Abstract
Blastocystis sp. is a common pathogen that infects the intestines of humans and animals, causing a threat to public health. However, little information on the prevalence and subtypes of Blastocystis sp. in diarrheic pigs in China is available. Herein, 1254 fecal samples were collected from diarrheic pigs in 37 intensive pig farms in Hunan, Jiangxi, and Fujian provinces in southern China, and the prevalence and subtypes of Blastocystis sp. were investigated. Blastocystis sp. was detected by PCR assay, which amplified the small subunit rRNA (SSU rRNA) gene. Overall prevalence of Blastocystis sp. was 31.4% (394/1254), including 21.5% (66/307), 33.1% (99/299), 58.9% (56/95), and 31.3% (173/553) in suckling piglets, weaned piglets, fattening pigs, and sows, respectively. Moreover, age and region factors were significantly related to prevalence of Blastocystis sp. (p < 0.05). Four Blastocystis sp. subtypes were identified, including ST1, ST3, ST5, and ST14. The preponderant subtype was ST5 (76.9%, 303/394). To our knowledge, ST14 was firstly found in pigs in China. The human-pathogenic subtypes (ST1, ST3, ST5, and ST14) that were observed in this study indicate a potential threat to public health. These findings provided a new sight for studying the genetic structure of Blastocystis sp.
1. Introduction
Blastocystis sp. is a zoonotic intestinal protozoan with a worldwide distribution. The host range of Blastocystis sp. is extensive, including humans, non-human primates, mammals, birds, fish, annelids, arthropods, reptiles, and amphibians [1]. Since the term “Blastocystis” was introduced by A. Alexieff in 1911, there has been a consensus that Blastocystis sp. is transmitted through the oral–fecal route, although its pathogenicity has been controversial [2,3]. Blastocystis sp. infection is in some cases thought to be associated with clinical symptoms, including abdominal pain, diarrhea, nausea, irritable bowel syndrome (IBS), and inflammatory bowel disease (IBD), which cause significant physical discomfort to human and animals [4,5,6,7]. Furthermore, in an infected host, Blastocystis sp. infection may concur with other zoonotic parasites such as Giardia duodenalis and Cryptosporidium spp. [8,9,10]. Hence, investigation of the prevalence and subtypes of Blastocystis sp. plays an important role in tracking and preventing the transmission of this protist.
Currently, based on molecular analysis of the small subunit ribosomal RNA (SSU rRNA), 27 subtypes of Blastocystis sp. have been identified, 12 of which (ST1–ST10, ST12, and ST14) were found in humans and animals, while the others were present in specific animals [9,10,11,12,13]. The prevalence of Blastocystis sp. varied considerably among different animals [14,15,16,17,18]. Furthermore, the dominant subtype of Blastocystis sp. in pigs is ST5, and ST10 is the dominant Blastocystis subtype in alpacas, while ST3 is the dominant Blastocystis subtype in humans [5,9,19]. ST14 appears more frequently in cattle and sheep [20]. The prevalence of Blastocystis sp. in pigs is varied in different regions [14,21].
Previous studies have reported high prevalence of Blastocystis sp. in humans, domestic animals, or wild animals in several provinces of China [22]. Prevalence of Blastocystis sp. in pigs has been reported in Shaanxi, Guangdong, Zhejiang, Heilongjiang, Jiangxi, and Yunnan provinces and Xinjiang Hui Autonomous Region in China [6,7,9,17,23,24]. However, there is no report of Blastocystis sp. infection in pigs in Hunan and Fujian provinces in China. Although there was a previous report of pig infection with Blastocystis sp. in Jiangxi Province [23], the sample size was too small, and might not have reflected the true situation of pigs infected with Blastocystis sp. Therefore, this study examined the prevalence of Blastocystis sp. and its subtypes in diarrheic pigs of different age groups and regions in three southern provinces of China.
