Regulation of Antigenic Variation by Trypanosoma brucei Telomere Proteins Depends on Their Unique DNA Binding Activities
Abstract
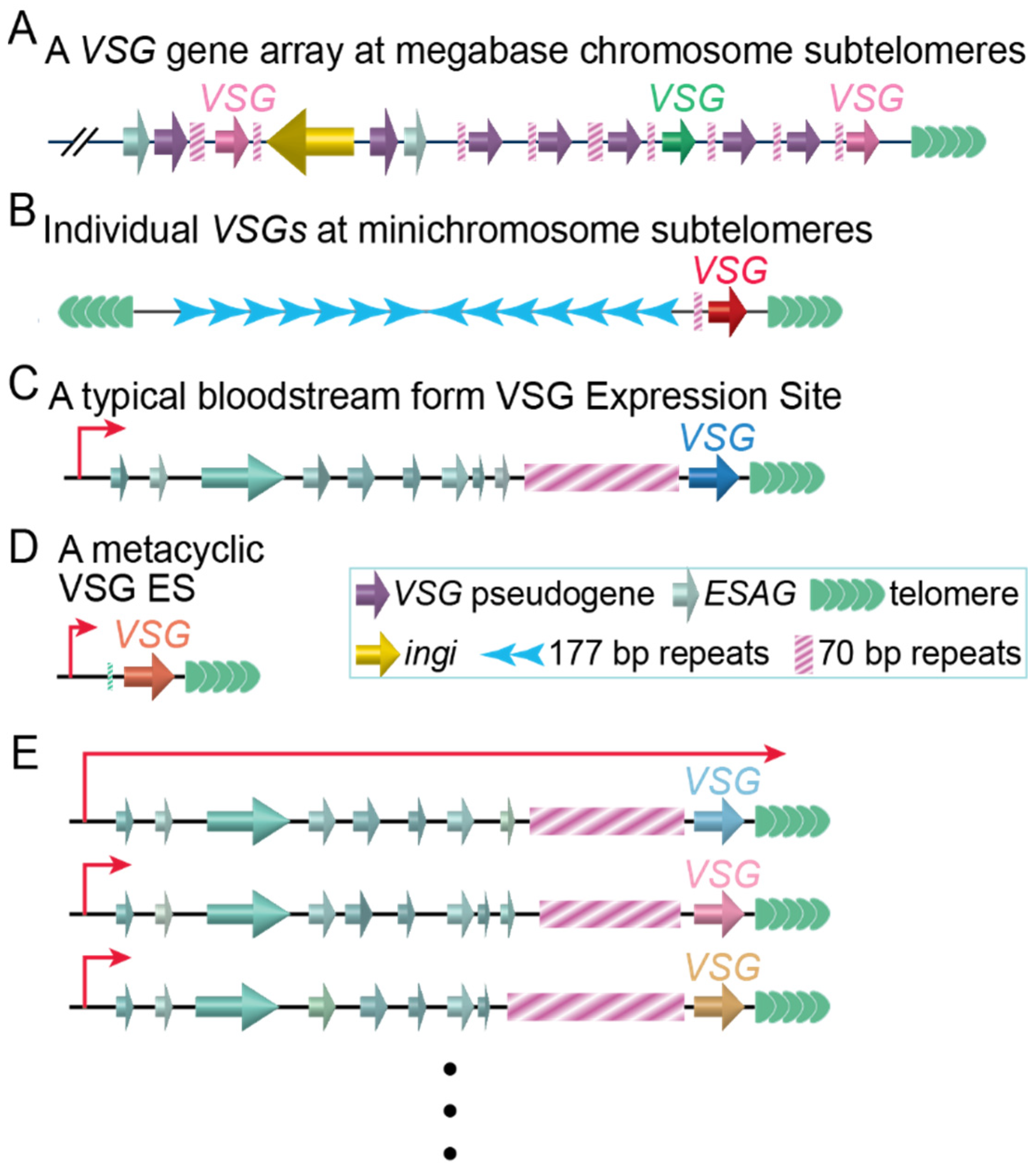
1. Antigenic Variation in T. brucei Involves Its Major Surface Antigen, Variant SURFACE Glycoprotein (VSG)
1.1. VSG Is Expressed in a Monoallelic Manner from a Single VSG Expression Site (ES)
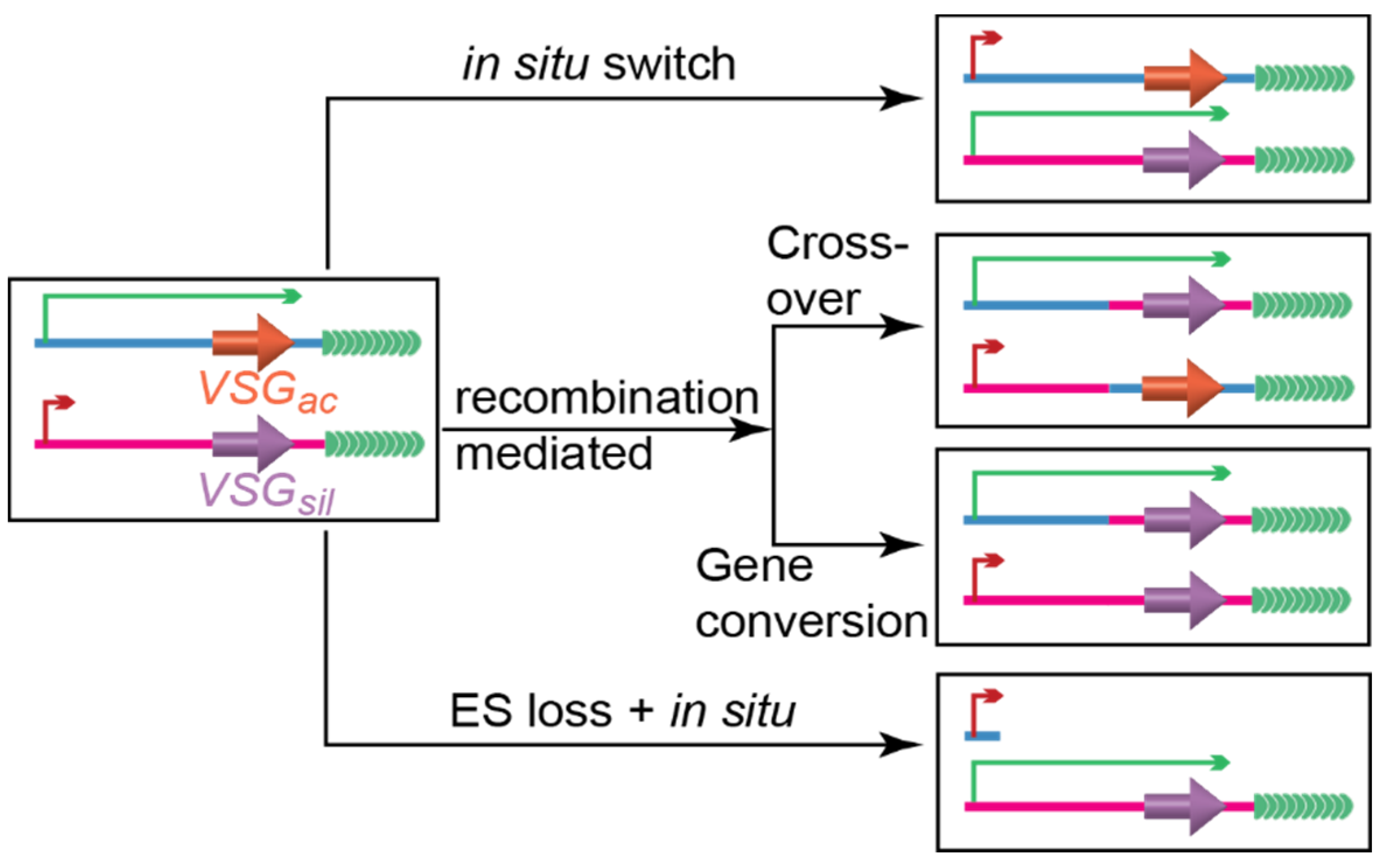
1.2. Several Pathways Mediate VSG Switching
2. Telomere Functions in Antigenic Variation
2.1. Telomeres Are Essential for Genome Integrity and Affect Nearby Gene Expression
2.2. DNA Binding Activities of Telomere Proteins Are Critical for Their Essential Functions
2.3. T. brucei TRF and RAP1 Play Crucial Roles in Antigenic Variation
2.3.1. The TbTRF Myb Domain Has Sequence-Specific Duplex Telomere DNA and TERRA Binding Activities That Are Critical for Maintaining the Telomere Integrity and for Suppressing VSG Switching
2.3.2. The Electrostatics-Based Sequence-Nonspecific dsDNA Binding Activity of TbRAP1 Is Essential for VSG Silencing and Telomere Integrity
3. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sutherland, C.S.; Yukich, J.; Goeree, R.; Tediosi, F. A Literature Review of Economic Evaluations for a Neglected Tropical Disease: Human African Trypanosomiasis (“Sleeping Sickness”). PLoS Negl. Trop. Dis. 2015, 9, e0003397. [Google Scholar] [CrossRef]
- WHO. Investing to Overcome the Global Impact of Neglected Tropical Diseases. Third WHO Report on Neglected Tropical DiseasesWHO. 2015. Available online: https://apps.who.int/iris/handle/10665/152781 (accessed on 29 July 2021).
