Accuracy and Impact on Patient Management of New Tools for Diagnosis of Sepsis: Experience with the T2 Magnetic Resonance Bacteria Panel
Abstract
:1. Introduction
2. Results
2.1. Study Population and Antimicrobial Therapy at the Time of T2 Sample Collection
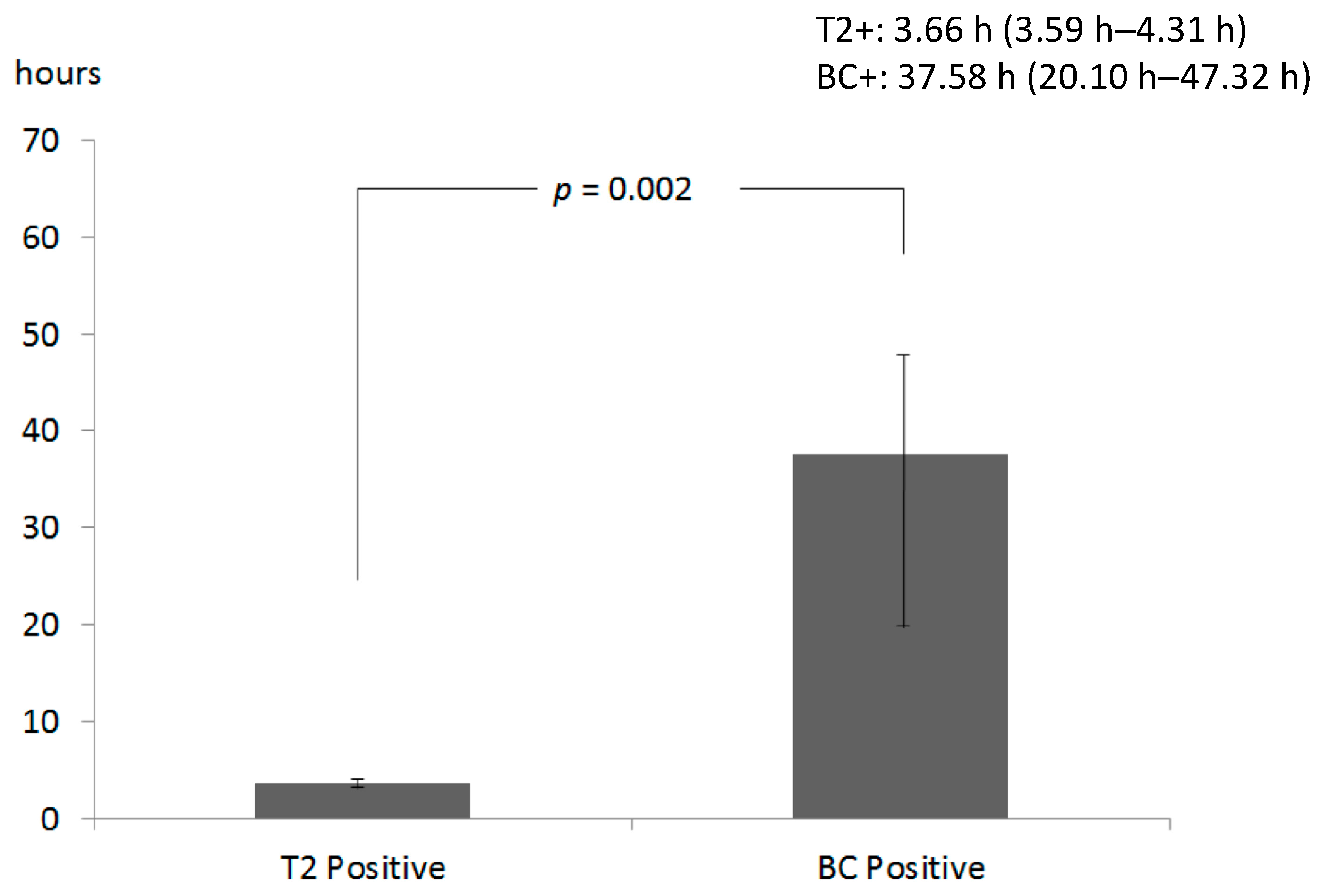
2.2. Comparison of T2 and BC Results
2.3. Clinical and Laboratory Details in Patients with Discordant T2+/BC− Results
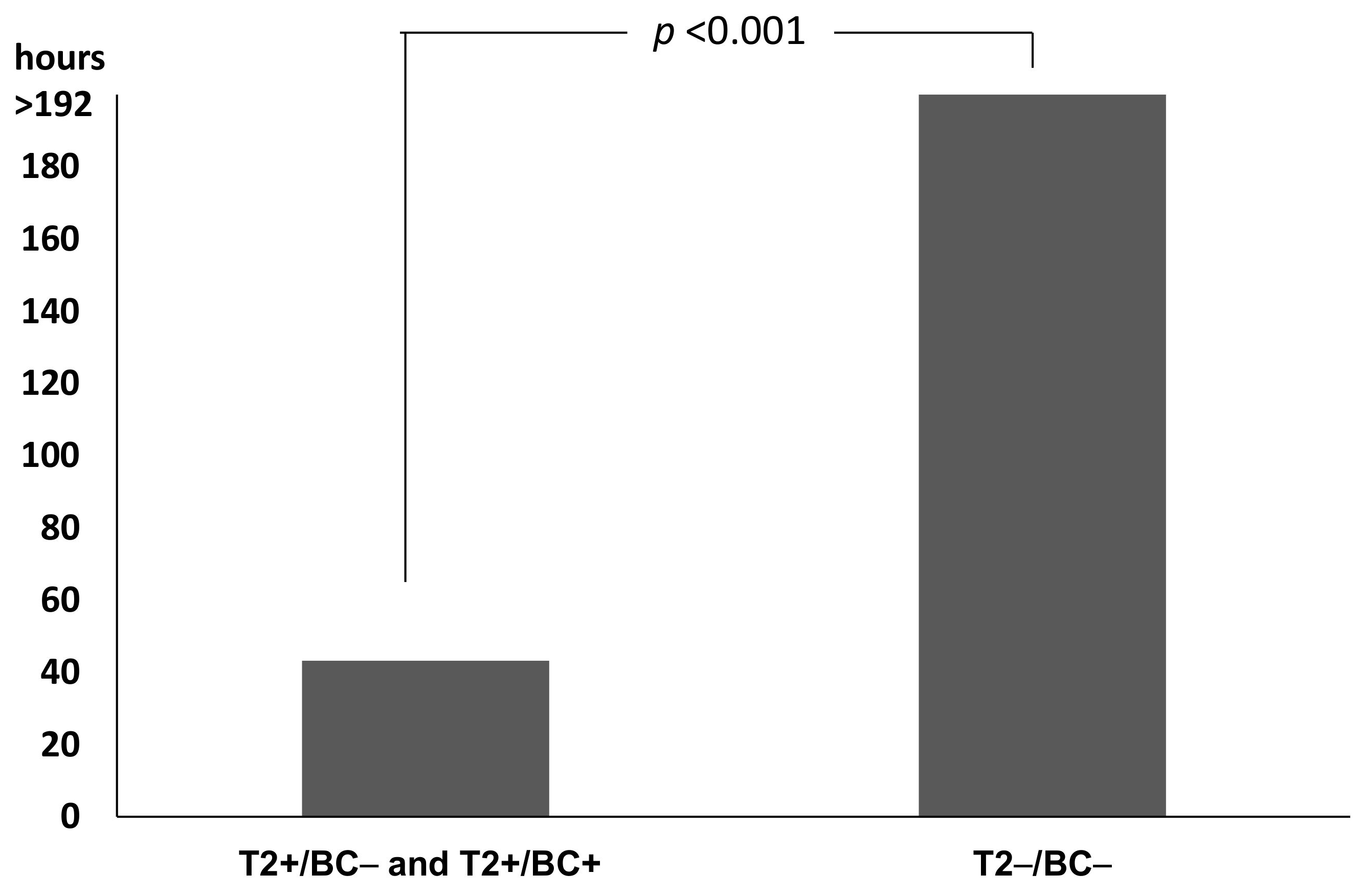
2.4. Impact on Time to Results and Therapy
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. Blood Culture and Standard Protocol
4.3. T2 Sampling and Assay
4.4. Analysis of Clinical Impact
4.5. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.D.; Coopersmith, C.M.; et al. The third international consensus definitions for sepsis and septic shock (sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef]