2. Results
2.1. Prevalence of Blastocystis sp. in Diarrheic Pigs
In the present study, 31.4% (394/1254, 95% CI 28.85–33.99) of the 1254 fecal samples of diarrheic pigs were positive for Blastocystis sp. The prevalence of Blastocystis sp. in pigs in Fujian Province (43.7%, 59/135, 95% CI 35.34–52.07) and Jiangxi Province (30.5%, 316/1036, 95% CI 27.70–33.31) was higher than in Hunan Province (22.9%, 19/83, 95% CI 13.85–31.93), with ORs of 2.62 (95% CI 1.41–4.84) and 1.47 (95% CI 0.86–2.49), respectively (Table 1). In Fujian Province, the highest prevalence of Blastocystis sp. was found in pigs in Sanming City, with a positive rate of 45.2% (56/124, 95% CI 36.40–53.92). In Hunan Province, the highest Blastocystis prevalence was found in Zhuzhou City, with a positive rate of 41.7% (10/24, 95% CI 21.94–61.39). In Jiangxi Province, the highest Blastocystis prevalence was found in Fuzhou, with a positive rate of 47.2% (42/89, 95% CI 36.82–57.56) (Table 2). Significant differences (p < 0.05) in the prevalence of Blastocystis sp. in pigs in the three investigated provinces and in different cities of Jiangxi province were observed (Table 1 and Table 2).

Table 1.
Factors associated with prevalence and subtype distribution of Blastocystis sp. in pigs with diarrhea in southern China.

Table 2.
Distribution of Blastocystis sp. subtype in different sampling cities.
Among different growing stages of pigs, fattening pigs had the highest prevalence (58.9%, 56/95, 95% CI 49.06–68.84) of Blastocystis sp., followed by weaned piglets (33.1%, 99/299, 95% CI 27.78–38.44), sows (31.3%, 173/553, 95% CI 27.42–35.15), and suckling piglets (21.5%, 66/307, 95% CI 16.90–26.09), and the differences were statistically significant (p < 0.001) (Table 1). Furthermore, fattening pigs had 5.24 times (95% CI 3.21–8.57) more risk of infection with Blastocystis than that of suckling piglets.
2.2. Subtype Distribution of Blastocystis sp. in Diarrheic Pigs
A total of four subtypes (ST1, ST3, ST5, and ST14) of Blastocystis sp. were identified in all growing stages of pigs in this study. Of these subtypes, ST5 was the predominant subtype of Blastocystis sp., with an infection rate of 76.9% (303/394), followed by ST3 (11.9%, 47/394), ST1 (6.6%, 26/394), and ST14 (4.6%, 18/394). Moreover, the ST14 was first identified in pigs in Fujian (3/18) and Jiangxi Provinces (15/18) (Table 1).
2.3. Phylogenetic Analysis of Blastocystis sp.
Phylogenetic analyses showed that the sequences of the four subtypes (ST1, ST3, ST5, and ST14) obtained from pigs in this study clustered with other ST1, ST3, ST5, and ST14 sequences obtained from other animals or humans into one branch, with high bootstrap values (Figure 1). Notably, The ST1 sequences (MW767060–MW767062) obtained from pigs in this study were closely related to the ST1 sequence (MK719635) obtained from humans (Figure 1). Furthermore, the ST14 sequences obtained from pigs in this study showed a closer genetic relationship with other ST14 sequences from ruminants (Figure 2).
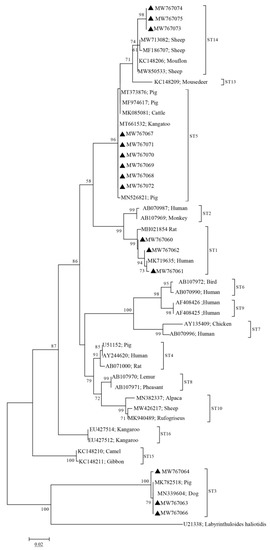
Figure 1.
