Structural and Functional Genomics of the Resistance of Cacao to Phytophthora palmivora
Abstract
1. Introduction
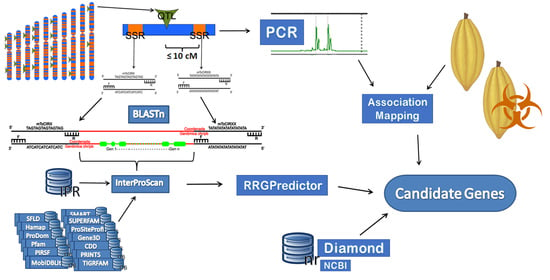
2. Materials and Methods
2.1. Plant Materials
2.2. DNA Extraction and PCR Amplification
2.3. SSR Analysis
2.4. Genome Alignment
2.5. Analyses of Population Structure
2.6. Association Mapping
2.7. Retrieving of the Protein Sequences
2.8. Alignment and Functional Annotation of Protein Sequences Based on Domains
2.9. Classification of Genes Associated with Resistance to Pathogens in Plants
2.10. Alignment and Functional Annotation of Complete Protein Sequences
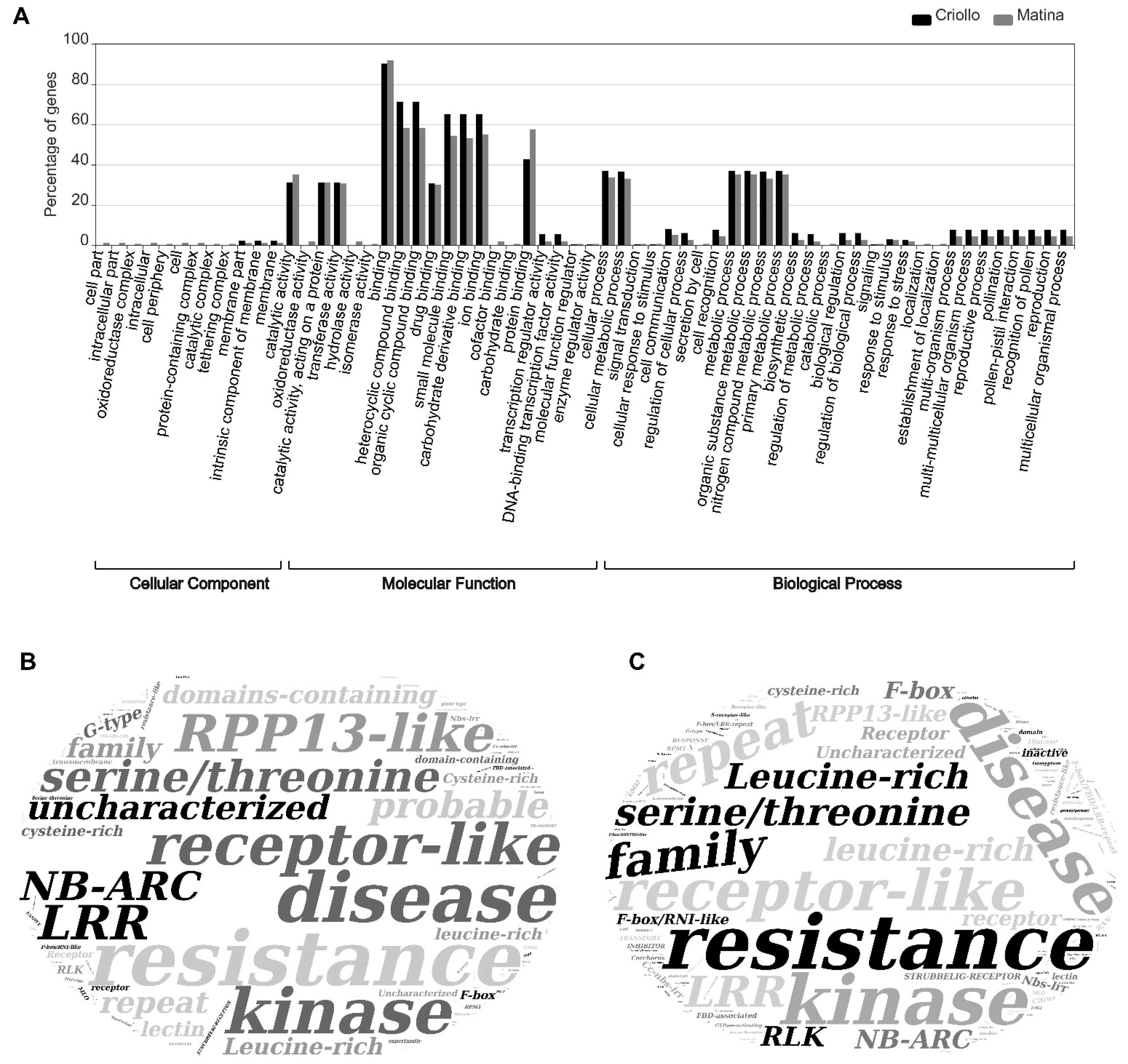
2.11. Functional Annotation in Gene Ontology Language
3. Results and Discussion
3.1. SSR Analysis
3.2. Structure and Relatedness
3.3. Association Mapping
3.4. Genomic Localization
3.5. Selection of Candidate Genes and Functional Annotation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chase, M.W.; Christenhusz, M.J.M.; Fay, M.F.; Byng, J.W.; Judd, W.S.; Soltis, D.E.; Mabberley, D.J.; Sennikov, A.N.; Soltis, P.S.; Stevens, P.F.; et al. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar] [CrossRef]
- Big Chocolate. Brings in $83 Billion per Year. Where Does All This Money End up? Available online: https://www.greenamerica.org/blog/big-chocolate-brings-83-billion-year-where-does-all-money-end (accessed on 29 November 2020).