- Cecconi, M.; Evans, L.; Levy, M.; Rhodes, A. Sepsis and septic shock. Lancet 2018, 392, 75–87. [Google Scholar] [CrossRef]
- Reinhart, K.; Daniels, R.; Kissoon, N.; Machado, F.R.; Schachter, R.D.; Finfer, S. Recognizing sepsis as a global health priority—A WHO resolution. N. Engl. J. Med. 2017, 377, 414–417. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Roberts, D.; Wood, K.E.; Light, B.; Parrillo, J.E.; Sharma, S.; Suppes, R.; Feinstein, D.; Zanotti, S.; Taiberg, L.; et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med. 2006, 34, 1589–1596. [Google Scholar] [CrossRef] [PubMed]
- Paul, M.; Kariv, G.; Goldberg, E.; Raskin, M.; Shaked, H.; Hazzan, R.; Samra, Z.; Paghis, D.; Bishara, J.; Leibovici, L. Importance of appropriate empirical antibiotic therapy for methicillin resistant Staphylococcus aureus bacteraemia. J. Antimicrob. Chemother. 2010, 65, 2658–2665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, M.; Shani, V.; Muchtar, E.; Kariv, G.; Robenshtok, E.; Leibovici, L. Systematic review and meta-analysis of the efficacy of appropriate empiric antibiotic therapy for sepsis. Antimicrob. Agents Chemother. 2010, 54, 4851–4863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rhodes, A.; Evan, L.E.; Alhazzani, W.; Levy, M.M.; Antonelli, M.; Ferrer, R.; Kumar, A.; Sevransky, J.E.; Sprung, C.L.; Nunnally, M.E.; et al. Surviving sepsis campaign: International guidelines for management of sepsis and septic shock: 2016. Intensive Care Med. 2017, 43, 304–377. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention (CDC). Antibiotic Resistance Threats in the United States. 2013. Available online: http://www.cdc.gov/drugresistance/threatreport-2013/pdf/ar-threats-2013-508 (accessed on 28 August 2021).