Phylogenetic analysis of Blastocystis subtypes based on sequences of the SSU rRNA gene using the maximum likelihood (ML) method. A bootstrap algorithm was used to assess the branch reliability with 1000 replicates. Only bootstrap values above 50% are shown. Sequences marked with black triangles indicate the sequences obtained in this study, and the GenBank accession numbers of the sequences are shown to the right of the triangle.
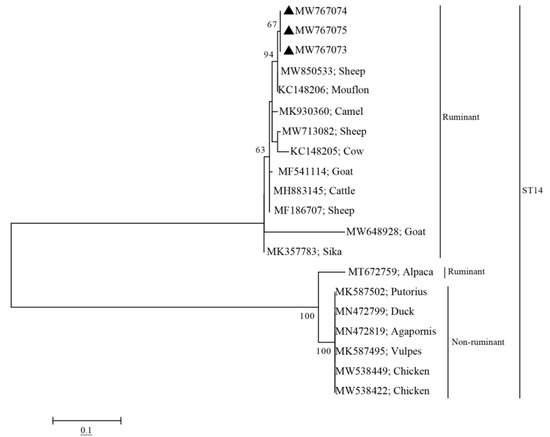
Figure 2.
Phylogenetic analysis of Blastocystis ST14 based on the SSU rRNA gene. The model and parameter settings for constructing the phylogenetic evolutionary tree were same as those used for constructing Figure 1. Sequences marked with black triangles indicate the sequences isolated in this study, and the GenBank accession numbers of the sequences are shown to the right of the triangle.
3. Discussion
Although Blastocystis sp. has been researched for more than a century, its pathogenicity remains controversial [6,7]. There is not enough evidence for the clinical importance of Blastocystis sp., but its potential pathogenicity has long been studied [9]. Therefore, extensive investigation of Blastocystis sp. may improve the understanding of its pathogenicity and lead to effective prevention and control.
The overall prevalence of Blastocystis sp. in diarrheic pigs in Hunan, Jiangxi, and Fujian provinces was 31.4% (394/1254, 95% CI 28.85–33.99). Compared with the prevalence of Blastocystis sp. in healthy pigs in other studies, it was lower than that in Zimbabwe (82.8%) [25], Thailand (76.9%) [26], the Philippines (100%) [27], the USA (100%) [28], Australia (76.7%) [5], Cambodia (45.2%) [29], Indonesia (87.1%) [30], Brazil (44.4%) [31], Korea (59.3%) [32], the Czech Republic (89.8%) [25], and Germany (60.0%) [33]. It also was lower than that in Jiangxi Province (100%) [23], Shaanxi Province (74.8%) [9], Guangdong Province (55.6%) [7], and Yunnan Province (50.0%) [7,24] in China, but higher than that in Spain (7.5%) [14], the UK (28.5%) [34], Xinjiang Hui Autonomous Region (21.7%) [6], and Heilongjiang Province (8.80%) [35]. However, we did not collect fecal samples from healthy pigs in this study, making it impossible to directly compare the prevalence of Blastocystis sp. in healthy and diarrheic pigs. It is possible that no difference of prevalence could be found in health and diarrheic pigs. This finding reveals that diarrhea may not be associated with prevalence of Blastocystis sp., but more evidence is needed explain this finding. In addition, many factors may also contribute to the varying prevalence of Blastocystis sp., such as different detection methods, breeds, geography, or sample sizes. However, these pigs’ samples were previously tested for viruses and other pathogens, including PEDV, PDCoV, and Escherichia coli [36,37]. Several pathogens, especially helminths and protozoa, which are included among the most frequent causes of diarrhea in pigs, were not investigated, which was a limitation of the current study.
In previous reports, the prevalence of Blastocystis sp. in sows was generally significantly higher than that in fattening pigs, but in this study, the Blastocystis prevalence in fattening pigs was higher than that in other growing stages of pigs, which was consistent with the results of other studies [7,9,32]. This difference may have been caused by the raising condition. Furthermore, the infection rate of Blastocystis sp. was the lowest in suckling piglets compared to other growing stages of pigs, which might have been related to the important role of maternal antibodies.