- ICCO. The International Cocoa Organization. Annual Report 2015/2016; The International Cocoa Organization: London, UK, 2019.
- World Cocoa Foundation History of Cocoa. Available online: https://www.worldcocoafoundation.org/blog/history-of-cocoa/ (accessed on 2 December 2020).
- IBGE. Sistema IBGE de Recuperação Automática—SIDRA, Brasil; Instituto Brasileiro de Geografia e Estatística: Rio de Janeiro, Brazil, 2020.
- Biosfera, D.A.; Mata, D.A.; Araujo, M. A mata atlântica do sul da bahia. Série Cad. Reserv. Biosf. Mata Atlântica 1998, 8, 20. [Google Scholar]
- PIRES, J.L. Avaliação Quantitativa e Molecular de Germoplasma Para o Melhoramento do Cacaueiro com Ênfase na Produtividade, Qualidade de Frutos e Resistência a Doenças; University of Viçosa: Viçosa, Brazil, 2003. [Google Scholar]
- Santos, E.S.L.; Cerqueira-Silva, C.B.M.; Mori, G.M.; Ahnert, D.; Mello, D.L.N.; Pires, J.L.; Corrêa, R.X.; De Souza, A.P. Genetic structure and molecular diversity of cacao plants established as local varieties for more than two centuries: The genetic history of cacao plantations in Bahia, Brazil. PLoS ONE 2015, 10, e0145276. [Google Scholar] [CrossRef]
- Bartley, B.G.D. The Genetic Diversity of Cacao and Its Utilization; Basil, G.D., Ed.; CABI: Oxfordshire, UK, 2005; ISBN 0851996191. [Google Scholar]
- Leite, P.B.; Maciel, L.F.; Opretzka, L.C.F.; Soares, S.E.; da Silva Bispo, E. Phenolic Compounds, Methylxanthines and Antioxidant Activity in Cocoa Mass and Chocolates Produced from “Witch Broom Disease” Resistant and Non Resistant Cocoa Cultivars. Ciênc. Agrotec. 2013, 3, 244–250. [Google Scholar] [CrossRef]
- Monteiro, W.R.; Ahnert, D. Melhoramento Genético do Cacaueiro. In Ciência, Tecnologia e Manejo do Cacaueiro; Valle, R.R., Ed.; Vital LTDA: Itabuna, Brazil, 2007; pp. 1–16. [Google Scholar]
- Luz, E.D.M.N.; Silva, S.D.V.M. Podridão-Parda dos Frutos, Cancro e Outras Doenças Causadas por Phytophthora no Cacaueiro. In Doenças Causadas por Phytophthora no Brasil; Luz, E.D.M.N., Santos, A.F., Matsuoka, K., Bezerra, J.L., Eds.; Rural: Campinas, Brazil, 2001; pp. 175–265. [Google Scholar]
- Lanaud, C.; Fouet, O.; Clément, D.; Boccara, M.; Risterucci, A.M.; Surujdeo-Maharaj, S.; Legavre, T.; Argout, X. A meta-QTL analysis of disease resistance traits of Theobroma cacao L. Mol. Breed. 2009, 24, 361–374. [Google Scholar] [CrossRef]
- Akaza, M.J.; Kouassi, A.B.; Akaffou, D.S.; Fouet, O.; N’guetta, A.S.-P.; Lanaud, C. resistance in three hybrid progenies of cocoa (Theobroma cacao L.) using SSR markers. Int. J. Sci. Res. Publ. 2016, 6, 298–311. [Google Scholar]
- Akrofi, A.Y.; Appiah, A.A.; Opoku, I.Y. Management of Phytophthora pod rot disease on cocoa farms in Ghana. Crop Prot. 2003, 22, 469–477. [Google Scholar] [CrossRef]
- Dakwa, J.T. A serious outbreak of black pod disease in a marginal area of Ghana. In Proceedings of the Xth International Cocoa Research Conference, Santo Domingo, Dominican Republic, 17 May 1987; pp. 447–452. [Google Scholar]
- Ploetz, R.C. Cacao Diseases: Important Threats to Chocolate Production Worldwide. Phytopathology 2007, 97, 1634–1639. [Google Scholar] [CrossRef] [PubMed]
- Pokou, N.D.; N’Goran, J.A.K.; Kébé, I.; Eskes, A.; Tahi, M.; Sangaré, A. Levels of resistance to Phytophthora pod rot in cocoa accessions selected on-farm in Côte d’Ivoire. Crop Prot. 2008, 27, 302–309. [Google Scholar] [CrossRef]
- Luz, E.D.M.N.; Sgrillo, R.B.; Santos Filho, L. Estimativas de danos e perdas causadas por doencas no cacaueiro. In Proceedings of the WorkShop de Epidemiologia de doencas de Plantas, Viçosa, Brazil, 15 June 2004; pp. 67–79. [Google Scholar]
- Guest, D. Black pod: Diverse pathogens with a global impact on cocoa yield. Proc. Phytopathol. 2007, 97, 1650–1653. [Google Scholar] [CrossRef]