- Burnham, J.P.; Olsen, M.A.; Kollef, M.H. Re-estimating annual deaths due to multidrug-resistant organism infections. Infect. Control Hosp. Epidemiol. 2019, 40, 112–113. [Google Scholar] [CrossRef] [Green Version]
- Pogue, J.M.; Kaye, K.S.; Cohen, D.A.; Marchaim, D. Appropriate antimicrobial therapy in the era of multidrug-resistant human pathogens. Clin. Microbiol. Infect. 2015, 21, 302–312. [Google Scholar] [CrossRef] [Green Version]
- Santoro, A.; Franceschini, E.; Meschiari, M.; Menozzi, M.; Zona, S.; Venturelli, C.; Digaetano, M.; Rogati, C.; Guaraldi, G.; Paul, M.; et al. Epidemiology and Risk Factors Associated With Mortality in Consecutive Patients With Bacterial Bloodstream Infection: Impact of MDR and XDR Bacteria. Open Forum Infect. Dis. 2020, 7, ofaa461. [Google Scholar] [CrossRef]
- Holmes, C.L.; Anderson, M.T.; Mobley, H.L.T.; Bachman, M.A. Pathogenesis of Gram-negative bacteremia. Clin. Microbiol. Rev. 2021, 34, e00234-20. [Google Scholar] [CrossRef]
- Peker, N.; Couto, N.; Sinha, B.; Rossen, J.W. Diagnosis of bloodstream infections from positive blood cultures and directly from blood samples: Recent developments in molecular approaches. Clin. Microbiol. Infect. 2018, 24, 944–955. [Google Scholar] [CrossRef] [Green Version]
- Dubourg, G.; Raoult, D. Emerging methodologies for pathogen identification in positive blood culture testing. Expert Rev. Mol. Diagn. 2016, 16, 97–111. [Google Scholar] [CrossRef] [PubMed]
- Werner, A.S.; Cobbs, C.G.; Kaye, D.; Hook, E.W. Studies on the bacteremia of bacterial endocarditis. JAMA 1967, 202, 199–203. [Google Scholar] [CrossRef] [PubMed]
- Grumaz, S.; Stevens, P.; Grumaz, C.; Decker, S.O.; Weigand, M.A.; Hofer, S.; Brenner, T.; von Haeseler, A.; Sohn, K. Next-generation sequencing diagnostics of bacteremia in septic patients. Genome Med. 2016, 8, 73. [Google Scholar] [CrossRef]
- Grumaz, S.; Grumaz, C.; Vainshtein, Y.; Stevens, P.; Glanz, K.; Decker, S.O.; Hofer, S.; Weigand, M.A.; Brenner, T.; Sohn, K. Enhanced Performance of Next-Generation Sequencing Diagnostics Compared with Standard of Care Microbiological Diagnostics in Patients Suffering From Septic Shock. Crit. Care Med. 2019, 47, e394–e402. [Google Scholar] [CrossRef]
- Opota, O.; Jaton, K.; Greub, G. Microbial diagnosis of bloodstream infection: Towards molecular diagnosis directly from blood. Clin. Microbiol. Infect. 2015, 21, 323–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bacconi, A.; Richmond, G.S.; Baroldi, M.A.; Laffler, T.G.; Blyn, L.B.; Carolan, H.E.; Frinder, M.R.; Toleno, D.M.; Metzgar, D.; Gutierrez, J.R.; et al. Improved sensitivity for molecular detection of bacteria and Candida in blood. J. Clin. Microbiol. 2014, 52, 3164–3174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Onofrio, V.; Salimans, L.; Bedenić, B.; Cartuyvels, R.; Barišić, I.; Gyssens, I.C. The Clinical Impact of Rapid Molecular Microbiological Diagnostics for Pathogen and Resistance Gene Identification in Patients with Sepsis: A Systematic Review. Open Forum Infect. Dis. 2020, 7, ofaa352. [Google Scholar] [CrossRef] [PubMed]