Comparing the infection rate of ST3 with previous reports [5,9,15,21,32], we found that ST3 had the highest infection rate in this study (Table 1). According to previous studies, ST5 was widely distributed in all age groups of pigs, and only ST5 was detected in sows [6,9]. However, we found ST1, ST3, ST5, and ST14 in all age groups. Diarrhea often destroys the structure of the intestinal flora and alters the intestinal environment, which might lead to the change in dominant subtype [32]. This might explain the higher positive rate of ST3 compared to ST1 in diarrheic pigs in this study. Although the available data for ST14 in pigs is very limited [20], ST14 has been reported in humans [11], suggesting that ST14 has zoonotic potential, and that pigs may be a link in the transmission of ST14. Mixed infections were not detected in this investigation. While primers for subtype-specific detection are now available, only a limited number of subtypes are currently available for detection [17].
Phylogenetic analysis revealed that sequences of ST1 obtained from pigs in this study were closely related to human-derived sequences of ST1 (MK719635) (Figure 1), which further proved that pigs could be a possible reservoir for human infection with Blastocystis sp. The sequences of ST14 obtained in pigs and sequences of ST14 isolated in ruminants were clustered together (Figure 2). Since ST14 is more prevalent in ruminants, we speculated that the source of ST14 infection in pigs might be from ruminants. These findings should be enhanced in future molecular epidemiological studies.
4. Materials and Methods
4.1. Study Sites
The fecal samples of diarrheic pigs were collected from 37 intensive pig farms located in three southern provinces of China: Hunan (location: 24°38′–30°08′ N, 108°47′–114°15′ E), Jiangxi (location: 24°29′–30°04′ N, 113°34′–118°28′ E), and Fujian (location: 23°30′–28°22′ N, 115°50′–120°40′ E).
4.2. Sampling
From 2015 to 2019, a total of 1254 fresh fecal samples from pigs with diarrhea were collected from Jiangxi Province (n = 1036), Hunan Province (n = 83), and Fujian Province (n = 135) (Table 1 and Figure 3). Only pigs with dilute feces or watery diarrhea were sampled, and all samples were collected by anal swab. Among these fecal samples, 307 fecal samples were from suckling piglets (<21 days), 299 fecal samples were from weaned piglets (21–70 days), 95 fecal samples were from fattening pigs (71–180 days), and 553 fecal samples were from sows (>180 days) (Table 1). All fecal samples were placed in a cryopreservation box with an adequate amount of ice bags immediately after sampling, and then were stored in a refrigerator at −80 ℃ before DNA extraction.
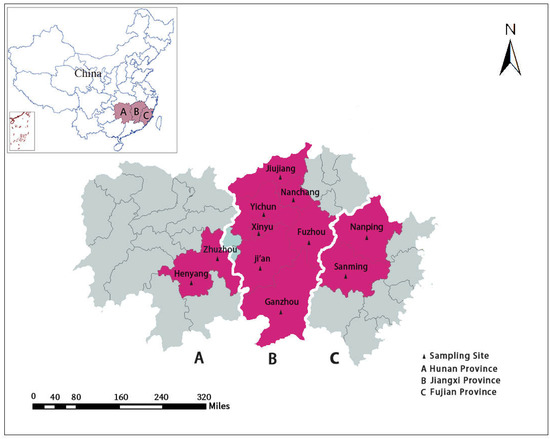
Figure 3.
The map of the sample collection in this study; the red areas represent the sampling locations.