- Barreto, M.A.; Rosa, J.R.B.F.; Holanda, I.S.A.; Cardoso-Silva, C.B.; Vildoso, C.I.A.; Ahnert, D.; Souza, M.M.; Corrêa, R.X.; Royaert, S.; Marelli, J.; et al. QTL mapping and identification of corresponding genomic regions for black pod disease resistance to three Phytophthora species in Theobroma cacao L. Euphytica 2018, 214. [Google Scholar] [CrossRef]
- Brown, J.S.; Phillips-Mora, W.; Power, E.J.; Krol, C.; Cervantes-Martinez, C.; Motamayor, J.C.; Schnell, R.J. Mapping QTLs for resistance to frosty pod and black pod diseases and horticultural traits in Theobroma cacao L. Crop Sci. 2007, 47, 1851–1858. [Google Scholar] [CrossRef]
- Clement, D.; Risterucci, A.M.; Motamayor, J.C.; N’Goran, J.; Lanaud, C. Mapping QTL for yield components, vigor, and resistance to Phytophthora palmivora in Theobroma cacao L. Genome 2003, 46, 204–212. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Crouzillat, D.; Menard, B.; Mora, A.; Phillips, W.; Petiard, V. Quantitative trait analysis in Theobroma cacao using molecular markers. Euphytica 2000, 114, 13–23. [Google Scholar] [CrossRef]
- Flament, M.H.; Kebe, I.; Clément, D.; Pieretti, I.; Risterucci, A.M.; N’Goran, J.A.K.; Cilas, C.; Despréaux, D.; Lanaud, C. Genetic mapping of resistance factors to Phytophthora palmivora in cocoa. Genome 2001, 44, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Lanaud, C.; Risterucci, A.M.; Pieretti, I.; N’Goran, J.A.K.; Fargeas, D. Characterisation and genetic mapping of resistance and defence gene analogs in cocoa (Theobroma cacao L.). Mol. Breed. 2004, 13, 211–227. [Google Scholar] [CrossRef]
- Risterucci, A.M.; Paulin, D.; Ducamp, M.; N’Goran, J.A.K.; Lanaud, C. Identification of QTLs related to cocoa resistance to three species of Phytophthora. Theor. Appl. Genet. 2003, 108, 168–174. [Google Scholar] [CrossRef]
- Tong, C.; Zhang, B.; Shi, J. A hidden Markov model approach to multilocus linkage analysis in a full-sib family. Tree Genet. Genomes 2010, 6, 651–662. [Google Scholar] [CrossRef]
- Wu, R.; Ma, C.X.; Painter, I.; Zeng, Z.B. Simultaneous Maximum Likelihood Estimation of Linkage and Linkage Phases in Outcrossing Species. Theor. Popul. Biol. 2002, 61, 349–363. [Google Scholar] [CrossRef]
- Nyassé, S.; Cilas, C.; Herail, C.; Blaha, G. Leaf inoculation as an early screening test for cocoa (Theobroma cacao L.) resistance to Phytophthora black pod disease. Crop Prot. 1995, 14, 657–663. [Google Scholar] [CrossRef]
- Collard, B.C.Y.; Mackill, D.J. Marker-assisted selection: An approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. B Biol. Sci. 2008, 363, 557–572. [Google Scholar] [CrossRef] [PubMed]
- Boller, T.; Felix, G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009, 60, 379–407. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Denoux, C.; Galletti, R.; Mammarella, N.; Gopalan, S.; Werck, D.; De Lorenzo, G.; Ferrari, S.; Ausubel, F.M.; Dewdney, J. Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol. Plant 2008, 1, 423–445. [Google Scholar] [CrossRef]
- Hogenhout, S.A.; Van Der Hoorn, R.A.L.; Terauchi, R.; Kamoun, S. Emerging concepts in effector biology of plant-associated organisms. Mol. Plant-Microbe Interact. 2009, 22, 115–122. [Google Scholar] [CrossRef]
- Roux, F.; Voisin, D.; Badet, T.; Balagué, C.; Barlet, X.; Huard-Chauveau, C.; Roby, D.; Raffaele, S. Resistance to phytopathogens e tutti quanti: Placing plant quantitative disease resistance on the map. Mol. Plant Pathol. 2014, 15, 427–432. [Google Scholar] [CrossRef]
- Zipfel, C.; Robatzek, S.; Navarro, L.; Oakeley, E.J.; Jones, J.D.G.; Felix, G.; Boller, T. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 2004, 428, 764–767. [Google Scholar] [CrossRef]
- Birch, P.R.; Boevink, P.C.; Gilroy, E.M.; Hein, I.; Pritchard, L.; Whisson, S.C. Oomycete RXLR effectors: Delivery, functional redundancy and durable disease resistance. Curr. Opin. Plant Biol. 2008, 11, 373–379. [Google Scholar] [CrossRef]