- Weny, G.; Okwee-Acai, J.; Okech, S.G.; Tumwine, G.; Ndyanabo, S.; Abigaba, S.; Goldberg, T.L. Prevalence and Risk Factors Associated with Hemoparasites in Cattle and Goats at the Edge of Kibale National Park, Western Uganda. J. Parasitol. 2017, 103, 69–74. [Google Scholar] [CrossRef]
- Matthews, K.R. The developmental cell biology of Trypanosoma brucei. J. Cell Sci. 2005, 118, 283–290. [Google Scholar] [CrossRef]
- Acosta-Serrano, A.; Vassella, E.; Liniger, M.; Renggli, C.K.; Brun, R.; Roditi, I.; Englund, P.T. The surface coat of procyclic Trypanosoma brucei: Programmed expression and proteolytic cleavage of procyclin in the tsetse fly. Proc. Natl. Acad. Sci. USA 2001, 98, 1513–1518. [Google Scholar] [CrossRef] [PubMed]
- Quintana, J.F.; Zoltner, M.; Field, M.C. Evolving Differentiation in African Trypanosomes. Trends Parasitol. 2021, 37, 296–303. [Google Scholar] [CrossRef]
- Diffley, P. Trypanosoma brucei: Immunogenicity of the variant surface coat glycoprotein of virulent and avirulent subspecies. Exp. Parasitol. 1985, 59, 98–107. [Google Scholar] [CrossRef]
- Deitsch, K.W.; Lukehart, S.A.; Stringer, J.R. Common strategies for antigenic variation by bacterial, fungal and protozoan pathogens. Nat. Rev. Genet. 2009, 7, 493–503. [Google Scholar] [CrossRef]
- Cross, G.A.; Kim, H.-S.; Wickstead, B. Capturing the variant surface glycoprotein repertoire (the VSGnome) of Trypanosoma brucei Lister 427. Mol. Biochem. Parasitol. 2014, 195, 59–73. [Google Scholar] [CrossRef] [PubMed]
- Müller, L.S.; Cosentino, R.O.; Förstner, K.U.; Guizetti, J.; Wedel, C.; Kaplan, N.; Janzen, C.J.; Arampatzi, P.; Vogel, J.; Steinbiss, S.; et al. Genome organization and DNA accessibility control antigenic variation in trypanosomes. Nat. Cell Biol. 2018, 563, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Melville, S.E.; Leech, V.; Navarro, M.; Cross, G. The molecular karyotype of the megabase chromosomes of Trypanosoma brucei stock Mol. Biochem. Parasitol. 2000, 111, 261–273. [Google Scholar] [CrossRef]
- El-Sayed, N.M.; Hegde, P.; Quackenbush, J.; Melville, S.E.; Donelson, J.E. The African trypanosome genome. Int. J. Parasitol. 2000, 30, 329–345. [Google Scholar] [CrossRef]
- Alsford, S.; Wickstead, B.; Ersfeld, K.; Gull, K. Diversity and dynamics of the minichromosomal karyotype in Trypanosoma brucei. Mol. Biochem. Parasitol. 2001, 113, 79–88. [Google Scholar] [CrossRef]
- Günzl, A.; Bruderer, T.; Laufer, G.; Schimanski, B.; Tu, L.-C.; Chung, H.-M.; Lee, P.-T.; Lee, M.G.-S. RNA Polymerase I Transcribes Procyclin Genes and Variant Surface Glycoprotein Gene Expression Sites in Trypanosoma brucei. Eukaryot. Cell 2003, 2, 542–551. [Google Scholar] [CrossRef]
- Hertz-Fowler, C.; Figueiredo, L.M.; Quail, M.A.; Becker, M.; Jackson, A.; Bason, N.; Brooks, K.; Churcher, C.; Fahkro, S.; Goodhead, I.; et al. Telomeric Expression Sites Are Highly Conserved in Trypanosoma brucei. PLoS ONE 2008, 3, e3527. [Google Scholar] [CrossRef]
- De Lange, T.; Borst, P. Genomic environment of the expression-linked extra copies of genes for surface antigens of Trypanosoma brucei resembles the end of a chromosome. Nature 1982, 299, 451–453. [Google Scholar] [CrossRef] [PubMed]
- Barry, J.; Graham, S.; Fotheringham, M.; Graham, V.S.; Kobryn, K.; Wymer, B. VSG gene control and infectivity strategy of metacyclic stage Trypanosoma brucei. Mol. Biochem. Parasitol. 1998, 91, 93–105. [Google Scholar] [CrossRef]
- Cross, G. Identification, purification and properties of clone-specific glycoprotein antigens constituting the surface coat of Trypanosoma brucei. Parasitology 1975, 71, 393–417. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Figueiredo, L.M.; Espinal, A.; Okubo, E.; Li, B. RAP1 Is Essential for Silencing Telomeric Variant Surface Glycoprotein Genes in Trypanosoma brucei. Cell 2009, 137, 99–109. [Google Scholar] [CrossRef]
- Pandya, U.M.; Sandhu, R.; Li, B. Silencing subtelomeric VSGs by Trypanosoma brucei RAP1 at the insect stage involves chromatin structure changes. Nucleic Acids Res. 2013, 41, 7673–7682. [Google Scholar] [CrossRef]
- Afrin, M.; Gaurav, A.K.; Yang, X.; Pan, X.; Zhao, Y.; Li, B. TbRAP1 has an unusual duplex DNA binding activity required for its telomere localization and VSG silencing. Sci. Adv. 2020, 6, eabc4065. [Google Scholar] [CrossRef]
- Afrin, M.; Kishmiri, H.; Sandhu, R.; Rabbani, M.A.G.; Li, B. Trypanosoma brucei RAP1 Has Essential Functional Domains That Are Required for Different Protein Interactions. Msphere 2020, 5, e00027-20. [Google Scholar] [CrossRef] [PubMed]
- Günzl, A.; Kirkham, J.; Nguyen, T.N.; Badjatia, N.; Park, S.H. Mono-allelic VSG expression by RNA polymerase I in Trypanosoma brucei: Expression site control from both ends? Gene 2015, 556, 68–73. [Google Scholar] [CrossRef][Green Version]
- Rudenko, G. Epigenetics and transcriptional control in African trypanosomes. Essays Biochem. 2010, 48, 201–219. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Narayanan, M.S.; Rudenko, G. TDP1 is an HMG chromatin protein facilitating RNA polymerase I transcription in African trypanosomes. Nucleic Acids Res. 2013, 41, 2981–2992. [Google Scholar] [CrossRef]
- Povelones, M.L.; Gluenz, E.; Dembek, M.; Gull, K.; Rudenko, G. Histone H1 Plays a Role in Heterochromatin Formation and VSG Expression Site Silencing in Trypanosoma brucei. PLoS Pathog. 2012, 8, e1003010. [Google Scholar] [CrossRef][Green Version]
- Stanne, T.M.; Rudenko, G. Active VSG Expression Sites in Trypanosoma brucei are Depleted of Nucleosomes. Eukaryot. Cell 2010, 9, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, L.M.; Cross, G.A.M. Nucleosomes are Depleted at the VSG Expression Site Transcribed by RNA Polymerase I in African Trypanosomes. Eukaryot. Cell 2010, 9, 148–154. [Google Scholar] [CrossRef]
- Hughes, K.; Wand, M.; Foulston, L.; Young, R.; Harley, K.; Terry, S.; Ersfeld, K.; Rudenko, G. A novel ISWI is involved in VSG expression site downregulation in African trypanosomes. EMBO J. 2007, 26, 2400–2410. [Google Scholar] [CrossRef]
- Denninger, V.; Rudenko, G. FACT plays a major role in histone dynamics affecting VSG expression site control in Trypanosoma brucei. Mol. Microbiol. 2014, 94, 945–962. [Google Scholar] [CrossRef]
- Denninger, V.; Fullbrook, A.; Bessat, M.; Ersfeld, K.; Rudenko, G. The FACT subunit TbSpt16 is involved in cell cycle specific control of VSG expression sites in Trypanosoma brucei. Mol. Microbiol. 2010, 78, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Navarro, M.; Cross, G.A.; Wirtz, E. Trypanosoma brucei variant surface glycoprotein regulation involves coupled activation/inactivation and chromatin remodeling of expression sites. EMBO J. 1999, 18, 2265–2272. [Google Scholar] [CrossRef]
- Schulz, D.; Zaringhalam, M.; Papavasiliou, F.N.; Kim, H.-S. Base J and H3.V Regulate Transcriptional Termination in Trypanosoma brucei. PLoS Genet. 2016, 12, e1005762. [Google Scholar] [CrossRef]
- Kassem, A.; Pays, E.; Vanhamme, L. Transcription is initiated on silent variant surface glycoprotein expression sites despite monoallelic expression in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 2014, 111, 8943–8948. [Google Scholar] [CrossRef]
- Vanhamme, L.; Poelvoorde, P.; Pays, A.; Tebabi, P.; Van Xong, H.; Pays, E. Differential RNA elongation controls the variant surface glycoprotein gene expression sites of Trypanosoma brucei. Mol. Microbiol. 2000, 36, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Cestari, I.; McLeland-Wieser, H.; Stuart, K. Nuclear Phosphatidylinositol 5-Phosphatase Is Essential for Allelic Exclusion of Variant Surface Glycoprotein Genes in Trypanosomes. Mol. Cell. Biol. 2019, 39, 39. [Google Scholar] [CrossRef] [PubMed]
- Cestari, I.; Stuart, K. Inositol phosphate pathway controls transcription of telomeric expression sites in trypanosomes. Proc. Natl. Acad. Sci. USA 2015, 112, E2803–E2812. [Google Scholar] [CrossRef] [PubMed]