4.3. Genomic DNA Extraction and PCR Amplification
Approximately 300 mg of each fecal sample was washed 3 times with distilled water by centrifuging at 13,000× g for 5 min to remove the impurities. The remaining sediments were used to extract the genomic DNA using the E.Z.N.A.® Fecal DNA Kit (D4015-02, Omega Bio-Tek Inc., Norcross, GA, USA). The genomic DNA was stored at −20 ℃ for further analysis. The genomic DNA samples were screened for Blastocystis sp. by PCR amplification of the SSU rRNA genes with a target fragment size of roughly 600 using the primers BhRDr (5′-GAGCTTTTTAACTGCAACAACG-3′) and RD5 (5′-ATCTGGTTGATCCTGCCAGT-3′) [9]. The 25 μL reaction system contained 2 μL of genomic DNA, 0.2 mM of dNTP mixture, 1.5 mM of MgCl2, 2.5 μL of 10× Ex Taq buffer, 1.25 U of TaKaRa Ex Taq® (Takara Bio Inc., Dalian, China), and 0.25 μL of primers (10 mol/μL). The PCR reaction conditions were set as follows: initial denaturation at 94 °C for 5 min; 35 cycles at 94 °C for 45 s, 59 °C for 45 s, and 72 °C for 1 min; and an additional 72 °C extension for 3 min. Each reaction included a positive (DNA from Blastocystis sp.) and a negative control (reagent water). The final PCR products were identified by 2% (w/v) agarose gel electrophoresis and stained with ethidium bromide.
4.4. Sequence Analysis
Approximately 600 bp PCR product of each sample was recovered and purified by Tsingke Biotechnology Co., Ltd. for sequencing using the Sanger sequencing method. The subtypes of Blastocystis sp. were identified by aligning with obtained sequences and corresponding sequences available in the GenBank database (http://www.ncbi.lm.nih.gov/GenBank/, accessed on 21 March 2021) using Clustal X 2.1 (http://www.clustal.org/clustal2/, accessed on 28 March 2021) [26]. The maximum likelihood (ML) method with a Kimura 2-parameter model in MEGA 7.0 (http://www.megasoftware.net accessed on 1 September 2021) [27] was used to establish a phylogenetic tree with 1000 repeats under a bootstrap method, and U21338 was set as the out-group (Figure 1 and Figure 2).
4.5. Statistical Analysis
Data obtained in this study on the prevalence of Blastocystis sp. between different regions and age groups were systematically analyzed with a chi-square test (χ2) using SPSS version 25.0 (IBM SPSS Inc., Chicago, IL, USA), and significant differences were considered only when the obtained p value was less than 0.05.
5. Conclusions
In the present study, a total of 1254 fecal samples from diarrheic pigs in three provinces in southern China were examined for the prevalence and subtypes of Blastocystis sp. This was the first report on Blastocystis sp. infection in pigs in Hunan and Fujian provinces. Three zoonotic subtypes (ST1, ST3, and ST5) and one potential zoonotic subtype (ST14) were identified, and ST14 was detected for the first time in pigs in China. Compared to previous reports of healthy pigs infected with Blastocystis sp., the great differences found in the present study were reflected in the increased frequency of ST1 and ST3, the significantly higher Blastocystis sp. infection in fattening pigs rather than in sows, and the detection of ST14. These findings may help understand the genetic structure of Blastocystis sp. in pigs, providing useful data for effective prevention and control of Blastocystis sp. in southern China in the future.
Author Contributions
X.-Q.C. and D.-P.S. conceived and designed the study; P.W. (Pei Wang) performed the experiments, analyzed the data, and drafted the manuscript; S.L., Z.-W.H. and P.W. (Ping Wang) participated in the implementation of the study; X.-Q.C., D.-P.S., X.-Q.Z. and Y.Z. critically revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Science and Technology Program Funding of the Educational Commission of Jiangxi Province, China (Grant No. GJJ180185); the Fund for Shanxi “1331 Project” (Grant No. 20211331-13); the National Natural Science Foundation of China (31960711); the Natural Science Foundation of Jiangxi Province (20202BABL215024); the Agricultural Science and Technology Innovation Program (ASTIP) (Grant No. CAAS-ASTIP-2016-LVRI-03); and the Yunnan Expert Workstation (Grant No. 202005AF150041).
Institutional Review Board Statement
The study protocol was reviewed and approved by the Animal Ethics Committee of Jiangxi Agricultural University (protocol code: JXAU-LL-2013-03; date of approval: 11 December 2013).
