Real-Time Fluorometric Isothermal LAMP Assay for Detection of Chlamydia pecorum in Rapidly Processed Ovine Abortion Samples: A Veterinary Practitioner’s Perspective
Abstract
1. Introduction
2. Results
2.1. Limit of Detection of the C. pecorum LAMP Assay
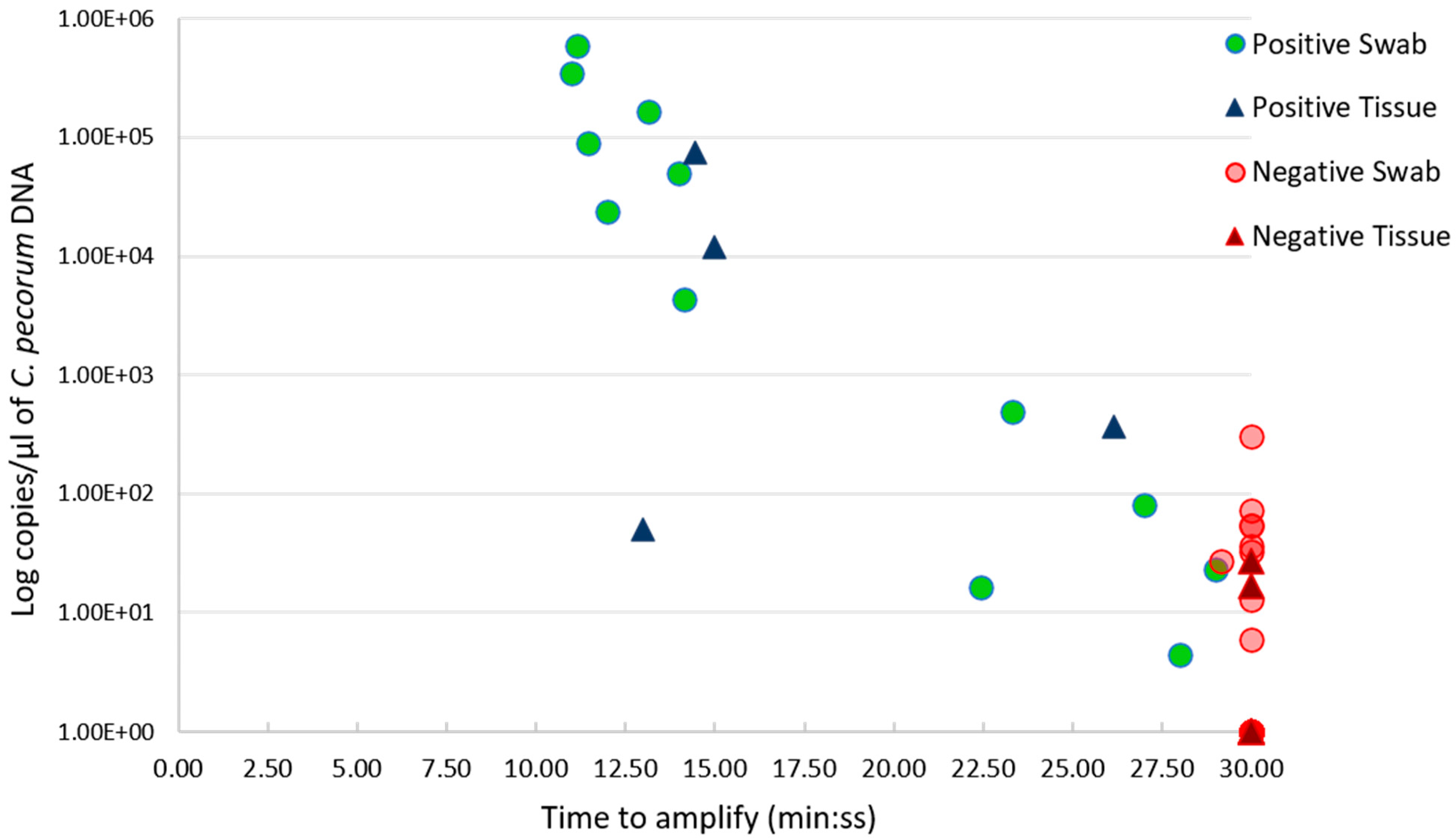
2.2. C. pecorum Detection in Rapidly Processed Clinical Samples Using LAMP Assays
2.3. C. pecorum LAMP Using Swab Suspensions
2.4. C. pecorum LAMP Using Tissue Lysates
2.5. C. pecorum LAMP Testing of Paired Swab and Tissue Samples
2.6. Overall C. pecorum LAMP and qPCR Agreement
2.7. Preliminary Evaluation of the Use of LAMP Assay and the Real-Time Fluorometer as a POC Diagnostic Tool
3. Discussion
4. Materials and Methods
4.1. Samples
4.2. Swab and Tissue Processing
4.3. C. pecorum Isothermal Assays
4.4. Testing of Spiked Samples
4.5. Test Congruence and Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Walker, E.; Moore, C.; Shearer, P.; Jelocnik, M.; Bommana, S.; Timms, P.; Polkinghorne, A. Clinical, diagnostic and pathologic features of presumptive cases of Chlamydia pecorum-associated arthritis in Australian sheep flocks. BMC Vet. Res. 2016, 12, 193. [Google Scholar] [CrossRef] [PubMed]
- Jelocnik, M.; Laurence, M.; Murdoch, F.R.; Polkinghorne, A. Detection of Chlamydiaceae in ocular swabs from Australian pre-export feedlot sheep. Aust. Vet. J. 2019, 97, 401–403. [Google Scholar] [CrossRef] [PubMed]
- Walker, E.; Jelocnik, M.; Bommana, S.; Timms, P.; Carver, S.; Polkinghorne, A. Understanding the health and production impacts of endemic Chlamydia pecorum infections in lambs. Vet. Microbiol. 2018, 217, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, J.; Yang, R.; Kessell, A.; Ryan, U.; Schröder, J.; Rutley, D. Detection of Chlamydia pecorum in joints trimmed from ovine carcases with arthritis at an abattoir in southern Australia. Small Rumin. Res. 2017, 150, 80–86. [Google Scholar] [CrossRef][Green Version]
- Ostfeld, N.; Islam, M.M.; Jelocnik, M.; Hilbe, M.; Sydler, T.; Hartnack, S.; Jacobson, C.; Clune, T.; Marsh, I.; Sales, N.; et al. Chlamydia pecorum-induced arthritis in experimentally and naturally infected sheep. Vet. Pathol. 2021, 58, 346–360. [Google Scholar] [CrossRef] [PubMed]
- Bommana, S.; Walker, E.; Desclozeaux, M.; Jelocnik, M.; Timms, P.; Polkinghorne, A.; Carver, S. Molecular and serological dynamics of Chlamydia pecorum infection in a longitudinal study of prime lamb production. PeerJ 2018, 6, e4296. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Jacobson, C.; Gardner, G.; Carmichael, I.; Campbell, A.J.D.; Ryan, U. Longitudinal prevalence and faecal shedding of Chlamydia pecorum in sheep. Vet. J. 2014, 201, 322–326. [Google Scholar] [CrossRef] [PubMed]
- Clune, T.; Besier, S.; Hair, S.; Hancock, S.; Lockwood, A.; Thompson, A.; Jelocnik, M.; Jacobson, C. Chlamydia pecorum detection in aborted and stillborn lambs from Western Australia. Vet. Res. 2021, 52, 84. [Google Scholar] [CrossRef] [PubMed]