- Dubois, K.N.; Alsford, S.; Holden, J.M.; Buisson, J.; Swiderski, M.; Bart, J.-M.; Ratushny, A.V.; Wan, Y.; Bastin, P.; Barry, J.D.; et al. NUP-1 Is a Large Coiled-Coil Nucleoskeletal Protein in Trypanosomes with Lamin-Like Functions. PLoS Biol. 2012, 10, e1001287. [Google Scholar] [CrossRef] [PubMed]
- Maishman, L.; Obado, S.O.; Alsford, S.; Bart, J.-M.; Chen, W.-M.; Ratushny, A.V.; Navarro, M.; Horn, D.; Aitchison, J.D.; Chait, B.T.; et al. Co-dependence between trypanosome nuclear lamina components in nuclear stability and control of gene expression. Nucleic Acids Res. 2016, 44, 10554–10570. [Google Scholar] [CrossRef]
- López-Farfán, D.C.; Bart, J.-M.; Rojas-Barros, D.I.; Navarro, M. SUMOylation by the E3 Ligase TbSIZ1/PIAS1 Positively Regulates VSG Expression in Trypanosoma brucei. PLoS Pathog. 2014, 10, e1004545. [Google Scholar] [CrossRef] [PubMed]
- Benmerzouga, I.; Concepcion-Acevedo, J.; Kim, H.S.; Vandoros, A.V.; Cross, G.A.; Klingbeil, M.M.; Li, B. Trypanosoma brucei Orc1 is essential for nuclear DNA replication and affects both VSG silencing and VSG switching. Mol. Microbiol. 2013, 87, 196–210. [Google Scholar] [CrossRef]
- Tiengwe, C.; Marcello, L.; Farr, H.; Dickens, N.; Kelly, S.; Swiderski, M.; Vaughan, D.; Gull, K.; Barry, J.D.; Bell, S.D.; et al. Genome-wide Analysis Reveals Extensive Functional Interaction between DNA Replication Initiation and Transcription in the Genome of Trypanosoma brucei. Cell Rep. 2012, 2, 185–197. [Google Scholar] [CrossRef]
- Kim, H.-S.; Park, S.H.; Günzl, A.; Cross, G.A.M. MCM-BP Is Required for Repression of Life-Cycle Specific Genes Transcribed by RNA Polymerase I in the Mammalian Infectious Form of Trypanosoma brucei. PLoS ONE 2013, 8, e57001. [Google Scholar] [CrossRef]
- Kim, H.-S. Genome-wide function of MCM-BP in Trypanosoma brucei DNA replication and transcription. Nucleic Acids Res. 2019, 47, 634–647. [Google Scholar] [CrossRef]
- Faria, J.; Glover, L.; Hutchinson, S.; Boehm, C.; Field, M.C.; Horn, D. Monoallelic expression and epigenetic inheritance sustained by a Trypanosoma brucei variant surface glycoprotein exclusion complex. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef]
- Glover, L.; Hutchinson, S.; Alsford, S.; Horn, D. VEX1 controls the allelic exclusion required for antigenic variation in trypanosomes. Proc. Natl. Acad. Sci. USA 2016, 113, 7225–7230. [Google Scholar] [CrossRef] [PubMed]
- Faria, J.; Luzak, V.; Müller, L.S.M.; Brink, B.G.; Hutchinson, S.; Glover, L.; Horn, D.; Siegel, T.N. Spatial integration of transcription and splicing in a dedicated compartment sustains monogenic antigen expression in African trypanosomes. Nat. Microbiol. 2021, 6, 289–300. [Google Scholar] [CrossRef]
- Aresta-Branco, F.; Sanches-Vaz, M.; Bento, F.; Rodrigues, J.A.; Figueiredo, L.M. African trypanosomes expressing multiple VSGs are rapidly eliminated by the host immune system. Proc. Natl. Acad. Sci. USA 2019, 116, 20725–20735. [Google Scholar] [CrossRef] [PubMed]
- Batram, C.; Jones, N.G.; Janzen, C.J.; Markert, S.M.; Engstler, M. Expression site attenuation mechanistically links antigenic variation and development in Trypanosoma brucei. Elife 2014, 3, e02324. [Google Scholar] [CrossRef]
- Myler, P.; Allison, J.; Agabian, N.; Stuart, K. Antigenic variation in African trypanosomes by gene replacement or activation of alternate telomeres. Cell 1984, 39, 203–211. [Google Scholar] [CrossRef]
- Myler, P.; Nelson, R.G.; Agabian, N.; Stuart, K. Two mechanisms of expression of a predominant variant antigen gene of Trypanosoma brucei. Nat. Cell Biol. 1984, 309, 282–284. [Google Scholar] [CrossRef]
- Kamper, S.M.; Barbet, A.F. Surface epitope variation via mosaic gene formation is potential key to long-term survival of Trypanosoma brucei. Mol. Biochem. Parasitol. 1992, 53, 33–44. [Google Scholar] [CrossRef]
- Marcello, L.; Barry, J.D. Analysis of the VSG gene silent archive in Trypanosoma brucei reveals that mosaic gene expression is prominent in antigenic variation and is favored by archive substructure. Genome Res. 2007, 17, 1344–1352. [Google Scholar] [CrossRef]
- Jehi, S.E.; Li, X.; Sandhu, R.; Ye, F.; Benmerzouga, I.; Zhang, M.; Zhao, Y.; Li, B. Suppression of subtelomeric VSG switching by Trypanosoma brucei TRF requires its TTAGGG repeat-binding activity. Nucleic Acids Res. 2014, 42, 12899–12911. [Google Scholar] [CrossRef]
- Jehi, S.E.; Wu, F.; Li, B. Trypanosoma brucei TIF2 suppresses VSG switching by maintaining subtelomere integrity. Cell Res. 2014, 24, 870–885. [Google Scholar] [CrossRef] [PubMed]
- Boothroyd, C.E.; Dreesen, O.; Leonova, T.; Ly, K.I.; Figueiredo, L.M.; Cross, G.; Papavasiliou, F.N. A yeast-endonuclease-generated DNA break induces antigenic switching in Trypanosoma brucei. Nat. Cell Biol. 2009, 459, 278–281. [Google Scholar] [CrossRef] [PubMed]
- Glover, L.; Alsford, S.; Horn, D. DNA Break Site at Fragile Subtelomeres Determines Probability and Mechanism of Antigenic Variation in African Trypanosomes. PLoS Pathog. 2013, 9, e1003260. [Google Scholar] [CrossRef]
- Cross, M.; Taylor, M.C.; Borst, P. Frequent Loss of the Active Site during Variant Surface Glycoprotein Expression Site Switching In Vitro in Trypanosoma brucei. Mol. Cell. Biol. 1998, 18, 198–205. [Google Scholar] [CrossRef][Green Version]
- Kim, H.-S.; Cross, G.A.M. Identification of Trypanosoma brucei RMI1/BLAP75 Homologue and Its Roles in Antigenic Variation. PLoS ONE 2011, 6, e25313. [Google Scholar] [CrossRef]
- Robinson, N.; Burman, N.; Melville, S.E.; Barry, J.D. Predominance of Duplicative VSG Gene Conversion in Antigenic Variation in African Trypanosomes. Mol. Cell. Biol. 1999, 19, 5839–5846. [Google Scholar] [CrossRef]
- Hovel-Miner, G.; Boothroyd, C.E.; Mugnier, M.; Dreesen, O.; Cross, G.; Papavasiliou, F.N. Telomere Length Affects the Frequency and Mechanism of Antigenic Variation in Trypanosoma brucei. PLoS Pathog. 2012, 8, e1002900. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-S.; Cross, G.A.M. TOPO3α Influences Antigenic Variation by Monitoring Expression-Site-Associated VSG Switching in Trypanosoma brucei. PLoS Pathog. 2010, 6, e1000992. [Google Scholar] [CrossRef][Green Version]
- Nanavaty, V.; Sandhu, R.; Jehi, S.E.; Pandya, U.; Li, B. Trypanosoma brucei RAP1 maintains telomere and subtelomere integrity by suppressing TERRA and telomeric RNA:DNA hybrids. Nucleic Acids Res. 2017, 45, 5785–5796. [Google Scholar] [CrossRef]
- McCulloch, R.; Barry, J.D. A role for RAD51 and homologous recombination in Trypanosoma brucei antigenic variation. Genes Dev. 1999, 13, 2875–2888. [Google Scholar] [CrossRef]
- Proudfoot, C.; McCulloch, R. Distinct roles for two RAD51-related genes in Trypanosoma brucei antigenic variation. Nucleic Acids Res. 2005, 33, 6906–6919. [Google Scholar] [CrossRef]
- Hartley, C.L.; McCulloch, R. Trypanosoma brucei BRCA2 acts in antigenic variation and has undergone a recent expansion in BRC repeat number that is important during homologous recombination. Mol. Microbiol. 2008, 68, 1237–1251. [Google Scholar] [CrossRef]
- Devlin, R.; Marques, C.A.; Paape, D.; Prorocic, M.; Zurita-Leal, A.; Campbell, S.J.; Lapsley, C.; Dickens, N.; McCulloch, R. Mapping replication dynamics in Trypanosoma brucei reveals a link with telomere transcription and antigenic variation. Elife 2016, 5, e12765. [Google Scholar] [CrossRef]
- Jehi, S.E.; Nanavaty, V.; Li, B. Trypanosoma brucei TIF2 and TRF Suppress VSG Switching Using Overlapping and Independent Mechanisms. PLoS ONE 2016, 11, e0156746. [Google Scholar] [CrossRef]
- Briggs, E.; Crouch, K.; Lemgruber, L.; Lapsley, C.; McCulloch, R. Ribonuclease H1-targeted R-loops in surface antigen gene expression sites can direct trypanosome immune evasion. PLoS Genet. 2018, 14, e1007729. [Google Scholar] [CrossRef]