- Kamoun, S. A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 2006, 44, 41–60. [Google Scholar] [CrossRef] [PubMed]
- Howden, A.J.M.; Huitema, E. Effector-triggered post-translational modifications and their role in suppression of plant immunity. Front. Plant Sci. 2012, 3, 160. [Google Scholar] [CrossRef]
- Flor, H.H. Inheritance of pathogenicity in Melampsora lini. Phytopathology 1942, 32, 653–669. [Google Scholar]
- Oßwald, W.; Fleischmann, F.; Rigling, D.; Coelho, A.C.; Cravador, A.; Diez, J.; Dalio, R.J.; Jung, M.H.; Pfanz, H.; Robin, C.; et al. Strategies of attack and defence in woody plant- Phytophthora interactions. For. Pathol. 2014, 44, 169–190. [Google Scholar] [CrossRef]
- Gabriëls, S.H.E.J.; Vossen, J.H.; Ekengren, S.K.; van Ooijen, G.; Abd-El-Haliem, A.M.; van den Berg, G.C.M.; Rainey, D.Y.; Martin, G.B.; Takken, F.L.W.; Wit, P.J.G.M.D.; et al. An NB-LRR protein required for HR signalling mediated by both extra- and intracellular resistance proteins. Plant J. 2007, 50, 14–28. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Lim, C.J.; Lee, B.W.; Choi, J.P.; Oh, S.K.; Ahmad, R.; Kwon, S.Y.; Ahn, J.; Hur, C.G. A genome-wide comparison of NB-LRR type of resistance gene analogs (RGA) in the plant Kingdom. Mol. Cells 2012, 33, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Sanseverino, W.; Ercolano, M.R. In silico approach to predict candidate R proteins and to define their domain architecture. BMC Res. Notes 2012, 5, 678. [Google Scholar] [CrossRef] [PubMed]
- Sanseverino, W.; Roma, G.; De Simone, M.; Faino, L.; Melito, S.; Stupka, E.; Frusciante, L.; Ercolano, M.R. PRGdb: A bioinformatics platform for plant resistance gene analysis. Nucleic Acids Res. 2009, 38, D814. [Google Scholar] [CrossRef]
- Sanseverino, W.; Hermoso, A.; D’Alessandro, R.; Vlasova, A.; Andolfo, G.; Frusciante, L.; Lowy, E.; Roma, G.; Ercolano, M.R. PRGdb 2.0: Towards a community-based database model for the analysis of R-genes in plants. Nucleic Acids Res. 2013, 41. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, M. Diversidade Genética e Variabilidade Morfológica de Frutos em Cultivares Locais de Cacau Resistentes à Podridão Parda Provenientes de Uruçuca, Bahia. Mater’s Thesis, Universidade Estadual de Santa Cruz, Ilhéus, Brazil, February 2017. [Google Scholar]
- Nyassé, S.; Despréaux, D.; Cilas, C. Validity of a leaf inoculation test to assess the resistance to Phytophthora megakarya in a Cocoa (Theobroma cacao L.) diallel mating design. Euphytica 2002, 123, 395–399. [Google Scholar] [CrossRef]
- de Cássia Bahia, C.; Aguilar-Vildoso, C.I.; Luz, E.D.M.N.; Lopes, U.V.; Machado, R.C.R.; Corrêa, R.X. Resistance to Black Pod Disease in a Segregating Cacao Tree Population. Trop. Plant Pathol. 2015, 40, 13–18. [Google Scholar] [CrossRef]
- Barreto, M.A.; Santos, J.C.S.; Corrêa, R.X.; Luz, E.D.M.N.; Marelli, J.; Souza, A.P. Detection of genetic resistance to cocoa black pod disease caused by three Phytophthora species. Euphytica 2015, 206, 677–687. [Google Scholar] [CrossRef]
- Luz, E.D.M.N.; Paim, M.C.A.; Silva, S.D.V.M.; Pires, J.L.; Santos, L.P.; Monteiro, W.R. Correlations of Cocoa Clones Reaction to Diferent Species of Phytophthora. In Global Appoaches to Cocoa Germolasm Utilization and Conservation. Final Report of the CFC/ICCO/IPGRI Project on “Cocoa Germplasm Utilization and Conservation: A Global Approach” (1998–2004); Eskes, A.B., Efroin, Y., Eds.; CFC: Amsterdam, The Netherlands; ICCO: London, UK; IPGRI: Rome, Italy, 2006; pp. 132–135. ISBN 92-9043-734-0. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Argout, X.; Martin, G.; Droc, G.; Fouet, O.; Labadie, K.; Rivals, E.; Aury, J.M.; Lanaud, C. The cacao Criollo genome v2.0: An improved version of the genome for genetic and functional genomic studies. BMC Genom. 2017, 18, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Motamayor, J.C.; Mockaitis, K.; Schmutz, J.; Haiminen, N.; Livingstone, D.; Cornejo, O.; Findley, S.D.; Zheng, P.; Utro, F.; Royaert, S.; et al. The genome sequence of the most widely cultivated cacao type and its use to identify candidate genes regulating pod color. Genome Biol. 2013, 14, r53. [Google Scholar] [CrossRef]