- Pasqualini, L.; Mencacci, A.; Leli, C.; Montagna, P.; Cardaccia, A.; Cenci, E.; Montecarlo, I.; Pirro, M.; di Filippo, F.; Cistaro, E.; et al. Diagnostic Performance of a Multiple Real-Time PCR Assay in Patients with Suspected Sepsis Hospitalized in an Internal Medicine Ward. J. Clin. Microbiol. 2012, 50, 1285. [Google Scholar] [CrossRef] [Green Version]
- Warhurst, G.; Dunn, G.; Chadwick, P.; Blackwood, B.; McAuley, D.; Perkins, G.D.; McMullan, R.; Gates, S.; Bentley, A.; Young, D.; et al. Rapid detection of health-care-associated bloodstream infection in critical care using Multipathogen real-time polymerase chain reaction technology: A diagnostic accuracy study and systematic review. Health Technol. Assess. 2015, 19, 1–141. [Google Scholar] [CrossRef]
- Casalta, J.P.; Gouriet, F.; Roux, V.; Thuny, F.; Habib, G.; Raoult, D. Evaluation of the LightCycler® SeptiFast test in the rapid etiologic diagnostic of infectious endocarditis. Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 569–573. [Google Scholar] [CrossRef]
- Mongelli, G.; Romeo, M.A.; Denaro, C.; Gennaro, M.; Fraggetta, F.; Stefani, S. Added value of multi-pathogen probe-based real-time PCR SeptiFast in the rapid diagnosis of bloodstream infections in patients with bacteraemia. J. Med. Microbiol. 2015, 64, 670–675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfaller, M.A.; Wolk, D.M.; Lowery, T.J. T2MR and T2Candida: Novel technology for the rapid diagnosis of candidemia and invasive candidiasis. Future Microbiol. 2015, 11, 103–117. [Google Scholar] [CrossRef] [PubMed]
- Clancy, C.J.; Pappas, P.G.; Vazquez, J.; Judson, M.A.; Kontoyiannis, D.P.; Thompson, G.R., 3rd; Garey, K.W.; Reboli, A.; Greenberg, R.N.; Apewokin, S.; et al. Detecting Infections Rapidly and Easily for Candidemia Trial, Part 2 (DIRECT2): A Prospective, Multicenter Study of the T2Candida Panel. Clin. Infect. Dis. 2018, 66, 1678–1686. [Google Scholar] [CrossRef]
- Sanguinetti, M.; Posteraro, B.; Beigelman-Aubry, C.; Lamoth, F.; Dunet, V.; Slavin, M.; Richardson, M.D. Diagnosis and treatment of invasive fungal infections: Looking ahead. J. Antimicrob. Chemother. 2019, 74 (Suppl. S2), ii27–ii37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muñoz, P.; Vena, A.; Machado, M. T2Candida MR as a predictor of outcome in patients with suspected invasive candidiasis starting empirical antifungal treatment: A prospective pilot study. J. Antimicrob. Chemother. 2018, 73, iv6–iv12. [Google Scholar] [CrossRef] [Green Version]
- Mylonakis, E.; Clancy, C.J.; Ostrosky-Zeichner, L.; Garey, K.W.; Alangaden, G.J.; Vazquez, J.A.; Groeger, J.S.; Judson, M.A.; Vinagre, Y.M.; Heard, S.O.; et al. T2 magnetic resonance assay for the rapid diagnosis of candidemia in whole blood: A clinical trial. Clin. Infect. Dis. 2015, 60, 892–899. [Google Scholar] [CrossRef] [Green Version]
- Steuber, T.D.; Butler, L.; Sawyer, A.; Chappell, R.; Edwards, J. Comparison of blood cultures versus T2 Candida Panel in management of candidemia at a large community hospital. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 997–1001. [Google Scholar] [CrossRef] [PubMed]