- Briggs, E.; Crouch, K.; Lemgruber, L.; Hamilton, G.; Lapsley, C.; McCulloch, R. Trypanosoma brucei ribonuclease H2A is an essential R-loop processing enzyme whose loss causes DNA damage during transcription initiation and antigenic variation. Nucleic Acids Res. 2019, 47, 9180–9197. [Google Scholar] [CrossRef]
- Da Silva, M.S.; Hovel-Miner, G.A.; Briggs, E.M.; Elias, M.C.; McCulloch, R. Evaluation of mechanisms that may generate DNA lesions triggering antigenic variation in African trypanosomes. PLoS Pathog. 2018, 14, e1007321. [Google Scholar] [CrossRef]
- McCulloch, R.; Cobbold, C.A.; Figueiredo, L.; Jackson, A.; Morrison, L.J.; Mugnier, M.R.; Papavasiliou, N.; Schnaufer, A.; Matthews, K. Emerging challenges in understanding trypanosome antigenic variation. Emerg. Top. Life Sci. 2017, 1, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Ridewood, S.; Ooi, C.; Hall, B.; Trenaman, A.; Wand, N.V.; Sioutas, G.; Scherwitzl, I.; Rudenko, G. The role of genomic location and flanking 3′UTR in the generation of functional levels of variant surface glycoprotein in Trypanosoma brucei. Mol. Microbiol. 2017, 106, 614–634. [Google Scholar] [CrossRef]
- Haber, J.E. Partners and pathwaysrepairing a double-strand break. Trends Genet. 2000, 16, 259–264. [Google Scholar] [CrossRef]
- Saha, A.; Gaurav, A.K.; Pandya, U.M.; Afrin, M.; Sandhu, R.; Nanavaty, V.; Schnur, B.; Li, B. TbTRF suppresses the TERRA level and regulates the cell cycle-dependent TERRA foci number with a TERRA binding activity in its C-terminal Myb domain. Nucleic Acids Res. 2021, 49, 5637–5653. [Google Scholar] [CrossRef] [PubMed]
- Podlevsky, J.D.; Bley, C.J.; Omana, R.V.; Qi, X.; Chen, J.J.-L. The Telomerase Database. Nucleic Acids Res. 2007, 36, D339–D343. [Google Scholar] [CrossRef] [PubMed]
- Henderson, E.R.; Blackburn, E.H. An overhanging 3’ terminus is a conserved feature of telomeres. Mol. Cell Biol. 1989, 9, 345–348. [Google Scholar] [CrossRef] [PubMed]
- Wellinger, R.J.; Wolf, A.J.; Zakian, V.A. Saccharomyces telomeres acquire single-strand TG1–3 tails late in S phase. Cell 1993, 72, 51–60. [Google Scholar] [CrossRef]
- Makarov, V.L.; Hirose, Y.; Langmore, J.P. Long G Tails at Both Ends of Human Chromosomes Suggest a C Strand Degradation Mechanism for Telomere Shortening. Cell 1997, 88, 657–666. [Google Scholar] [CrossRef]
- Dionne, I.; Wellinger, R.J. Cell cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase. Proc. Natl. Acad. Sci. USA 1996, 93, 13902–13907. [Google Scholar] [CrossRef]
- Jacob, N.K.; Skopp, R.; Price, C.M. G-overhang dynamics at Tetrahymena telomeres. EMBO J. 2001, 20, 4299–4308. [Google Scholar] [CrossRef]
- Chai, W.; Du, Q.; Shay, J.W.; Wright, W.E. Human Telomeres Have Different Overhang Sizes at Leading versus Lagging Strands. Mol. Cell 2006, 21, 427–435. [Google Scholar] [CrossRef]
- Tomita, K.; Kibe, T.; Kang, H.-Y.; Seo, Y.-S.; Uritani, M.; Ushimaru, T.; Ueno, M. Fission Yeast Dna2 Is Required for Generation of the Telomeric Single-Strand Overhang. Mol. Cell. Biol. 2004, 24, 9557–9567. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, R.; Li, B. Examination of the Telomere G-overhang Structure in Trypanosoma brucei. J. Vis. Exp. 2011, 47, e1959. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, R.; Li, B. Telomerase activity is required for the telomere G-overhang structure in Trypanosoma brucei. Sci. Rep. 2017, 7, 15983. [Google Scholar] [CrossRef] [PubMed]
- Griffith, J.D.; Comeau, L.; Rosenfield, S.; Stansel, R.M.; Bianchi, A.; Moss, H.; de Lange, T. Mammalian Telomeres End in a Large Duplex Loop. Cell 1999, 97, 503–514. [Google Scholar] [CrossRef]
- Murti, K.G.; Prescott, D.M. Telomeres of polytene chromosomes in a ciliated protozoan terminate in duplex DNA loops. Proc. Natl. Acad. Sci. USA 1999, 96, 14436–14439. [Google Scholar] [CrossRef]
- Nikitina, T.; Woodcock, C.L. Closed chromatin loops at the ends of chromosomes. J. Cell Biol. 2004, 166, 161–165. [Google Scholar] [CrossRef]
- Muñoz-Jordán, J.L.; Cross, G.; De Lange, T.; Griffith, J.D. t-loops at trypanosome telomeres. EMBO J. 2001, 20, 579–588. [Google Scholar] [CrossRef]
- Cesare, A.J.; Quinney, N.; Willcox, S.; Subramanian, D.; Griffith, J.D. Telomere looping in P. sativum (common garden pea). Plant J. 2003, 36, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Raices, M.; Verdun, R.E.; Compton, S.A.; Haggblom, C.I.; Griffith, J.D.; Dillin, A.; Karlseder, J. C. elegans Telomeres Contain G-Strand and C-Strand Overhangs that Are Bound by Distinct Proteins. Cell 2008, 132, 745–757. [Google Scholar] [CrossRef]
- Cesare, A.J.; Groff-Vindman, C.; Compton, S.A.; McEachern, M.J.; Griffith, J.D. Telomere Loops and Homologous Recombination-Dependent Telomeric Circles in a Kluyveromyces lactis Telomere Mutant Strain. Mol. Cell. Biol. 2008, 28, 20–29. [Google Scholar] [CrossRef]
- Wynford-Thomas, D.; Kipling, D. Telomerase. Cancer and the knockout mouse. Nature 1997, 389, 551–552. [Google Scholar] [PubMed]
- Autexier, C.; Lue, N.F. The Structure and Function of Telomerase Reverse Transcriptase. Annu. Rev. Biochem. 2006, 75, 493–517. [Google Scholar] [CrossRef] [PubMed]
- Greider, C.W.; Blackburn, E.H. Identification of a specific telomere terminal transferase activity in tetrahymena extracts. Cell 1985, 43, 405–413. [Google Scholar] [CrossRef]
- Greider, C.; Blackburn, E.H. The telomere terminal transferase of tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell 1987, 51, 887–898. [Google Scholar] [CrossRef]
- Greider, C.; Blackburn, E.H. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nat. Cell Biol. 1989, 337, 331–337. [Google Scholar] [CrossRef]
- Sobinoff, A.; Pickett, H.A. Alternative Lengthening of Telomeres: DNA Repair Pathways Converge. Trends Genet. 2017, 33, 921–932. [Google Scholar] [CrossRef]
- Sobinoff, A.; Pickett, H.A. Mechanisms that drive telomere maintenance and recombination in human cancers. Curr. Opin. Genet. Dev. 2020, 60, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Tang, H.-B.; Liu, N.-N.; Tong, X.-J.; Dang, W.; Duan, Y.-M.; Fu, X.-H.; Zhang, Y.; Peng, J.; Meng, F.; et al. Telomerase-Null Survivor Screening Identifies Novel Telomere Recombination Regulators. PLoS Genet. 2013, 9, e1003208. [Google Scholar] [CrossRef]
- Lundblad, V. Telomere maintenance without telomerase. Oncogene 2002, 21, 522–531. [Google Scholar] [CrossRef]
- De Lange, T. Shelterin-Mediated Telomere Protection. Annu. Rev. Genet. 2018, 52, 223–247. [Google Scholar] [CrossRef]
- Maciejowski, J.; De Lange, T. Telomeres in cancer: Tumour suppression and genome instability. Nat. Rev. Mol. Cell Biol. 2017, 18, 175–186. [Google Scholar] [CrossRef]
- Maciejowski, J.; Li, Y.; Bosco, N.; Campbell, P.J.; de Lange, T. Chromothripsis and Kataegis Induced by Telomere Crisis. Cell 2015, 163, 1641–1654. [Google Scholar] [CrossRef]
- Pellestor, F.; Gaillard, J.; Schneider, A.; Puechberty, J.; Gatinois, V. Chromoanagenesis, the mechanisms of a genomic chaos. Semin. Cell Dev. Biol. 2021, 1084. [Google Scholar] [CrossRef]
- Cleal, K.; Baird, D.M. Catastrophic Endgames: Emerging Mechanisms of Telomere-Driven Genomic Instability. Trends Genet. 2020, 36, 347–359. [Google Scholar] [CrossRef]
- De Lange, T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev. 2005, 19, 2100–2110. [Google Scholar] [CrossRef] [PubMed]
- Chong, L.; Van Steensel, B.; Broccoli, D.; Erdjument-Bromage, H.; Hanish, J.; Tempst, P.; De Lange, T. A Human Telomeric Protein. Science 1995, 270, 1663–1667. [Google Scholar] [CrossRef] [PubMed]
- Broccoli, D.; Smogorzewska, A.; Chong, L.; de Lange, T. Human telomeres contain two distinct Myb–related proteins, TRF1 and TRF1 and TRF2. Nat. Genet. 1997, 17, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Bilaud, T.; Brun, C.; Ancelin, K.; Koering, C.E.; Laroche, T.; Gilson, E. Telomeric localization of TRF2, a novel human telobox protein. Nat. Genet. 1997, 17, 236–239. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Oestreich, S.; de Lange, T. Identification of Human Rap1: Implications for Telomere Evolution. Cell 2000, 101, 471–483. [Google Scholar] [CrossRef]