- Favre, H.A.; Powell, W.H. Nomenclature of Organic Chemistry; Royal Society of Chemistry: Cambridge, UK, 2013; ISBN 978-0-85404-182-4. [Google Scholar]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.J.; Ter Braak, C.J.F. Permutation tests for multi-factorial analysis of variance. J. Stat. Comput. Simul. 2003, 73, 85–113. [Google Scholar] [CrossRef]
- Skinner, M.E.; Uzilov, A.V.; Stein, L.D.; Mungall, C.J.; Holmes, I.H. JBrowse: A next-generation genome browser. Genome Res. 2009, 19, 1630–1638. [Google Scholar] [CrossRef]
- Stein, L.D.; Mungall, C.; Shu, S.; Caudy, M.; Mangone, M.; Day, A.; Nickerson, E.; Stajich, J.E.; Harris, T.W.; Arva, A.; et al. The generic genome browser: A building block for a model organism system database. Genome Res. 2002, 12, 1599–1610. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Santana Silva, R.J.; Micheli, F. RRGPredictor, a set-theory-based tool for predicting pathogen-associated molecular pattern receptors (PRRs) and resistance (R) proteins from plants. Genomics 2020, 112, 2666–2676. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2014, 12, 59–60. [Google Scholar] [CrossRef]
- Ye, J.; Zhang, Y.; Cui, H.; Liu, J.; Wu, Y.; Cheng, Y.; Xu, H.; Huang, X.; Li, S.; Zhou, A.; et al. WEGO 2.0: A web tool for analyzing and plotting GO annotations, 2018 update. Nucleic Acids Res. 2018, 46, W71–W75. [Google Scholar] [CrossRef] [PubMed]
- Flores-Rentería, L.; Krohn, A. Scoring microsatellite loci. Methods Mol. Biol. 2013, 1006, 319–336. [Google Scholar] [CrossRef] [PubMed]
- Bennett, A.B. Out of the Amazon: Theobroma cacao enters the genomic era. Trends Plant Sci. 2003, 8, 561–563. [Google Scholar] [CrossRef] [PubMed]
- Motamayor, J.C.; Lachenaud, P.; da Silva e Mota, J.W.; Loor, R.; Kuhn, D.N.; Brown, J.S.; Schnell, R.J. Geographic and Genetic Population Differentiation of the Amazonian Chocolate Tree (Theobroma cacao L). PLoS ONE 2008, 3, e3311. [Google Scholar] [CrossRef]
- Lanaud, C.; Sounigo, O.; Amefia, Y.K.; Paulin, D.; Lachenau, P.; Clément, D. Nouvelles données sur le fonctionnement du système d’incompatibilité du cocoayer et ses conséquences pour la élection. Café Cocoa Thé 1987, 31, 267–277. [Google Scholar]
- Motamayor, J.C.; Risterucci, A.M.; Heath, M.; Lanaud, C. Cacao domestication II: Progenitor germplasm of the Trinitario cacao cultivar. Heredity 2003, 91, 322–330. [Google Scholar] [CrossRef]
- Aikpokpodion, P.O.; Motamayor, J.C.; Adetimirin, V.O.; Adu-Ampomah, Y.; Ingelbrecht, I.; Eskes, A.B.; Schnell, R.J.; Kolesnikova-Allen, M. Genetic diversity assessment of sub-samples of cacao, Theobroma cacao L. collections in West Africa using simple sequence repeats marker. Tree Genet. Genomes 2009, 5, 699–711. [Google Scholar] [CrossRef]
- Motilal, L.A.; Zhang, D.; Mischke, S.; Meinhardt, L.W.; Umaharan, P. Microsatellite-aided detection of genetic redundancy improves management of the International Cocoa Genebank, Trinidad. Tree Genet. Genomes 2013, 9, 1395–1411. [Google Scholar] [CrossRef]
- Sereno, M.L.; Albuquerque, P.S.B.; Vencovsky, R.; Figueira, A. Genetic diversity and natural population structure of cacao (Theobroma cacao L.) from the Brazilian Amazon evaluated by microsatellite markers. Conserv. Genet. 2006, 7, 13–24. [Google Scholar] [CrossRef]