Informed Consent Statement
Not applicable.
Data Availability Statement
For reasonable requests, the data obtained for this study can be obtained by contacting the corresponding author. The sequences of the Blastocystis sp. obtained from this study were deposited in the NCBI GenBank database under the accession numbers MW767060–MW767075.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wang, W.; Bielefeldt-Ohmann, H.; Traub, R.J.; Cuttell, L.; Owen, H. Location and pathogenic potential of Blastocystis in the porcine intestine. PLoS ONE 2014, 9, e103962. [Google Scholar] [CrossRef] [Green Version]
- Clark, C.G.; van der Giezen, M.; Alfellani, M.A.; Stensvold, C.R. Recent developments in Blastocystis research. Adv. Parasitol. 2013, 82, 1–32. [Google Scholar]
- Pintong, A.R.; Sunyanusin, S.; Prasertbun, R.; Mahittikorn, A.; Mori, H.; Changbunjong, T.; Komalamisra, C.; Sukthana, Y.; Popruk, S. Blastocystis subtype 5: Predominant subtype on pig farms, Thailand. Parasitol. Int. 2018, 67, 824–828. [Google Scholar] [CrossRef]
- Graczyk, T.K.; Shiff, C.K.; Tamang, L.; Munsaka, F.; Beitin, A.M.; Moss, W.J. The association of Blastocystis hominis and Endolimax nana with diarrheal stools in Zambian school-age children. Parasitol. Res. 2005, 98, 38–43. [Google Scholar] [CrossRef]
- Wang, W.; Owen, H.; Traub, R.J.; Cuttell, L.; Inpankaew, T.; Bielefeldt-Ohmann, H. Molecular epidemiology of Blastocystis in pigs and their in-contact humans in Southeast Queensland, Australia, and Cambodia. Vet. Parasitol. 2014, 203, 264–269. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, Y.; Jiang, Y.; Xing, J.; Tao, D.; Qi, M. First report of Blastocystis infection in pigs from large farms in Xinjiang, China. J. Eukaryot. Microbiol. 2020, 67, 642–647. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Yang, W.B.; Zou, F.C.; Lin, R.Q.; Zhu, X.Q.; Hou, J.L. Molecular detection and subtype distribution of Blastocystis in farmed pigs in southern China. Microb. Pathog. 2021, 151, 104751. [Google Scholar] [CrossRef] [PubMed]
- Haider, S.S.; Baqai, R.; Qureshi, F.M.; Boorom, K. Blastocystis spp., Cryptosporidium spp., and Entamoeba histolytica exhibit similar symptomatic and epidemiological patterns in healthcare-seeking patients in Karachi. Parasitol. Res. 2012, 111, 1357–1368. [Google Scholar] [CrossRef] [PubMed]
- Song, J.K.; Hu, R.S.; Fan, X.C.; Wang, S.S.; Zhang, H.J.; Zhao, G.H. Molecular characterization of Blastocystis from pigs in Shaanxi province of China. Acta Trop. 2017, 173, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Fayer, R.; Santin, M.; Macarisin, D. Detection of concurrent infection of dairy cattle with Blastocystis, Cryptosporidium, Giardia, and Enterocytozoon by molecular and microscopic methods. Parasitol. Res. 2012, 111, 1349–1355. [Google Scholar] [CrossRef]
- Khaled, S.; Gantois, N.; Ly, A.T.; Senghor, S.; Even, G.; Dautel, E.; Dejager, R.; Sawant, M.; Baydoun, M.; Benamrouz-Vanneste, S.; et al. Prevalence and subtype distribution of Blastocystis sp. in senegalese school children. Microorganisms 2020, 8, 1408. [Google Scholar] [CrossRef] [PubMed]