- Kim, S.-H.; Kaminker, P.; Campisi, J. TIN2, a new regulator of telomere length in human cells. Nat. Genet. 1999, 23, 405–412. [Google Scholar] [CrossRef]
- Houghtaling, B.R.; Cuttonaro, L.; Chang, W.; Smith, S. A Dynamic Molecular Link between the Telomere Length Regulator TRF1 and the Chromosome End Protector TRF2. Curr. Biol. 2004, 14, 1621–1631. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Safari, A.; O’Connor, M.S.; Chan, D.W.; Laegeler, A.; Qin, J.; Songyang, Z. PTOP interacts with POT1 and regulates its localization to telomeres. Nat. Cell Biol. 2004, 6, 673–680. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.Z.-S.; Hockemeyer, D.; Krutchinsky, A.N.; Loayza, D.; Hooper, S.M.; Chait, B.T.; De Lange, T. POT1-interacting protein PIP1: A telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 2004, 18, 1649–1654. [Google Scholar] [CrossRef]
- Baumann, P.; Cech, T.R. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 2001, 292, 1171–1175. [Google Scholar] [CrossRef] [PubMed]
- Loayza, D.; De Lange, T. POT1 as a terminal transducer of TRF1 telomere length control. Nature 2003, 424, 1013–1018. [Google Scholar] [CrossRef]
- Surovtseva, Y.V.; Churikov, D.; Boltz, K.A.; Song, X.; Lamb, J.C.; Warrington, R.; Leehy, K.; Heacock, M.; Price, C.M.; Shippen, D.E. Conserved Telomere Maintenance Component 1 Interacts with STN1 and Maintains Chromosome Ends in Higher Eukaryotes. Mol. Cell 2009, 36, 207–218. [Google Scholar] [CrossRef]
- Feng, X.; Hsu, S.-J.; Bhattacharjee, A.; Wang, Y.; Diao, J.; Price, C.M. CTC1-STN1 terminates telomerase while STN1-TEN1 enables C-strand synthesis during telomere replication in colon cancer cells. Nat. Commun. 2018, 9, 2827. [Google Scholar] [CrossRef]
- Zhong, Z.; Shiue, L.; Kaplan, S.; De Lange, T. A mammalian factor that binds telomeric TTAGGG repeats in vitro. Mol. Cell Biol. 1992, 12, 4834–4843. [Google Scholar] [CrossRef]
- Bianchi, A.; Smith, S.; Chong, L.; Elias, P.; De Lange, T. TRF1 is a dimer and bends telomeric DNA. EMBO J. 1997, 16, 1785–1794. [Google Scholar] [CrossRef]
- Bianchi, A.; Stansel, R.M.; Fairall, L.; Griffith, J.D.; Rhodes, D.; de Lange, T. TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J. 1999, 18, 5735–5744. [Google Scholar] [CrossRef]
- Hanaoka, S.; Nagadoi, A.; Nishimura, Y. Comparison between TRF2 and TRF1 of their telomeric DNA-bound structures and DNA-binding activities. Protein Sci. 2009, 14, 119–130. [Google Scholar] [CrossRef]
- Loayza, D.; Parsons, H.; Donigian, J.; Hoke, K.; De Lange, T. DNA binding features of human POT1: A nonamer 5’-TAGGGTTAG-3’ minimal binding site, sequence specificity, and internal binding to multimeric sites. J. Biol. Chem. 2004, 279, 13241–13248. [Google Scholar] [CrossRef] [PubMed]
- Rice, C.; Skordalakes, E. Structure and function of the telomeric CST complex. Comput. Struct. Biotechnol. J. 2016, 14, 161–167. [Google Scholar] [CrossRef]
- Ye, J.Z.-S.; Donigian, J.R.; van Overbeek, M.; Loayza, D.; Luo, Y.; Krutchinsky, A.N.; Chait, B.T.; de Lange, T. TIN2 Binds TRF1 and TRF2 Simultaneously and Stabilizes the TRF2 Complex on Telomeres. J. Biol. Chem. 2004, 279, 47264–47271. [Google Scholar] [CrossRef] [PubMed]
- Smogorzewska, A.; Karlseder, J.; Holtgreve-Grez, H.; Jauch, A.; de Lange, T. DNA Ligase IV-Dependent NHEJ of Deprotected Mammalian Telomeres in G1 and G2. Curr. Biol. 2002, 12, 1635–1644. [Google Scholar] [CrossRef]
- Celli, G.B.; De Lange, T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 2005, 7, 712–718. [Google Scholar] [CrossRef]
- Deng, Y.; Guo, X.; Ferguson, D.O.; Chang, S. Multiple roles for MRE11 at uncapped telomeres. Nat. Cell Biol. 2009, 460, 914–918. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.C.; Smogorzewska, A.; de Lange, T. Homologous Recombination Generates T-Loop-Sized Deletions at Human Telomeres. Cell 2004, 119, 355–368. [Google Scholar] [CrossRef] [PubMed]
- Sfeir, A.; Kabir, S.; Van Overbeek, M.; Celli, G.B.; De Lange, T. Loss of Rap1 Induces Telomere Recombination in the Absence of NHEJ or a DNA Damage Signal. Science 2010, 327, 1657–1661. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Rai, R.; Zhou, Z.-R.; Kanoh, J.; Ribeyre, C.; Yang, Y.; Zheng, H.; Damay, P.; Wang, F.; Tsujii, H.; et al. A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms. Nat. Struct. Mol. Biol. 2011, 18, 213–221. [Google Scholar] [CrossRef]
- Guo, X.; Deng, Y.; Lin, Y.; Cosme-Blanco, W.; Chan, S.; He, H.; Yuan, G.; Brown, E.J.; Chang, S. Dysfunctional telomeres activate an ATM-ATR-dependent DNA damage response to suppress tumorigenesis. EMBO J. 2007, 26, 4709–4719. [Google Scholar] [CrossRef] [PubMed]
- Rai, R.; Zheng, H.; He, H.; Luo, Y.; Multani, A.; Carpenter, P.B.; Chang, S. The function of classical and alternative non-homologous end-joining pathways in the fusion of dysfunctional telomeres. EMBO J. 2010, 29, 2598–2610. [Google Scholar] [CrossRef]
- Sfeir, A.; De Lange, T. Removal of shelterin reveals the telomere end-protection problem. Science 2012, 336, 593–597. [Google Scholar] [CrossRef] [PubMed]
- Van Ly, D.; Low, R.R.J.; Frölich, S.; Bartolec, T.; Kafer, G.; Pickett, H.A.; Gaus, K.; Cesare, A.J. Telomere Loop Dynamics in Chromosome End Protection. Mol. Cell 2018, 71, 510–525.e6. [Google Scholar] [CrossRef]
- Ottaviani, A.; Gilson, E.; Magdinier, F. Telomeric position effect: From the yeast paradigm to human pathologies? Biochimie 2008, 90, 93–107. [Google Scholar] [CrossRef]
- O’Kane, C.J.; Hyland, E.M. Yeast epigenetics: The inheritance of histone modification states. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Longtine, M.S.; Wilson, N.M.; Petracek, M.E.; Berman, J. A yeast telomere binding activity binds to two related telomere sequence motifs and is indistinguishable from RAPT. Curr. Genet. 1989, 16, 225–239. [Google Scholar] [CrossRef]
- König, P.; Giraldo, R.; Chapman, L.; Rhodes, D. The Crystal Structure of the DNA-Binding Domain of Yeast RAP1 in Complex with Telomeric DNA. Cell 1996, 85, 125–136. [Google Scholar] [CrossRef]
- König, P.; Rhodes, D. Recognition of telomeric DNA. Trends Biochem. Sci. 1997, 22, 43–47. [Google Scholar] [CrossRef]
- Luo, K.; Vega-Palas, M.A.; Grunstein, M. Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev. 2002, 16, 1528–1539. [Google Scholar] [CrossRef] [PubMed]
- Moretti, P.; Shore, D. Multiple Interactions in Sir Protein Recruitment by Rap1p at Silencers and Telomeres in Yeast. Mol. Cell. Biol. 2001, 21, 8082–8094. [Google Scholar] [CrossRef]
- Cockell, M.; Palladino, F.; Laroche, T.; Kyrion, G.; Liu, C.; Lustig, A.J.; Gasser, S. The carboxy termini of Sir4 and Rap1 affect Sir3 localization: Evidence for a multicomponent complex required for yeast telomeric silencing. J. Cell Biol. 1995, 129, 909–924. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Lustig, A.J. Genetic Analysis of Rap1p/Sir3p Interactions in Telomeric and HML Silencing in Saccharomyces cerevisiae. Genetics 1996, 143, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Lustig, A.J.; Liu, C.; Zhang, C.; Hanish, J.P. Tethered Sir3p nucleates silencing at telomeres and internal loci in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996, 16, 2483–2495. [Google Scholar] [CrossRef][Green Version]
- Smith, J.S.; Brachmann, C.B.; Celic, I.; Kenna, M.A.; Muhammad, S.; Starai, V.J.; Avalos, J.L.; Escalante-Semerena, J.C.; Grubmeyer, C.; Wolberger, C.; et al. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. USA 2000, 97, 6658–6663. [Google Scholar] [CrossRef]
- Gartenberg, M.; Smith, J.S. The Nuts and Bolts of Transcriptionally Silent Chromatin in Saccharomyces cerevisiae. Genetics 2016, 203, 1563–1599. [Google Scholar] [CrossRef]
- Kyrion, G.; Liu, K.; Liu, C.; Lustig, A.J. RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev. 1993, 7, 1146–1159. [Google Scholar] [CrossRef] [PubMed]