- Dievart, A.; Clark, S.E. LRR-containing receptors regulating plant development and defense. Development 2004, 131, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Afzal, A.J.; Wood, A.J.; Lightfoot, D.A. Plant receptor-like serine threonine kinases: Roles in signaling and plant defense. Mol. Plant-Microbe Interact. 2008, 21, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Sawano, Y.; Miyakawa, T.; Yamazaki, H.; Tanokura, M.; Hatano, K.I. Purification, characterization, and molecular gene cloning of an antifungal protein from Ginkgo biloba seeds. Biol. Chem. 2007, 388, 273–280. [Google Scholar] [CrossRef]
- Wang, H.H.; Ng, T.B. Ginkbilobin, a novel antifungal protein from Ginkgo biloba seeds with sequence similarity to embryo-abundant protein. Biochem. Biophys. Res. Commun. 2000, 279, 407–411. [Google Scholar] [CrossRef] [PubMed]
- Miyakawa, T.; Hatano, K.I.; Miyauchi, Y.; Suwa, Y.I.; Sawano, Y.; Tanokura, M. A secreted protein with plant-specific cysteine-rich motif functions as a mannose-binding lectin that exhibits antifungal activity. Plant Physiol. 2014, 166, 766–778. [Google Scholar] [CrossRef] [PubMed]
- Gao, N.; Wadhwani, P.; Mühlhäuser, P.; Liu, Q.; Riemann, M.; Ulrich, A.S.; Nick, P. An antifungal protein from Ginkgo biloba binds actin and can trigger cell death. Protoplasma 2016, 253, 1159–1174. [Google Scholar] [CrossRef]
- Santos, C.; Duarte, S.; Tedesco, S.; Fevereiro, P.; Costa, R.L. Expression profiling of castanea genes during resistant and susceptible interactions with the oomycete pathogen phytophthora cinnamomi reveal possible mechanisms of immunity. Front. Plant Sci. 2017, 8, 1–12. [Google Scholar] [CrossRef]
- Bittner-Eddy, P.; Can, C.; Gunn, N.; Pinel, M.; Tör, M.; Crute, I.; Holub, E.B.; Beynon, J. Genetic and physical mapping of the RPP13 locus, in arabidopsis, responsible for specific recognition of several Peronospora parasitica (downy mildew) isolates. Mol. Plant-Microbe Interact. 1999, 12, 792–802. [Google Scholar] [CrossRef]
- Bittner-Eddy, P.D.; Crute, I.R.; Holub, E.B.; Beynon, J.L. RPP13 is a simple locus in Arabidopsis thaliana for alleles that specify downy mildew resistance to different avirulence determinants in Peronospora parasitica. Plant J. 2000, 21, 177–188. [Google Scholar] [CrossRef]
- Motilal, L.A.; Zhang, D.; Umaharan, P.; Mischke, S.; Mooleedhar, V.; Meinhardt, L.W. The relic Criollo cacao in Belize—Genetic diversity and relationship with Trinitario and other cacao clones held in the International Cocoa Genebank, Trinidad. Plant Genet. Resour. Characterisation Util. 2010, 8, 106–115. [Google Scholar] [CrossRef]
- Iwaro, A.D.; Bekele, F.L.; Butler, D.R. Evaluation and utilisation of cacao (Theobroma cacao L.) germplasm at the International Cocoa Genebank, Trinidad. Euphytica 2003, 130, 207–221. [Google Scholar] [CrossRef]
- Schnell, R.J.; Kuhn, D.N.; Brown, J.S.; Olano, C.T.; Phillips-Mora, W.; Amores, F.M.; Motamayor, J.C. Development of a marker assisted selection program for cacao. Phytopathology 2007, 97, 1664–1669. [Google Scholar] [CrossRef] [PubMed]
- Tahi, G.M.; Kébé, B.I.; N’Goran, J.A.K.; Sangaré, A.; Mondeil, F.; Cilas, C.; Eskes, A.B. Expected selection efficiency for resistance to cacao pod rot (Phytophthora palmivora) comparing leaf disc inoculations with field observations. Euphytica 2006, 149, 35–44. [Google Scholar] [CrossRef]


| Marker | Major Allele Frequency | Minor Allele Frequency | Allele Number | Number Heterozygous | Proportion Heterozygous |
|---|---|---|---|---|---|
| mTcCIR184 | 0.9375 | 0.0625 | 2 | 5 | 0.125 |
| mTcCIR118 | 0.6 | 0.325 | 4 | 30 | 0.75 |
| mTcCIR273 | 0.975 | 0.025 | 2 | 0 | 0 |
| mTcCIR422 | 0.5625 | 0.375 | 5 | 29 | 0.725 |
| mTcCIR275 | 0.7625 | 0.1625 | 4 | 17 | 0.425 |
| mTcCIR240 | 0.95 | 0.05 | 2 | 4 | 0.1 |
| mTcCIR268 | 0.6625 | 0.2 | 5 | 11 | 0.275 |
| mTcCIR152 | 0.55 | 0.35 | 4 | 32 | 0.8 |
| mTcCIR176 | 0.7375 | 0.2375 | 3 | 19 | 0.475 |
| mTcCIR410 | 0.775 | 0.1125 | 6 | 6 | 0.15 |
| mTcCIR131 | 0.6625 | 0.225 | 4 | 21 | 0.525 |
| mTcCIR81 | 0.6625 | 0.3125 | 3 | 9 | 0.225 |
| mTcCIR168 | 0.4375 | 0.3375 | 3 | 31 | 0.775 |
| mTcCIR213 | 0.625 | 0.175 | 4 | 13 | 0.325 |