- Maloney, J.G.; Santin, M. Mind the gap: New full-length sequences of Blastocystis subtypes generated via Oxford Nanopore Minion sequencing allow for comparisons between full-length and partial sequences of the small subunit of the ribosomal RNA gene. Microorganisms 2021, 9, 997. [Google Scholar] [CrossRef] [PubMed]
- Maloney, J.G.; Jang, Y.; Molokin, A.; George, N.S.; Santin, M. Wide genetic diversity of Blastocystis in white-tailed deer (Odocoileus virginianus) from Maryland, USA. Microorganisms 2021, 9, 1343. [Google Scholar] [CrossRef]
- Quilez, J.; Clavel, A.; Sanchez-Acedo, C.; Causape, A.C. Detection of Blastocystis sp. in pigs in Aragon (Spain). Vet. Parasitol. 1995, 56, 345–348. [Google Scholar] [CrossRef]
- Silberman, J.D.; Sogin, M.L.; Leipe, D.D.; Clark, C.G. Human parasite finds taxonomic home. Nature 1996, 380, 398. [Google Scholar] [CrossRef] [PubMed]
- Zanzani, S.A.; Gazzonis, A.L.; Epis, S.; Manfredi, M.T. Study of the gastrointestinal parasitic fauna of captive non-human primates (Macaca fascicularis). Parasitol. Res. 2016, 115, 307–312. [Google Scholar] [CrossRef]
- Wang, J.; Gong, B.; Liu, X.; Zhao, W.; Bu, T.; Zhang, W.; Liu, A.; Yang, F. Distribution and genetic diversity of Blastocystis subtypes in various mammal and bird species in northeastern China. Parasites Vectors 2018, 11, 522. [Google Scholar] [CrossRef] [Green Version]
- Gabrielli, S.; Palomba, M.; Furzi, F.; Brianti, E.; Gaglio, G.; Napoli, E.; Rinaldi, L.; Alburqueque, R.A.; Mattiucci, S. Molecular subtyping of Blastocystis sp. isolated from farmed animals in southern Italy. Microorganisms 2021, 9, 1656. [Google Scholar] [CrossRef]
- Ma, Y.T.; Liu, Q.; Xie, S.C.; Li, X.D.; Ma, Y.Y.; Li, T.S.; Gao, W.W.; Zhu, X.Q. Prevalence and subtypes of Blastocystis in alpacas, vicugna pacos in Shanxi province, China. Korean J. Parasitol. 2020, 58, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Rauff-Adedotun, A.A.; Mohd Zain, S.N.; Farah Haziqah, M.T. Current status of Blastocystis sp. in animals from Southeast Asia: A review. Parasitol. Res. 2020, 119, 3559–3570. [Google Scholar] [CrossRef]
- Navarro, C.; Domínguez-Márquez, M.V.; Garijo-Toledo, M.M.; Vega-García, S.; Fernández-Barredo, S.; Pérez-Gracia, M.T.; García, A.; Borrás, R.; Gómez-Muñoz, M.T. High prevalence of Blastocystis sp. in pigs reared under intensive growing systems: Frequency of ribotypes and associated risk factors. Vet. Parasitol. 2008, 153, 347–358. [Google Scholar] [CrossRef]
- Deng, L.; Chai, Y.; Zhou, Z.; Liu, H.; Zhong, Z.; Hu, Y.; Fu, H.; Yue, C.; Peng, G. Epidemiology of Blastocystis sp. infection in China: A systematic review. Parasite 2019, 26, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Y.; Su, S.; Ye, J.; Lai, X.; Lai, R.; Liao, H.; Chen, G.; Zhang, R.; Hou, Z.; Luo, X. Blastocystis sp. subtype 5: A possibly zoonotic genotype. Parasitol. Res. 2007, 101, 1527–1532. [Google Scholar] [CrossRef] [PubMed]
- Han, J.Q.; Li, Z.; Zou, Y.; Pu, L.H.; Zhu, X.Q.; Zou, F.C.; Huang, C.Q. Prevalence, molecular characterization and risk factors of Blastocystis sp. from farmed pigs in Yunnan province, southwestern China. Acta Parasitol. 2020, 65, 1005–1010. [Google Scholar] [CrossRef]
