google Scholar] [CrossRef] [Green Version]
- Xing, L.; Li, T.C.; Mayazaki, N.; Simon, M.N.; Wall, J.S.; Moore, M.; Wang, C.Y.; Takeda, N.; Wakita, T.; Miyamura, T.; et al. Structure of hepatitis E virion-sized particle reveals an RNA-dependent viral assembly pathway. J. Biol. Chem. 2010, 285, 33175–33183. [Google Scholar] [CrossRef] [Green Version]
- Mori, Y.; Matsuura, Y. Structure of hepatitis E viral particle. Virus Res. 2011, 161, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Shiota, T.; Li, T.C.; Yoshizaki, S.; Kato, T.; Wakita, T.; Ishii, K. The hepatitis E virus capsid C-terminal region is essential for the viral life cycle: Implication for viral genome encapsidation and particle stabilization. J. Virol. 2013, 87, 6031–6036. [Google Scholar] [CrossRef] [Green Version]
- Tam, A.W.; Smith, M.M.; Guerra, M.E.; Huang, C.C.; Bradley, D.W.; Fry, K.E.; Reyes, G.R. Hepatitis E virus (HEV): Molecular cloning and sequencing of the full-length viral genome. Virology 1991, 185, 120–131. [Google Scholar] [CrossRef]
- Surjit, M.; Jameel, S.; Lal, S.K. The ORF2 protein of hepatitis E virus binds the 5’ region of viral RNA. J. Virol. 2004, 78, 320–328. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Sirkis, D.W.; Schekman, R. Protein sorting at the trans-Golgi network. Annu. Rev. Cell Dev. Biol. 2014, 30, 169–206. [Google Scholar] [CrossRef]
- Nagashima, S.; Takahashi, M.; Jirintai, S.; Tanggis; Kobayashi, T.; Nishizawa, T.; Okamoto, H. The membrane on the surface of hepatitis E virus particles is derived from the intracellular membrane and contains trans-Golgi network protein 2. Arch. Virol. 2014, 159, 979–991. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, S.; Surjit, M.; Roy, A.K.; Jameel, S.; Lal, S.K. The ORF3 protein of hepatitis E virus interacts with liver-specific alpha1-microglobulin and its precursor alpha1-microglobulin/bikunin precursor (AMBP) and expedites their export from the hepatocyte. J. Biol. Chem. 2004, 279, 29308–29319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Primadharsini, P.P.; Nagashima, S.; Takahashi, M.; Kobayashi, T.; Nishiyama, T.; Nishizawa, T.; Yasuda, J.; Mulyanto; Okamoto, H. Multivesicular body sorting and the exosomal pathway are required for the release of rat hepatitis E virus from infected cells. Virus Res. 2020, 278, 197868. [Google Scholar] [CrossRef]
- Nagashima, S.; Jirintai, S.; Takahashi, M.; Kobayashi, T.; Tanggis; Nishizawa, T.; Kouki, T.; Yashiro, T.; Okamoto, H. Hepatitis E virus egress depends on the exosomal pathway, with secretory exosomes derived from multivesicular bodies. J. Gen. Virol. 2014, 95, 2166–2175. [Google Scholar] [CrossRef] [PubMed]
- Surjit, M.; Oberoi, R.; Kumar, R.; Lal, S.K. Enhanced alpha1 microglobulin secretion from Hepatitis E virus ORF3-expressing human hepatoma cells is mediated by the tumor susceptibility gene 101. J. Biol. Chem. 2006, 281, 8135–8142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Wang, M.; Yin, X. Cellular Organelles Involved in Hepatitis E Virus Infection. Pathogens 2021, 10, 1206. https://doi.org/10.3390/pathogens10091206
Liu X, Wang M, Yin X. Cellular Organelles Involved in Hepatitis E Virus Infection. Pathogens. 2021; 10(9):1206. https://doi.org/10.3390/pathogens10091206
Chicago/Turabian StyleLiu, Xing, Menghang Wang, and Xin Yin. 2021. "Cellular Organelles Involved in Hepatitis E Virus Infection" Pathogens 10, no. 9: 1206. https://doi.org/10.3390/pathogens10091206
APA StyleLiu, X., Wang, M., & Yin, X. (2021). Cellular Organelles Involved in Hepatitis E Virus Infection. Pathogens, 10(9), 1206. https://doi.org/10.3390/pathogens10091206