- Buck, S.W.; Shore, D. Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev. 1995, 9, 370–384. [Google Scholar] [CrossRef]
- Rudenko, G.; Van Der Ploeg, L. Transcription of telomere repeats in protozoa. EMBO J. 1989, 8, 2633–2638. [Google Scholar] [CrossRef]
- Damasceno, J.D.; La Silva, G.; Tschudi, C.; Tosi, L.R. Evidence for regulated expression of Telomeric Repeat-containing RNAs (TERRA) in parasitic trypanosomatids. Memórias Do Inst. Oswaldo Cruz 2017, 112, 572–576. [Google Scholar] [CrossRef]
- Saha, A.; Nanavaty, V.P.; Li, B. Telomere and Subtelomere R-loops and Antigenic Variation in Trypanosomes. J. Mol. Biol. 2020, 432, 4167–4185. [Google Scholar] [CrossRef]
- Morea, E.G.O.; Vasconcelos, E.J.R.; de Santis Alves, C.; Giorgio, S.; Myler, P.J.; Langoni, H.; Azzalin, C.M.; Cano, M.I.N. Exploring TERRA during Leishmania major developmental cycle and continuous in vitro passages. Int. J. Biol. Macromol. 2021, 174, 573–586. [Google Scholar] [CrossRef]
- Azzalin, C.M.; Reichenbach, P.; Khoriauli, L.; Giulotto, E.; Lingner, J. Telomeric Repeat Containing RNA and RNA Surveillance Factors at Mammalian Chromosome Ends. Science 2007, 318, 798–801. [Google Scholar] [CrossRef]
- Schoeftner, S.; Blasco, M.A. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 2007, 10, 228–236. [Google Scholar] [CrossRef]
- Bah, A.; Wischnewski, H.; Shchepachev, V.; Azzalin, C.M. The telomeric transcriptome of Schizosaccharomyces pombe. Nucleic Acids Res. 2011, 40, 2995–3005. [Google Scholar] [CrossRef]
- Luke, B.; Panza, A.; Redon, S.; Iglesias, N.; Li, Z.; Lingner, J. The Rat1p 5′ to 3′ Exonuclease Degrades Telomeric Repeat-Containing RNA and Promotes Telomere Elongation in Saccharomyces cerevisiae. Mol. Cell 2008, 32, 465–477. [Google Scholar] [CrossRef]
- Solovei, I.; Gaginskaya, E.; MacGregor, H.C. The arrangement and transcription of telomere DNA sequences at the ends of lampbrush chromosomes of birds. Chromosom. Res. 1994, 2, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Toubiana, S.; Selig, S. DNA: RNA hybrids at telomeres–When it is better to be out of the (R) loop. FEBS J. 2018, 285, 2552–2566. [Google Scholar] [CrossRef] [PubMed]
- Li, B. Keeping Balance Between Genetic Stability and Plasticity at the Telomere and Subtelomere of Trypanosoma brucei. Front. Cell Dev. Biol. 2021, 9, 1722. [Google Scholar] [CrossRef] [PubMed]
- Nergadze, S.G.; Farnung, B.O.; Wischnewski, H.; Khoriauli, L.; Vitelli, V.; Chawla, R.; Giulotto, E.; Azzalin, C.M. CpG-island promoters drive transcription of human telomeres. RNA 2009, 15, 2186–2194. [Google Scholar] [CrossRef]
- Iglesias, N.; Redon, S.; Pfeiffer, V.; Dees, M.; Lingner, J.; Luke, B. Subtelomeric repetitive elements determine TERRA regulation by Rap1/Rif and Rap1/Sir complexes in yeast. EMBO Rep. 2011, 12, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Porro, A.; Feuerhahn, S.; Delafontaine, J.; Riethman, H.; Rougemont, J.; Lingner, J. Functional characterization of the TERRA transcriptome at damaged telomeres. Nat. Commun. 2014, 5, 1–13. [Google Scholar] [CrossRef]
- Feretzaki, M.; Nunes, P.R.; Lingner, J. Expression and differential regulation of human TERRA at several chromosome ends. RNA 2019, 25, 1470–1480. [Google Scholar] [CrossRef]
- Deng, Z.; Wang, Z.; Stong, N.; Plasschaert, R.; Moczan, A.; Chen, H.-S.; Hu, S.; Wikramasinghe, P.; Davuluri, R.V.; Bartolomei, M.S.; et al. A role for CTCF and cohesin in subtelomere chromatin organization, TERRA transcription, and telomere end protection. EMBO J. 2012, 31, 4165–4178. [Google Scholar] [CrossRef]
- Feretzaki, M.; Lingner, J. A practical qPCR approach to detect TERRA, the elusive telomeric repeat-containing RNA. Methods 2017, 114, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Arora, R.; Brun, C.M.; Azzalin, C.M. Transcription regulates telomere dynamics in human cancer cells. RNA 2012, 18, 684–693. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Peška, V.; Schrumpfová, P.P.; Fajkus, J. Using the telobox to search for plant telomere binding proteins. Curr. Protein Pept. Sci. 2011, 12, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Lue, N.F. Duplex Telomere-Binding Proteins in Fungi with Canonical Telomere Repeats: New Lessons in the Rapid Evolution of Telomere Proteins. Front. Genet. 2021, 12, 238. [Google Scholar] [CrossRef]
- Horvath, M.P. Structural anatomy of telomere OB proteins. Crit. Rev. Biochem. Mol. Biol. 2011, 46, 409–435. [Google Scholar] [CrossRef]
- Amir, M.; Khan, P.; Queen, A.; Dohare, R.; Alajmi, M.F.; Hussain, A.; Islam, A.; Ahmad, F.; Hassan, I. Structural Features of Nucleoprotein CST/Shelterin Complex Involved in the Telomere Maintenance and Its Association with Disease Mutations. Cells 2020, 9, 359. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K.A.; Wuttke, D.S. Telomerase and Telomere-Associated Proteins: Structural Insights into Mechanism and Evolution. Structure 2012, 20, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Ogata, K.; Morikawa, S.; Nakamura, H.; Sekikawa, A.; Inoue, T.; Kanai, H.; Sarai, A.; Ishii, S.; Nishimura, Y. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell 1994, 79, 639–648. [Google Scholar] [CrossRef]
- Van Steensel, B.; de Lange, T. Control of telomere length by the human telomeric protein TRF1. Nat. Cell Biol. 1997, 385, 740–743. [Google Scholar] [CrossRef]
- Van Steensel, B.; Smogorzewska, A.; de Lange, T. TRF2 Protects Human Telomeres from End-to-End Fusions. Cell 1998, 92, 401–413. [Google Scholar] [CrossRef]
- Cooper, J.P.; Nimmo, E.R.; Allshire, R.; Cech, T.R. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nat. Cell Biol. 1997, 385, 744–747. [Google Scholar] [CrossRef]
- Spink, K.G.; Evans, R.J.; Chambers, A. Sequence-specific binding of Taz1p dimers to fission yeast telomeric DNA. Nucleic Acids Res 2000, 28, 527–533. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Koering, C.E.; Fourel, G.; Binet-Brasselet, E.; Laroche, T.; Klein, F.; Gilson, E. Identification of high affinity Tbf1p-binding sites within the budding yeast genome. Nucleic Acids Res. 2000, 28, 2519–2526. [Google Scholar] [CrossRef] [PubMed]
- Berthiau, A.-S.; Yankulov, K.; Bah, A.; Revardel, E.; Luciano, P.; Wellinger, R.J.; Géli, V.; Gilson, E. Subtelomeric proteins negatively regulate telomere elongation in budding yeast. EMBO J. 2006, 25, 846–856. [Google Scholar] [CrossRef]
- Gottschling, D.E.; Zakian, V.A. Telomere proteins: Specific recognition and protection of the natural termini of Oxytricha macronuclear DNA. Cell 1986, 47, 195–205. [Google Scholar] [CrossRef]
- Gray, J.T.; Celander, D.W.; Price, C.; Cech, T.R. Cloning and expression of genes for the Oxytricha telomere-binding protein: Specific subunit interactions in the telomeric complex. Cell 1991, 67, 807–814. [Google Scholar] [CrossRef]
- Horvath, M.P.; Schweiker, V.L.; Bevilacqua, J.M.; Ruggles, J.A.; Schultz, S.C. Crystal Structure of the Oxytricha nova Telomere End Binding Protein Complexed with Single Strand DNA. Cell 1998, 95, 963–974. [Google Scholar] [CrossRef]
- Classen, S.; Ruggles, J.A.; Schultz, S.C. Crystal structure of the N-terminal domain of Oxytricha nova telomere end-binding protein α subunit both uncomplexed and complexed with telomeric ssDNA. J. Mol. Biol. 2001, 314, 1113–1125. [Google Scholar] [CrossRef]
- Buczek, P.; Horvath, M.P. Structural Reorganization and the Cooperative Binding of Single-stranded Telomere DNA in Sterkiella nova. J. Biol. Chem. 2006, 281, 40124–40134. [Google Scholar] [CrossRef] [PubMed]
- Lei, M.; Podell, E.R.; Cech, T.R. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 2004, 11, 1223–1229. [Google Scholar] [CrossRef]
- Nandakumar, J.; Podell, E.R.; Cech, T.R. How telomeric protein POT1 avoids RNA to achieve specificity for single-stranded DNA. Proc. Natl. Acad. Sci. USA 2010, 107, 651–656. [Google Scholar] [CrossRef] [PubMed]