- Westermann, T.; Jenkins, C.; Onizawa, E.; Gestier, S.; McNally, J.; Kirkland, P.; Zhang, J.; Bogema, D.; Manning, L.K.; Walker, K.; et al. Chlamydia pecorum–associated sporadic ovine abortion. Vet. Pat. 2021, 58, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Sachse, K.; Vretou, E.; Livingstone, M.; Borel, N.; Pospischil, A.; Longbottom, D. Recent developments in the laboratory diagnosis of chlamydial infections. Vet. Microbiol. 2009, 135, 2–21. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fan, P.; Zhou, S.; Zhang, L. Loop-mediated isothermal amplification (LAMP): A novel rapid detection platform for pathogens. Microb. Pathog. 2017, 107, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Jelocnik, M.; Islam, M.M.; Madden, D.; Jenkins, C.; Branley, J.; Carver, S.; Polkinghorne, A. Development and evaluation of rapid novel isothermal amplification assays for important veterinary pathogens: Chlamydia psittaci and Chlamydia pecorum. PeerJ 2017, 5, e3799. [Google Scholar] [CrossRef] [PubMed]
- Hulse, L.S.; McDonald, S.; Johnston, S.D.; Beagley, K.W. Rapid point-ofpcare diagnostics for the detection of Chlamydia pecorum in koalas (Phascolarctos cinereus) using loop-mediated isothermal amplification without nucleic acid purification. Microbiol. Open 2019, 8, e916. [Google Scholar] [CrossRef] [PubMed]
- Jelocnik, M.; Nyari, S.; Anstey, S.; Playford, N.; Fraser, T.A.; Mitchell, K.; Blishen, A.; Pollak, N.M.; Carrick, J.; Chicken, C.; et al. Real-time fluorometric and End-point colorimetric isothermal assays for detection of equine pathogens Chlamydia psittaci and Equine Herpes Virus 1: Validation, comparison and application at the Point of Care. BMC Vet. Res. 2021, 17, 279. [Google Scholar] [CrossRef] [PubMed]
- Williams, S. The challenges of an unusual abortion outbreak in a ewe flock. In 2019 Sheep and Beef Proceedings, Wellington, New Zealand, 22–24 May 2019; The society of Sheep and Beef Cattle Veterinarians of the New Zealand Veterinary Association: Wellington, New Zealand, 2019; pp. 69–74. [Google Scholar]
- Best, N.; Rawlin, G.; Suter, R.; Rodoni, B.; Beddoe, T. Optimization of a loop mediated isothermal amplification (LAMP) assay for in-field detection of Dichelobacter nodosus with aprV2 (VDN LAMP) in Victorian sheep flocks. Front. Vet. Sci. 2019, 6, 67. [Google Scholar] [CrossRef] [PubMed]
- Best, N.; Rodoni, B.; Rawlin, G.; Beddoe, T. Evaluation of loop-mediated isothermal amplification (LAMP) assay for detection of aprV2 positive Dichelobacter nodosus in-field by secondary users. BMC Res. Notes 2019, 12, 534. [Google Scholar] [CrossRef] [PubMed]
- Jelocnik, M.; Forshaw, D.; Cotter, J.; Roberts, D.; Timms, P.; Polkinghorne, A. Molecular and pathological insights into Chlamydia pecorum-associated sporadic bovine encephalomyelitis (SBE) in Western Australia. BMC Vet. Res. 2014, 10, 121. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kaneko, H.; Kawana, T.; Fukushima, E.; Suzutani, T. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J. Biochem. Biophys. Methods 2007, 70, 499–501. [Google Scholar] [CrossRef] [PubMed]