- Diekema, D.J.; Hsueh, P.R.; Mendes, R.E.; Pfaller, M.A.; Rolston, K.V.; Sader, H.S.; Jones, R.N. The microbiology of bloodstream infection: 20-year trends from the SENTRY Antimicrobial Surveillance Program. Antimicrob. Agents Chemother. 2019, 63, e00355-19. [Google Scholar] [CrossRef] [Green Version]
- Giannella, M.; Pankey, G.A.; Pascale, P.; Miller, V.M.; Miller, L.E.; Seitz, T. Antimicrobial and Resource Utilization with T2 Magnetic Resonance for Rapid Diagnosis of Bloodstream Infections: Systematic Review with Meta-analysis of Controlled Studies. Expert Rev. Med. Devices 2021, 18, 473–482. [Google Scholar] [CrossRef]
- Voigt, C.; Silbert, S.; Widen, R.H.; Marturano, J.E.; Lowery, T.J.; Ashcraft, D.; Pankey, G. The T2Bacteria Assay Is a Sensitive and Rapid Detector of Bacteremia That Can Be Initiated in the Emergency Department and Has Potential to Favorably Influence Subsequent Therapy. J. Emerg. Med. 2020, 58, 785–796. [Google Scholar] [CrossRef]
- De Angelis, G.; Posteraro, B.; De Carolis, E.; Menchinelli, G.; Franceschi, F.; Tumbarello, M.; De Pascale, G.; Spanu, T.; Sanguinetti, M. T2Bacteria magnetic resonance assay for the rapid detection of ESKAPEc pathogens directly in whole blood. J. Antimicrob. Chemother. 2018, 73, iv20–iv26. [Google Scholar] [CrossRef]
- Maki, D.G. The T2Bacteria Panel had 90% sensitivity for detecting targeted organisms, 43% for any bloodstream infection organism. Ann. Intern. Med. 2019, 171, JC34. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.H.; Clancy, C.J.; Pasculle, A.W.; Pappas, P.G.; Alangaden, G.; Pankey, G.A.; Schmitt, B.H.; Rasool, A.; Weinstein, M.P.; Widen, R.; et al. Performance of the T2Bacteria Panel for Diagnosing Bloodstream Infections: A Diagnostic Accuracy Study. Ann. Intern. Med. 2019, 170, 845–852. [Google Scholar] [CrossRef] [PubMed]
- Drevinek, P.; Hurych, J.; Antuskova, M.; Tkadlec, J.; Berousek, J.; Prikrylova, Z.; Bures, J.; Vajter, J.; Soucek, M.; Masopust, J.; et al. Direct detection of ESKAPEc pathogens from whole blood using the T2Bacteria Panel allows early antimicrobial stewardship intervention in patients with sepsis. Microbiologyopen 2021, 10, e1210. [Google Scholar] [CrossRef]
- Farrell, J.J.; Hujer, A.M.; Sampath, R.; Bonomo, R.A. Salvage microbiology: Opportunities and challenges in the detection of bacterial pathogens following initiation of antimicrobial treatment. Expert Rev. Mol. Diagn. 2015, 15, 349–360. [Google Scholar] [CrossRef] [Green Version]
- Lodes, U.; Bohmeier, B.; Lippert, H.; König, B.; Meyer, F. PCR-based rapid sepsis diagnosis effectively guides clinical treatment in patients with new onset of SIRS. Langenbeck’s Arch. Surg. 2012, 397, 447–455. [Google Scholar] [CrossRef]
- Kalligeros, M.; Zacharioudakis, I.M.; Tansarli, G.S.; Tori, K.; Shehadeh, F.; Mylonakis, E. In-depth analysis of T2Bacteria positive results in patients with concurrent negative blood culture: A case series. BMC Infect. Dis. 2020, 20, 326. [Google Scholar] [CrossRef]
- Hall, K.K.; Lyman, J.A. Updated review of blood culture contamination. Clin. Microbiol. Rev. 2006, 19, 788–802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holland, T.L.; Baddour, L.M.; Bayer, A.S.; Hoen, B.; Miro, J.M.; Fowler, V.G. Infective endocarditis. Nat. Rev. Dis. Primers 2011, 2, 16059. [Google Scholar] [CrossRef]