- Hockemeyer, D.; Palm, W.; Else, T.; Daniels, J.-P.; Takai, K.K.; Ye, J.Z.-S.; Keegan, C.E.; de Lange, T.; Hammer, G.D. Telomere protection by mammalian Pot1 requires interaction with Tpp1. Nat. Struct. Mol. Biol. 2007, 14, 754–761. [Google Scholar] [CrossRef]
- Xin, H.; Liu, D.; Wan, M.; Safari, A.; Kim, H.; Sun, W.; O’Connor, M.S.; Songyang, Z. TPP1 is a homologue of ciliate TEBP-β and interacts with POT1 to recruit telomerase. Nat. Cell Biol. 2007, 445, 559–562. [Google Scholar] [CrossRef]
- Wang, F.; Podell, E.R.; Zaug, A.J.; Yang, Y.; Baciu, P.; Cech, T.R.; Lei, M. The POT1–TPP1 telomere complex is a telomerase processivity factor. Nat. Cell Biol. 2007, 445, 506–510. [Google Scholar] [CrossRef]
- Gao, H.; Cervantes, R.B.; Mandell, E.K.; Otero, J.H.; Lundblad, V. RPA-like proteins mediate yeast telomere function. Nat. Struct. Mol. Biol. 2007, 14, 208–214. [Google Scholar] [CrossRef]
- Sun, J.; Yu, E.Y.; Yang, Y.; Confer, L.A.; Sun, S.H.; Wan, K.; Lue, N.F.; Lei, M. Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres. Genes Dev. 2009, 23, 2900–2914. [Google Scholar] [CrossRef]
- Bryan, C.; Rice, C.; Harkisheimer, M.; Schultz, D.C.; Skordalakes, E. Structure of the Human Telomeric Stn1-Ten1 Capping Complex. PLoS ONE 2013, 8, e66756. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.M.; Halsey, W.A.; Wuttke, D.S. Delineation of the high-affinity single-stranded telomeric DNA-binding domain of Saccharomyces cerevisiae Cdc13. Nucleic Acids Res. 2002, 30, 4305–4313. [Google Scholar] [CrossRef]
- Eldridge, A.M.; Wuttke, D.S. Probing the mechanism of recognition of ssDNA by the Cdc13-DBD. Nucleic Acids Res. 2008, 36, 1624–1633. [Google Scholar] [CrossRef] [PubMed]
- Mitton-Fry, R.M.; Anderson, E.M.; Hughes, T.R.; Lundblad, V.; Wuttke, D.S. Conserved Structure for Single-Stranded Telomeric DNA Recognition. Science 2002, 296, 145–147. [Google Scholar] [CrossRef] [PubMed]
- Mitton-Fry, R.M.; Anderson, E.M.; Theobald, D.L.; Glustrom, L.W.; Wuttke, D.S. Structural Basis for Telomeric Single-stranded DNA Recognition by Yeast Cdc13. J. Mol. Biol. 2004, 338, 241–255. [Google Scholar] [CrossRef]
- Theobald, D.; Wuttke, D.S. Prediction of Multiple Tandem OB-Fold Domains in Telomere End-Binding Proteins Pot1 and Cdc13. Structure 2004, 12, 1877–1879. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Yang, Y.; Wan, K.; Mao, N.; Yu, T.-Y.; Lin, Y.-C.; DeZwaan, D.C.; Freeman, B.C.; Lin, J.-J.; Lue, N.F.; et al. Structural bases of dimerization of yeast telomere protein Cdc13 and its interaction with the catalytic subunit of DNA polymerase α. Cell Res. 2010, 21, 258–274. [Google Scholar] [CrossRef]
- Yu, E.Y.; Sun, J.; Lei, M.; Lue, N.F. Analyses of Candida Cdc13 Orthologues Revealed a Novel OB Fold Dimer Arrangement, Dimerization-Assisted DNA Binding, and Substantial Structural Differences between Cdc13 and RPA. Mol. Cell. Biol. 2012, 32, 186–198. [Google Scholar] [CrossRef]
- Bhattacharjee, A.; Stewart, J.; Chaiken, M.; Price, C.M. STN1 OB Fold Mutation Alters DNA Binding and Affects Selective Aspects of CST Function. PLoS Genet. 2016, 12, e1006342. [Google Scholar] [CrossRef]
- Bhattacharjee, A.; Wang, Y.; Diao, J.; Price, C.M. Dynamic DNA binding, junction recognition and G4 melting activity underlie the telomeric and genome-wide roles of human CST. Nucleic Acids Res. 2017, 45, 12311–12324. [Google Scholar] [CrossRef]
- Hom, R.A.; Wuttke, D.S. Human CST Prefers G-Rich but Not Necessarily Telomeric Sequences. Biochemistry 2017, 56, 4210–4218. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Espinal, A.; Cross, G.A.M. Trypanosome Telomeres Are Protected by a Homologue of Mammalian TRF. Mol. Cell. Biol. 2005, 25, 5011–5021. [Google Scholar] [CrossRef]
- Reis, H.; Schwebs, M.; Dietz, S.; Janzen, C.J.; Butter, F. TelAP1 links telomere complexes with developmental expression site silencing in African trypanosomes. Nucleic Acids Res. 2018, 46, 2820–2833. [Google Scholar] [CrossRef]
- Dreesen, O.; Li, B.; Cross, G.A.M. Telomere structure and shortening in telomerase-deficient Trypanosoma brucei. Nucleic Acids Res. 2005, 33, 4536–4543. [Google Scholar] [CrossRef]
- Sandhu, R.; Sanford, S.; Basu, S.; Park, M.; Pandya, U.M.; Li, B.; Chakrabarti, K. A trans-spliced telomerase RNA dictates telomere synthesis in Trypanosoma brucei. Cell Res. 2013, 23, 537–551. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Kolet, L.; Doniger, T.; Biswas, V.K.; Unger, R.; Tzfati, Y.; Michaeli, S. The Trypanosoma brucei telomerase RNA (TER) homologue binds core proteins of the C/D snoRNA family. FEBS Lett. 2013, 587, 1399–1404. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, M.S.; Perez, A.M.; Rita de Cássia, V.; de Moraes, C.E.; Siqueira-Neto, J.L.; Freitas-Junior, L.H.; Cano, M.I.N. The Leishmania amazonensis TRF (TTAGGG repeat-binding factor) homologue binds and co-localizes with telomeres. BMC Microbiol. 2010, 10, 136. [Google Scholar] [CrossRef] [PubMed]
- Fairall, L.; Chapman, L.; Moss, H.; de Lange, T.; Rhodes, D. Structure of the TRFH Dimerization Domain of the Human Telomeric Proteins TRF1 and TRF2. Mol. Cell 2001, 8, 351–361. [Google Scholar] [CrossRef]
- Crossley, M.P.; Bocek, M.; Cimprich, K.A. R-Loops as Cellular Regulators and Genomic Threats. Mol. Cell 2019, 73, 398–411. [Google Scholar] [CrossRef] [PubMed]
- Hegazy, Y.A.; Fernando, C.M.; Tran, E.J. The balancing act of R-loop biology: The good, the bad, and the ugly. J. Biol. Chem. 2020, 295, 905–913. [Google Scholar] [CrossRef]
- Brambati, A.; Zardoni, L.; Nardini, E.; Pellicioli, A.; Liberi, G. The dark side of RNA: DNA hybrids. Mutat. Res. Rev. Mutat. Res. 2020, 784, 108300. [Google Scholar] [CrossRef]
- König, P.; Fairall, L.; Rhodes, D. Sequence-specific DNA recognition by the Myb-like domain of the human telomere binding protein TRF1: A model for the protein-DNA complex. Nucleic Acids Res. 1998, 26, 1731–1740. [Google Scholar] [CrossRef]
- Nishikawa, T.; Nagadoi, A.; Yoshimura, S.; Aimoto, S.; Nishimura, Y. Solution structure of the DNA-binding domain of human telomeric protein, hTRF1. Structure 1998, 6, 1057–1065. [Google Scholar] [CrossRef]
- Court, R.; Chapman, L.; Fairall, L.; Rhodes, D. How the human telomeric proteins TRF1 and TRF2 recognize telomeric DNA: A view from high-resolution crystal structures. EMBO Rep. 2005, 6, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Borst, P.; Sabatini, R. Base J: Discovery, Biosynthesis, and Possible Functions. Annu. Rev. Microbiol. 2008, 62, 235–251. [Google Scholar] [CrossRef] [PubMed]
- Gommers-Ampt, J.H.; Van Leeuwen, F.; De Beer, A.L.; Vliegenthart, J.F.; Dizdaroglu, M.; Kowalak, J.A.; Crain, P.F.; Borst, P. β-d-glucosyl-hydroxymethyluracil: A novel modified base present in the DNA of the parasitic protozoan T. brucei. Cell 1993, 75, 1129–1136. [Google Scholar] [CrossRef]
- Van Leeuwen, F.; Wijsman, E.R.; Kuyl-Yeheskiely, E.; Van Der Marel, G.A.; Van Boom, J.H.; Borst, P. The telomeric GGGTTA repeats of Trypanosoma brucei contain the hypermodified base J in both strands. Nucleic Acids Res. 1996, 24, 2476–2482. [Google Scholar] [CrossRef]
- Van Leeuwen, F.; Wijsman, E.R.; Kieft, R.; Van Der Marel, G.A.; Van Boom, J.H.; Borst, P. Localization of the modified base J in telomeric VSG gene expression sites of Trypanosoma brucei. Genes Dev. 1997, 11, 3232–3241. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Borst, P.; Van Leeuwen, F. ß-D-glucosyl-hydroxymethyluracil, a novel base in African trypanosomes and other Kinetoplastida. Mol. Biochem. Parasitol. 1997, 90, 1–8. [Google Scholar] [CrossRef]
- Van Leeuwen, F.; Taylor, M.C.; Mondragon, A.; Moreau, H.; Gibson, W.; Kieft, R.; Borst, P. ß-d-glucosyl-hydroxymethyluracil is a conserved DNA modification in kinetoplastid protozoans and is abundant in their telomeres. Proc. Natl. Acad. Sci. USA 1998, 95, 2366–2371. [Google Scholar] [CrossRef] [PubMed]