- Clune, T.; Beetson, S.; Besier, S.; Knowles, G.; Paskin, R.; Rawlin, G.; Suter, R.; Jacobson, C. Ovine abortion and stillbirth investigations in Australia. Aust. Vet. J. 2021, 99, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Ganguli, A.; Nguyen, J.; Brisbin, R.; Shanmugam, K.; Hirschberg, D.L.; Wheeler, M.B.; Bashir, R.; Nash, D.M.; Cunningham, B.T. Smartphone-based multiplex 30-min nucleic acid test of live virus from nasal swab extract. Lab Chip 2020, 20, 1621–1627. [Google Scholar] [CrossRef] [PubMed]
| C. pecorum LAMP testing swabs | Reference C. pecorum qPCR with Swabs | |||
| Positive | Negative | Total | ||
| Positive | 12 | 0 | 12 | |
| Negative | 9 | 25 | 34 | |
| Total | 21 | 25 | 46 | |
| Kappa (95% CI; p-value) | 0.592 (0.3733–0.8101; 0.00) | |||
| McNemar’s Chi square (p-value) | 7.111 (0.008) | |||
| Overall agreement | 80.4% (72.73% PA; 84.75%NA) | |||
| C. pecorum LAMP testing tissue lysates | Reference C. pecorum qPCR with Lysed Tissue | |||
| Positive | Negative | Total | ||
| Positive | 4 | 0 | 4 | |
| Negative | 2 | 8 | 10 | |
| Total | 6 | 8 | 14 | |
| Kappa (95% CI; p-value) | 0.696 (0.3237–1.0676; 0.0031) | |||
| McNemar’s Chi square (p-value) | 0.5 (0.480) | |||
| Overall agreement | 85.71% (80% PA; 88.89% NA) | |||
| C. pecorum LAMP with lysed tissue | C. pecorum LAMP with Swabs | |||
| Positive | Negative | Total | ||
| Positive | 3 | 1 | 4 | |
| Negative | 1 | 9 | 10 | |
| Total | 4 | 10 | 14 | |
| Kappa (95% CI; p-value) | 0.65 (0.2068–1.0932; 0.0075) | |||
| McNemar’s Chi square (p-value) | 0.5 (0.480) | |||
| Overall agreement | 85.71% (75% PA; 90% NA) | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clune, T.; Anstey, S.; Kasimov, V.; Jacobson, C.; Jelocnik, M. Real-Time Fluorometric Isothermal LAMP Assay for Detection of Chlamydia pecorum in Rapidly Processed Ovine Abortion Samples: A Veterinary Practitioner’s Perspective. Pathogens 2021, 10, 1157. https://doi.org/10.3390/pathogens10091157
Clune T, Anstey S, Kasimov V, Jacobson C, Jelocnik M. Real-Time Fluorometric Isothermal LAMP Assay for Detection of Chlamydia pecorum in Rapidly Processed Ovine Abortion Samples: A Veterinary Practitioner’s Perspective. Pathogens. 2021; 10(9):1157. https://doi.org/10.3390/pathogens10091157
Chicago/Turabian StyleClune, Tom, Susan Anstey, Vasilli Kasimov, Caroline Jacobson, and Martina Jelocnik. 2021. "Real-Time Fluorometric Isothermal LAMP Assay for Detection of Chlamydia pecorum in Rapidly Processed Ovine Abortion Samples: A Veterinary Practitioner’s Perspective" Pathogens 10, no. 9: 1157. https://doi.org/10.3390/pathogens10091157
APA StyleClune, T., Anstey, S., Kasimov, V., Jacobson, C., & Jelocnik, M. (2021). Real-Time Fluorometric Isothermal LAMP Assay for Detection of Chlamydia pecorum in Rapidly Processed Ovine Abortion Samples: A Veterinary Practitioner’s Perspective. Pathogens, 10(9), 1157. https://doi.org/10.3390/pathogens10091157