- Pakandl, M. Occurrence of Blastocystis sp. in pigs. Folia Parasitol. 1991, 38, 297–301. [Google Scholar]
- Thathaisong, U.; Worapong, J.; Mungthin, M.; Tan-Ariya, P.; Viputtigul, K.; Sudatis, A.; Noonai, A.; Leelayoova, S. Blastocystis isolates from a pig and a horse are closely related to Blastocystis hominis. J. Clin. Microbiol. 2003, 41, 967–975. [Google Scholar] [CrossRef] [Green Version]
- Rivera, W.L.; Tan, M.A. Molecular characterization of Blastocystis isolates in the Philippines by riboprinting. Parasitol. Res. 2005, 96, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Santín, M.; Gómez-Muñoz, M.T.; Solano-Aguilar, G.; Fayer, R. Development of a new PCR protocol to detect and subtype Blastocystis spp. from humans and animals. Parasitol. Res. 2011, 109, 205–212. [Google Scholar] [CrossRef]
- Wang, W.; Cuttell, L.; Traub, R.J.; Owen, H.; Bielefeldt-Ohmann, H. Characterization of the Blastocystis-specific faecal IgA immune response in pigs. Parasite Immunol. 2014, 36, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, H.; Tokoro, M.; Nagamoto, T.; Arayama, S.; Asih, P.B.; Rozi, I.E.; Syafruddin, D. Molecular survey of Blastocystis sp. from humans and associated animals in an Indonesian community with poor hygiene. Parasitol. Int. 2016, 65, 780–784. [Google Scholar] [CrossRef]
- Moura, R.G.F.; Oliveira-Silva, M.B.; Pedrosa, A.L.; Nascentes, G.A.N.; Cabrine-Santos, M. Occurrence of Blastocystis spp. in domestic animals in Triângulo Mineiro area of Brazil. Rev. Soc. Bras. Med. Trop. 2018, 51, 240–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paik, S.; Jung, B.Y.; Lee, H.; Hwang, M.H.; Han, J.E.; Rhee, M.H.; Kim, T.H.; Kwon, O.D.; Kwak, D. Molecular detection and subtyping of Blastocystis in Korean pigs. Korean J. Parasitol. 2019, 57, 525–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- König, G.; Müller, H.E. Blastocystis hominis in animals: Incidence of four serogroups. Zentralbl. Bakteriol. 1997, 286, 435–440. [Google Scholar] [CrossRef]
- Alfellani, M.A.; Taner-Mulla, D.; Jacob, A.S.; Imeede, C.A.; Yoshikawa, H.; Stensvold, C.R.; Clark, C.G. Genetic diversity of Blastocystis in livestock and zoo animals. Protist 2013, 164, 497–509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Gong, B.; Yang, F.; Zhang, W.; Zheng, Y.; Liu, A. Subtype distribution and genetic characterizations of Blastocystis in pigs, cattle, sheep and goats in northeastern China’s Heilongjiang Province. Infect. Genet. Evol. 2018, 57, 171–176. [Google Scholar] [CrossRef]
- Zhang, F.; Luo, S.; Gu, J.; Li, Z.; Li, K.; Yuan, W.; Ye, Y.; Li, H.; Ding, Z.; Song, D.; et al. Prevalence and phylogenetic analysis of porcine diarrhea associated viruses in southern China from 2012 to 2018. BMC Vet. Res. 2019, 15, 470. [Google Scholar] [CrossRef] [Green Version]
- Song, D.; Zhou, X.; Peng, Q.; Chen, Y.; Zhang, F.; Huang, T.; Zhang, T.; Li, A.; Huang, D.; Wu, Q.; et al. Newly emerged porcine deltacoronavirus associated with diarrhoea in swine in China: Identification, prevalence and full-Length genome sequence analysis. Transbound. Emerg. Dis. 2015, 62, 575–580. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).