- Kavanagh, N.; Ryan, E.J.; Widaa, A.; Sexton, G.; Fennell, J.; O’Rourke, S.; Cahill, K.C.; Kearney, C.J.; O’Brien, F.J.; Kerrigan, S.W. Staphylococcal osteomyelitis: Disease progression, treatment challenges, and future directions. Clin. Microbiol. Rev. 2018, 31, e00084-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reers, Y.; Idelevich, E.A.; Patkau, H.; Sauerland, M.C.; Tafelski, S.; Nachtigall, I.; Berdel, W.E.; Peters, G.; Silling, G.; Becker, K.; et al. Multiplex PCR assay underreports true bloodstream infections with coagulase-negative staphylococci in hematological patients with febrile neutropenia. Diagn. Microbiol. Infect. Dis. 2016, 85, 413–415. [Google Scholar] [CrossRef] [PubMed]
- Bogan, C.; Marchaim, D. The role of antimicrobial stewardship in curbing carbapenem resistance. Future Microbiol. 2013, 8, 979–991. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.J.; Mencacci, A.; Paggi, R.; Douka, E.; Vrettou, C. Prospective study of T2 resistance system for detection of resistance genes in bacterial bloodsteam infections. In Proceedings of the ePoster 31th ECCMID 2021, Online, 9–12 July 2021. [Google Scholar]
- De Socio, G.; Di Donato, F.; Paggi, R.; Gabrielli, C.; Belati, A.; Rizza, G.; Savoia, M.; Repetto, A.; Cenci, E.; Mencacci, A. Laboratory automation reduces time to report of positive blood cultures and improves management of patients with bloodstream infection. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 2313–2322. [Google Scholar] [CrossRef]

| Variable | Value |
|---|---|
| Patients | |
| Years, median (IQR) | 62.5 (42.0–71.0) |
| Men | 57 (69.5%) |
| Hospital ward | |
| Medicine | 45 (54.9%) |
| Surgical | 3 (3.7%) |
| Intensive Care | 34 (41.5%) |
| Laboratory parameters | |
| Leucocytes, cells × 103/mL, Median (IQR) | 12.93 (3.90–18.39) |
| Leukocytosis, >12 × 103/mL | 35 (42.7%) |
| Leucopenia, <4 × 103/mL | 21 (25.6%) |
| Neutrophils percentage, Median (IQR) | 73.8 (66.6–87.0) |
| CRP, mg/dL, Median (IQR) | 12.1 (6.3–19.8) |
| PCT, ng/mL, Median (IQR) | 1.18 (0.40–7.36) |
| Lactate, mM/L, Median (IQR) | 1.50 (1.00–2.73) |
| Clinical data | |
| Body Temperature, °C, Median (IQR) | 37.5 (36.5–38.3) |
| Temperature > 38 °C | 23 (28.0%) |
| Temperature < 36 °C | 3 (3.6%) |
| Heart Rate, beats/min, Median (IQR) | 98.0 (80.0–115.0) |
| Mean Arterial Pressure, mmHg, Median (IQR) | 85.0 (73.0–93.0) |
| Mean Respiratory Rate, acts/min, Median (IQR) | 20.0 (16.0–24.0) |
| Glasgow Coma Scale Median (IQR) | 14.0 (11.0–15.0) |
| Septic Shock | 24 (29.3%) |
| Sequential Organ Failure Assessment Score, Median (IQR) | 5 (3–9) |
| 30-Days Mortality | 25 (30.5%) |
| Concomitant diseases | |
| Hypertension 18 | 33 (40%) |
| History of Cardiovascular Disease | 19 (23%) |
| Chronic renal failure | 14 (15%) |
| Solid Malignancy | 15 (18%) |
| Diabetes | 18 (22%) |
| Chronic Lung Disease | 11 (13%) |
| Transplant | 14 (17%) |
| Chronic Liver Disease | 8 (10/%) |
| Hematological Disease | 27 (33%) |
| BC Positive | BC Negative | Total | |
|---|---|---|---|
| T2 positive | 10 1 | 14 2 | 24 |
| T2 negative | 6 | 52 | 58 |
| Total | 16 | 66 | 82 |