| mTcCIR183 | 0.5125 | 0.2375 | 4 | 24 | 0.6 |
| mTcCIR95 | 0.475 | 0.4 | 5 | 31 | 0.775 |
| mTcCIR237 | 0.9875 | 0.0125 | 2 | 1 | 0.025 |
| mTcCIR343 | 0.5 | 0.45 | 4 | 8 | 0.2 |
| mTcCIR6 | 0.5375 | 0.3 | 4 | 3 | 0.075 |
| mTcCIR136 | 0.45 | 0.3625 | 4 | 26 | 0.65 |
| mTcCIR255 | 0.85 | 0.1 | 3 | 9 | 0.225 |
| mTcCIR337 | 0.5 | 0.425 | 3 | 36 | 0.9 |
| mTcCIR9 | 0.725 | 0.125 | 5 | 6 | 0.15 |
| mTcCIR291 | 0.675 | 0.1625 | 4 | 18 | 0.45 |
| mTcCIR282 | 0.5125 | 0.2125 | 5 | 29 | 0.725 |
| mTcCIR444 | 0.2375 | 0.225 | 5 | 17 | 0.425 |
| mTcCIR200 | 0.5 | 0.325 | 4 | 34 | 0.85 |
| mTcCIR61 | 0.7125 | 0.125 | 4 | 5 | 0.125 |
| mTcCIR37 | 0.65 | 0.125 | 6 | 8 | 0.2 |
| Marker Name | LG | Probability of Marker (Marker p) | Perm p | R2 (%) | add_p | dom_p | marker_df | minorObs |
|---|---|---|---|---|---|---|---|---|
| mTcCIR444 | 8 | 0.00015268 | 0.001 | 7.4312 | 2.6313 × 10−4 | 3.5095 × 10−6 | 7 | 7 |
| mTcCIR200 | 8 | 0.005 | 0.069 | 3.7166 | 0.08077 | 0.00748 | 5 | 10 |
| mTcCIR268 | 2 | 0.04508 | 0.521 | 2.7114 | 0.02145 | 0.91435 | 5 | 8 |
| mTcCIR81 | 3 | 0.0489 | 0.556 | 1.9224 | 0.05314 | 0.15503 | 3 | 9 |
| Marker | LG | Obs | Allele | Estimate | Allele size (bp) |
|---|---|---|---|---|---|
| mTcCIR444 | 8 | 5 | A | −1.2851 | 194 |
| mTcCIR444 | 8 | 2 | C | −6.6662 × 10−1 | 213 |
| mTcCIR444 | 8 | 7 | G | 0.63938 | 230 |
| mTcCIR444 | 8 | 2 | T | −1.6195 | 206 |
| mTcCIR444 | 8 | 7 | - | −3.9552 × 10−1 | - |
| mTcCIR444 | 8 | 3 | S | 0.15355 | - |
| mTcCIR444 | 8 | 9 | W | −1.4686 | - |
| mTcCIR444 | 8 | 5 | Y | 0 | - |
| mTcCIR200 | 8 | 3 | A | −7.4174 × 10−1 | 293 |
| mTcCIR200 | 8 | 1 | C | −2.5248 | 302 |
| mTcCIR200 | 8 | 1 | T | 1.40417 | 282 |
| mTcCIR200 | 8 | 1 | + | −2.4 | - |
| mTcCIR200 | 8 | 10 | M | −1.2796 | - |
| mTcCIR200 | 8 | 24 | W | 0 | - |
| mTcCIR268 | 2 | 21 | A | −7.2632 × 10−1 | 367 |
| mTcCIR268 | 2 | 2 | G | 0.10466 | 371 |
| mTcCIR268 | 2 | 4 | T | −1.3351 | 350 |
| mTcCIR268 | 2 | 2 | + | −1.3891 | - |
| mTcCIR268 | 2 | 3 | M | −5.9507 × 10−2 | - |
| mTcCIR268 | 2 | 8 | W | 0 | - |
| mTcCIR81 | 3 | 22 | A | −8.7655 × 10−1 | 296 |
| mTcCIR81 | 3 | 8 | T | −7.0592 × 10−2 | 317 |
| mTcCIR81 | 3 | 1 | + | 0.66546 | - |
| mTcCIR81 | 3 | 9 | W | 0 | - |
| QTL | LG | G | Physical Position | CN | CNL | MLO | N | NL | RLK | RLKGNK2 | RLP | RPW8NL | T | UNKNOWN | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AKAfolICS100CHR10 | 10 | C | 2551520:4052040 | - | - | 1 | - | - | - | - | - | - | - | 2 | 3 |
| M | 16571900:18072415 | - | - | - | - | - | - | - | - | - | - | 1 | 1 | ||
| AKAfolICS100CHR1 | 1 | C | 30694434:33468108 | - | - | - | - | - | - | - | 1 | - | - | - | 1 |
| M | 28502617:31502617 | - | - | - | - | - | - | - | - | - | - | 5 | 5 | ||
| AKAfolICS100CHR3 | 3 | C | 35179055:36293379 | - | 2 | - | 2 | 3 | - | - | 1 | - | - | 3 | 11 |
| M | 29572563:30703073 | 1 | 3 | - | 2 | 1 | - | - | 1 | - | - | 3 | 11 | ||
| AKAprrHCHR1 | 1 | C | 871438:2417006 | - | 2 | - | - | - | - | - | 1 | 3 | - | 3 | 9 |
| M | 867922:2421710 | - | 2 | - | - | - | - | - | 1 | 2 | - | 3 | 8 | ||
| AKAprrHCHR6 | 6 | C | 25659781:26148133 | - | - | - | - | - | - | - | - | - | - | - | 0 |
| M | 16380796:16859704 | - | - | - | - | - | - | - | - | - | - | - | 0 | ||
| AKAprrHCHR8 | 8 | C | 1022182:2251917 | - | - | - | - | - | - | - | 3 | - | - | 1 | 4 |
| M | 7171923:8400711 | - | - | - | - | - | - | - | 3 | - | - | 1 | 4 | ||
| AKAprrICS100CHR4 | 4 | C | 30388244:30586498 | - | - | - | - | - | - | - | - | - | - | - | 0 |
| M | 25770441:25972979 | - | - | - | - | - | - | - | - | - | - | - | 0 | ||
| AKAprrICS100CHR61 | 6 | C | 20000000:21474437 | - | 1 | - | - | 1 | - | - | 6 | - | - | 1 | 9 |
| M | 7295362:8795362 | - | - | - | - | - | - | - | - | - | - | 4 | 4 | ||
| AKAprrICS100CHR62 | 6 | C | 3494620:5000000 | - | - | - | - | - | - | - | 1 | - | - | 2 | 3 |