- Ekanayake, D.; Sabatini, R. Epigenetic Regulation of Polymerase II Transcription Initiation in Trypanosoma cruzi: Modulation of Nucleosome Abundance, Histone Modification, and Polymerase Occupancy by O-Linked Thymine DNA Glucosylation. Eukaryot. Cell 2011, 10, 1465–1472. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, D.; Cliffe, L.; Förstner, K.U.; Hon, C.-C.; Siegel, T.N.; Sabatini, R. Regulation of transcription termination by glucosylated hydroxymethyluracil, base J, in Leishmania major and Trypanosoma brucei. Nucleic Acids Res. 2014, 42, 9717–9729. [Google Scholar] [CrossRef]
- Reynolds, D.; Hofmeister, B.T.; Cliffe, L.; Alabady, M.; Siegel, T.N.; Schmitz, R.J.; Sabatini, R. Histone H3 Variant Regulates RNA Polymerase II Transcription Termination and Dual Strand Transcription of siRNA Loci in Trypanosoma brucei. PLoS Genet. 2016, 12, e1005758. [Google Scholar] [CrossRef]
- Deng, Z.; Norseen, J.; Wiedmer, A.; Riethman, H.; Lieberman, P.M. TERRA RNA Binding to TRF2 Facilitates Heterochromatin Formation and ORC Recruitment at Telomeres. Mol. Cell 2009, 35, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Deng, Z.; Vladimirova, O.; Gulve, N.; Johnson, F.B.; Drosopoulos, W.C.; Schildkraut, C.L.; Lieberman, P.M. TERRA G-quadruplex RNA interaction with TRF2 GAR domain is required for telomere integrity. Sci. Rep. 2021, 11, 3509. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.W.; Arora, R.; Wischnewski, H.; Azzalin, C.M. TRF1 participates in chromosome end protection by averting TRF2-dependent telomeric R loops. Nat. Struct. Mol. Biol. 2018, 25, 147–153. [Google Scholar] [CrossRef]
- Prouse, M.B.; Campbell, M. The interaction between MYB proteins and their target DNA binding sites. Biochim. Biophys. Acta (BBA) Bioenerg. 2012, 1819, 67–77. [Google Scholar] [CrossRef]
- Navarro, M.; Gull, K. A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nat. Cell Biol. 2001, 414, 759–763. [Google Scholar] [CrossRef]
- Liu, C.; Mao, X.; Lustig, A.J. Mutational analysis defines a C-terminal tail domain of RAP1 essential for Telomeric silencing in Saccharomyces cerevisiae. Genetics 1994, 138, 1025–1040. [Google Scholar] [CrossRef]
- Kanoh, J.; Ishikawa, F. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr. Biol. 2001, 11, 1624–1630. [Google Scholar] [CrossRef]
- Park, M.J.; Jang, Y.K.; Choi, E.S.; Kim, H.S.; Park, S.D. Fission yeast Rap1 homolog is a telomere-specific silencing factor and interacts with Taz1p. Mol. Cells 2002, 13, 327–333. [Google Scholar]
- Pena, A.C.; Pimentel, M.R.; Manso, H.; Vaz-Drago, R.; Pinto-Neves, D.; Aresta-Branco, F.; Rijo-Ferreira, F.; Guegan, F.; Coelho, L.P.; Carmo-Fonseca, M. Trypanosoma brucei histone H1 inhibits RNA polymerase I transcription and is important for parasite fitness in vivo. Mol. Microbiol. 2014, 93, 645–663. [Google Scholar] [CrossRef]
- Alsford, S.; Horn, D. Cell-cycle-regulated control of VSG expression site silencing by histones and histone chaperones ASF1A and CAF-1b in Trypanosoma brucei. Nucleic Acids Res. 2012, 40, 10150–10160. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, L.M.; Janzen, C.J.; Cross, G.A.M. A Histone Methyltransferase Modulates Antigenic Variation in African Trypanosomes. PLoS Biol. 2008, 6, e161. [Google Scholar] [CrossRef]
- Shore, D.; Nasmyth, K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell 1987, 51, 721–732. [Google Scholar] [CrossRef]
- Azad, G.K.; Tomar, R.S. The multifunctional transcription factor Rap1: A regulator of yeast physiology. Front. Biosci. 2016, 21, 918–930. [Google Scholar]
- Martinez, P.; Thanasoula, M.; Carlos, A.R.; López, G.G.; Tejera, A.M.; Schoeftner, S.; Dominguez, O.; Pisano, D.; Tarsounas, M.; Blasco, M.A. Mammalian Rap1 controls telomere function and gene expression through binding to telomeric and extratelomeric sites. Nat. Cell Biol. 2010, 12, 768–780. [Google Scholar] [CrossRef] [PubMed]
- Yeung, F.; Ramírez, C.M.; Mateos-Gomez, P.A.; Pinzaru, A.; Ceccarini, G.; Kabir, S.; Fernández-Hernando, C.; Sfeir, A. Nontelomeric Role for Rap1 in Regulating Metabolism and Protecting against Obesity. Cell Rep. 2013, 3, 1847–1856. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Xiong, Y.; Kim, H.; He, Q.; Li, Y.; Chen, R.; Songyang, Z. Human telomeric proteins occupy selective interstitial sites. Cell Res. 2011, 21, 1013–1027. [Google Scholar] [CrossRef]
- Teo, H.; Ghosh, S.; Luesch, H.; Ghosh, A.; Wong, E.T.; Malik, N.; Orth, A.; De Jesus, P.; Pery, A.S.; Oliver, J.D. Telomere-independent Rap1 is an IKK adaptor and regulates NF-kappaB-dependent gene expression. Nat. Cell Biol. 2010, 12, 758–767. [Google Scholar] [CrossRef] [PubMed]
- Kabir, S.; Sfeir, A.; De Lange, T. Taking apart Rap1: An adaptor protein with telomeric and non-telomeric functions. Cell Cycle 2010, 9, 4061–4067. [Google Scholar] [CrossRef]
- Hanaoka, S.; Nagadoi, A.; Yoshimura, S.; Aimoto, S.; Li, B.; de Lange, T.; Nishimura, Y. NMR structure of the hRap1 myb motif reveals a canonical three-helix bundle lacking the positive surface charge typical of myb DNA-binding domains. J. Mol. Biol. 2001, 312, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Nett, I.R.E.; Martin, D.; Miranda-Saavedra, D.; Lamont, D.; Barber, J.D.; Mehlert, A.; Ferguson, M.A.J. The Phosphoproteome of Bloodstream Form Trypanosoma brucei, Causative Agent of African Sleeping Sickness. Mol. Cell. Proteom. 2009, 8, 1527–1538. [Google Scholar] [CrossRef]
- Urbaniak, M.; Martin, D.M.A.; Ferguson, M.A.J. Global Quantitative SILAC Phosphoproteomics Reveals Differential Phosphorylation Is Widespread between the Procyclic and Bloodstream Form Lifecycle Stages of Trypanosoma brucei. J. Proteome Res. 2013, 12, 2233–2244. [Google Scholar] [CrossRef] [PubMed]
- Barry, J.D.; McCulloch, R. Antigenic variation in trypanosomes: Enhanced phenotypic variation in a eukaryotic parasite. Adv. Parasitol. 2001, 49, 1–70. [Google Scholar] [CrossRef] [PubMed]

| TRF Homologs | |||||||
|---|---|---|---|---|---|---|---|
| Species | Protein | DNA Binding | TERRA Binding | Homodimerization | |||
| Duplex Telomere DNA Binding | Binding Domain | TERRA Binding | Binding Domain | Activity | Domain | ||
| Human | hTRF1 | binds ds(TTAGGG)n | C-terminal Myb | Not reported | N/A | Yes | TRFH |
| hTRF2 | binds ds(TTAGGG)n | C-terminal Myb | binds (UUAGGG)n | N-terminal GAR domain | Yes | TRFH | |
| Budding yeast | ScTBF1 | binds ds(TTAGGG)n | C-terminal Myb | Not reported | N/A | ||
| Fission yeast | SpTAZ1 | binds ds[G2–8TTAC(A)]n | C-terminal Myb | Not reported | N/A | Yes | |
| SpTBF1 | binds ds[G2–8TTAC(A)]n | C-terminal Myb | Not reported | N/A | |||
| T. brucei | TbTRF | binds ds(TTAGGG)n | C-terminal Myb | binds (UUAGGG)n | C-terminal Myb | Yes | TRFH |
| RAP1 Homologs | |||
|---|---|---|---|
| Species | Protein | DNA Binding | DNA Binding Domain |
| Human | hRAP1 | No | N/A |
| Budding yeast | ScRAP1 | binds ds(TG1–3)n | Myb and Myb-Like |
| Fission yeast | SpRAP1 | No | N/A |
| T. brucei | TbRAP1 | binds ds- & ssDNA | 737RKRRR741 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, B.; Zhao, Y. Regulation of Antigenic Variation by Trypanosoma brucei Telomere Proteins Depends on Their Unique DNA Binding Activities. Pathogens 2021, 10, 967. https://doi.org/10.3390/pathogens10080967
Li B, Zhao Y. Regulation of Antigenic Variation by Trypanosoma brucei Telomere Proteins Depends on Their Unique DNA Binding Activities. Pathogens. 2021; 10(8):967. https://doi.org/10.3390/pathogens10080967
Chicago/Turabian StyleLi, Bibo, and Yanxiang Zhao. 2021. "Regulation of Antigenic Variation by Trypanosoma brucei Telomere Proteins Depends on Their Unique DNA Binding Activities" Pathogens 10, no. 8: 967. https://doi.org/10.3390/pathogens10080967
APA StyleLi, B., & Zhao, Y. (2021). Regulation of Antigenic Variation by Trypanosoma brucei Telomere Proteins Depends on Their Unique DNA Binding Activities. Pathogens, 10(8), 967. https://doi.org/10.3390/pathogens10080967