| Age, Gender | Hospital Ward 1 | Sofa Score | WBC × 103 (% Neutrophils) | Diagnosis on Admission | Suspected Infectious Focus | T2BP Result | Same Pathogen Cultured <15 Days from T2 Collection | Empirical Therapy 2 | Switch to Directed Therapy | Hours of Empirical Therapy |
|---|---|---|---|---|---|---|---|---|---|---|
| 54, M | ICU | 2 | 10.910 (82.1) | Endocarditis | Cardiac valve | S. aureus | No | OXA, DAP | No | N.A. 3 |
| 67, M | BMT | 7 | 0.28 (7.5) | Sepsis | Unknown | E. coli | No | MEM | MEM, AN | 26.0 |
| 71, M | BMT | 9 | 1.28 (69.5) | Sepsis | Urinary tract | K. pneumoniae + P. aeruginosa | Yes (urine) | CAZ/AVI, TGC | MEM, CAZ/AVI | 6.3 |
| 75, F | ICU | 7 | 69.20 (73.3) | Sepsis | Knee Prosthetic Device | S. aureus | Yes (synovial fluid, blood, urine, rectal swab) | OXA, RIF | DAP | 61.7 |
| 86, F 4 | MED | 9 | 10.42 (78.2) | Septic Shock | Unknown | P. aeruginosa | No | AMP/SUL, TED | C/T, AN | 36.6 |
| 27, F | MED | 1 | 24.50 (87.2) | Sepsis | Unknown | E. coli | No | ERT, CRO | No | N.A. |
| 81, M | MED | 15 | 21.37 (86.6) | Tracheo-esophageal fistula | Pneumonia | P. aeruginosa | Yes (respiratory tract, Rectal swab) | CAZ/AVI, TGC | CAZ/AVI, AN | 8.7 |
| 75, F | ICU | 1 | 7.04 (58.6) | Wound infection | Limb Abscess | P. aeruginosa + S. aureus | Yes (rectal swab) | RIF, DAP, OXA, FEP | No | N.A. |
| 60, M 4 | MED | 5 | 27.61 (94.5) | Cellulitis | Wound | K. pneumoniae | No | CC, CRO, TEC | MER, TEC | 16.6 |
| 16, M | BMT | 10 | 0.02 (-) | Fever in BMT patient | Oral Mucositis | P. aeruginosa | Yes (Blood) | MEM, AN. TGC, C/T | No | N.A. |
| 92, F | MED | 5 | 7.53 (66.3) | Sepsis | Urinary tract | K. pneumoniae | Yes (urine, blood, rectal swab) | DAP, TZP, CAZ/AVI | CAZ/AVI, TZP | 24.0 |
| 42, M | MED | 6 | 36.39 (76.6) | Septic shock | Urinary tract | E. coli | No | DAP, FF, CAZ/AVI, TZP | MEM | 8.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paggi, R.; Cenci, E.; De Socio, G.V.; Belati, A.; Marini, D.; Gili, A.; Camilloni, B.; Mencacci, A. Accuracy and Impact on Patient Management of New Tools for Diagnosis of Sepsis: Experience with the T2 Magnetic Resonance Bacteria Panel. Pathogens 2021, 10, 1132. https://doi.org/10.3390/pathogens10091132
Paggi R, Cenci E, De Socio GV, Belati A, Marini D, Gili A, Camilloni B, Mencacci A. Accuracy and Impact on Patient Management of New Tools for Diagnosis of Sepsis: Experience with the T2 Magnetic Resonance Bacteria Panel. Pathogens. 2021; 10(9):1132. https://doi.org/10.3390/pathogens10091132
Chicago/Turabian StylePaggi, Riccardo, Elio Cenci, Giuseppe Vittorio De Socio, Alessandra Belati, Daniele Marini, Alessio Gili, Barbara Camilloni, and Antonella Mencacci. 2021. "Accuracy and Impact on Patient Management of New Tools for Diagnosis of Sepsis: Experience with the T2 Magnetic Resonance Bacteria Panel" Pathogens 10, no. 9: 1132. https://doi.org/10.3390/pathogens10091132
APA StylePaggi, R., Cenci, E., De Socio, G. V., Belati, A., Marini, D., Gili, A., Camilloni, B., & Mencacci, A. (2021). Accuracy and Impact on Patient Management of New Tools for Diagnosis of Sepsis: Experience with the T2 Magnetic Resonance Bacteria Panel. Pathogens, 10(9), 1132. https://doi.org/10.3390/pathogens10091132