| M | 14881286:16381286 | - | 2 | - | - | - | - | - | 2 | - | - | - | 4 | ||
| AKAprrICS95CHR2 | 2 | C | 703843:2174093 | - | 2 | - | - | - | - | - | 2 | - | - | 7 | 11 |
| M | 731519:1316407 | - | 1 | - | - | 2 | - | - | - | - | - | 7 | 10 | ||
| AKAprrICS95CHR4 | 4 | C | 19242045:19491718 | - | - | - | - | - | - | - | - | - | - | - | 0 |
| M | 11207385:11409170 | - | - | - | - | - | - | - | - | - | - | - | 0 | ||
| BARq1BPPcCHR1 | 1 | C | 30694434:35306336 | - | - | - | - | - | 1 | - | 1 * | - | - | - | 1 |
| M | 39928772:41428772 | - | 4 | - | - | - | - | - | 2 | - | - | 3 | 9 | ||
| BARq1BPPctCHR6 | 6 | C | 219626:689235 | 1 | 1 | - | - | - | - | - | 2 | - | - | 1 | 5 |
| M | 6827201:7295820 | - | 2 | - | - | - | - | - | 2 | - | - | 1 | 5 | ||
| BARq1BPPpCHR6 | 6 | C | 23520483:25804709 | - | - | - | - | - | 10 | 16 | 2 | - | 1 | 9 | 38 |
| M | 16717720:19056211 | - | - | - | - | - | 6 | 16 | 2 | - | 1 | 9 | 34 | ||
| BARq2BPPcCHR2 | 2 | C | 8244114:9182779 | - | - | - | - | - | - | - | - | - | - | 4 | 4 |
| M | 7336749:8276242 | - | - | - | - | - | - | 1 | - | - | - | 4 | 5 | ||
| BARq3BPPcCHR3 | 3 | C | 35366498:36293379 | - | 2 * | - | 2 * | 3 * | - | - | 1 * | - | - | 3 * | 0 |
| M | 29572563:30545307 | 1 * | 3 * | - | 2 * | 1 * | - | - | 1 * | - | - | 3 * | 0 | ||
| BARq4BPPcCHR4 | 4 | C | 27592136:28579412 | - | - | - | - | - | 1 | - | 1 | - | - | 3 | 5 |
| M | 21130572:22096856 | - | - | - | - | - | 1 | - | 1 | - | - | 3 | 5 | ||
| BROphy1CHR4 | 4 | C | 1:1000000 | - | - | - | - | - | - | 2 | 1 | - | - | - | 3 |
| M | 1:1000000 | - | - | - | - | - | - | 1 | 1 | - | - | - | 2 | ||
| BROphy2CHR8 | 8 | C | 2391280:4552442 | - | 3 | 2 | - | - | - | - | 3 | - | - | 2 | 10 |
| M | 4898997:7034634 | - | 3 | 2 | - | - | - | - | 3 | - | - | 2 | 10 | ||
| BROphy3CHR10 | 10 | C | 16000000:20882174 | 1 | 13 | - | 1 | 8 | - | 1 | 4 | - | - | 19 | 47 |
| M | 2511121:9990261 | - | 9 | - | - | 4 | - | 1 | 4 | - | - | 25 | 43 | ||
| Total | C | 2 | 24 | 3 | 3 | 12 | 12 | 19 | 28 | 3 | 1 | 57 | 164 | ||
| M | 1 | 26 | 2 | 2 | 7 | 7 | 19 | 22 | 2 | 1 | 71 | 160 |
| CRIOLLO | MATINA | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LG | RLP | CNL | MLO | NL | N | UN | RLP | CNL | MLO | NL | RLKGNK2 | CN | N | UN | |
| AKAprrHCHR8 | 8 | 3 | - | - | - | - | 1 | 3 | - | - | - | - | - | - | 1 |
| BROphy2CHR8 | 8 | 3 | 3 | 2 | - | - | 2 | 3 | 3 | 2 | - | - | - | - | 2 |
| AKAprrICS95CHR2 | 2 | 2 | 2 | - | - | - | 7 | - | 1 | - | 2 | - | - | - | 7 |
| BARq2BPPcCHR2 | 2 | - | - | - | - | - | 4 | - | - | - | - | 1 | - | - | 4 |
| AKAfolICS100CHR3 | 3 | 1 | 2 | - | 3 | 2 | 3 | 1 | 3 | - | 1 | - | 1 | 2 | 3 |
| BARq3BPPcCHR3 | 3 | 1 * | 2 * | - | 3 * | 2 * | 3 * | 1 * | 3 * | - | 1 * | - | 1 * | 2 * | 3 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mucherino Muñoz, J.J.; de Melo, C.A.F.; Santana Silva, R.J.; Luz, E.D.M.N.; Corrêa, R.X. Structural and Functional Genomics of the Resistance of Cacao to Phytophthora palmivora. Pathogens 2021, 10, 961. https://doi.org/10.3390/pathogens10080961
Mucherino Muñoz JJ, de Melo CAF, Santana Silva RJ, Luz EDMN, Corrêa RX. Structural and Functional Genomics of the Resistance of Cacao to Phytophthora palmivora. Pathogens. 2021; 10(8):961. https://doi.org/10.3390/pathogens10080961
Chicago/Turabian StyleMucherino Muñoz, Jonathan Javier, Cláusio Antônio Ferreira de Melo, Raner José Santana Silva, Edna Dora Martins Newman Luz, and Ronan Xavier Corrêa. 2021. "Structural and Functional Genomics of the Resistance of Cacao to Phytophthora palmivora" Pathogens 10, no. 8: 961. https://doi.org/10.3390/pathogens10080961
APA StyleMucherino Muñoz, J. J., de Melo, C. A. F., Santana Silva, R. J., Luz, E. D. M. N., & Corrêa, R. X. (2021). Structural and Functional Genomics of the Resistance of Cacao to Phytophthora palmivora. Pathogens, 10(8), 961. https://doi.org/10.3390/pathogens10080961